Abstract
The Agr quorum-sensing system of Staphylococcus aureus modulates the expression of virulence factors in response to autoinducing peptides (AIPs). The peptides are seven to nine residues in length and have the C-terminal five residues constrained in a thiolactone ring. We have developed a new method to generate AIP structures using an engineered DnaB mini-intein from Synechocystis sp. strain PCC6803. In the method, an oligonucleotide encoding the AIP is ligated to the intein and the fusion protein is expressed and purified by affinity chromatography. To produce the correct AIP structure, intein splicing is interrupted, allowing the cysteine side chain to catalyze thiolactone ring formation and release AIP from the resin. The technique is simple and robust, and we have successfully produced the three main classes of AIPs using the intein system. The intein-generated AIPs possessed the correct thiolactone ring modification based on biochemical analysis, and, importantly, all the samples were bioactive against S. aureus. The AIP activity was confirmed through Agr interference and activation profiling with developed S. aureus reporter strains. The simplicity of the method, benefits of DNA encoding, and scalable nature enable the production of S. aureus AIPs for many biological applications.
Staphylococcus aureus controls the expression of extracellular virulence factors through a quorum-sensing mechanism. This regulatory cascade, frequently referred to as the Agr (accessory gene regulator) system, responds to the extracellular presence of a secreted peptide signal (also called an autoinducing peptide or AIP). The AIP signals are seven to nine amino acids in length and have the C-terminal five residues constrained as a thiolactone ring through a cysteine side chain (Fig. 1). The genes required for the quorum-sensing system are located in the agr locus, a chromosomal region that has been investigated in detail and is known to contain two divergent transcripts, called RNAII and RNAIII (18, 24). The RNAII transcript encodes the majority of proteins necessary to generate and sense extracellular AIPs, while the RNAIII transcript is a regulatory RNA and the primary effector of the Agr system. Like other quorum-sensing molecules, AIPs are produced during growth and accumulate outside the cell until they reach a critical concentration, activating the Agr system. The regulatory cascade increases levels of the RNAII and RNAIII transcripts, leading to induction of virulence factor expression (24).
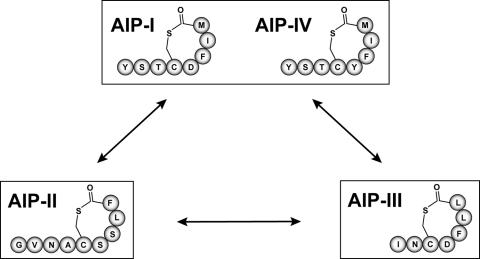
FIG. 1.
The four AIP signals of S. aureus and the cross-inhibitory groups. The amino acid sequence of each of the four AIPs is shown, and the signals are boxed into three inhibitory classes. AIP-I and AIP-IV differ by only one amino acid and function interchangeably.
An interesting feature of the Agr system is the variation among S. aureus strains (24). There are four different classes of Agr systems each recognizing a unique AIP structure (referred to as Agr-I, Agr-II, Agr-III, and Agr-IV; similarly, their cognate signals are termed AIP-I through AIP-IV). Through a fascinating mechanism of chemical communication, these different AIP signals cross-inhibit the activity of the others with surprising potency, presumably giving a competitive advantage to the producing S. aureus strain. Indeed, Agr interference has been observed with in vivo competition experiments (7), and the addition of an inhibitory AIP will block development of an acute infection (38).
Among the four AIP classes, the five-residue thiolactone ring structure is always conserved, while the other ring and tail residues differ (Fig. 1). Similarly, the proteins involved in signal biosynthesis and surface receptor binding also show variability (39, 42). In Agr interference, there are three classes of cross-inhibitory groups: AIP-I/IV, AIP-II, and AIP-III (Fig. 1). Since AIP-I and AIP-IV differ by only one amino acid and function interchangeably (13), they are grouped together. The three AIP groups all cross-inhibit each other with binding constants in the low nanomolar range (19, 20). Interestingly, the typing of the four Agr systems roughly correlates with specific classes of diseases (13, 14), although the significance of this observation is unclear.
Studies that have relied on extracellular addition of AIPs have required chemical synthesis of the signal (33, 38). While the strategy has been effective, it is prohibitive for many laboratories, impeding research on the AIP molecules. The AIPs can be purified from culture supernatants (15), but the yields are low and the procedures are labor-intensive, making this approach unattractive. In this report, we have devised a convenient, enzymatic approach to generating AIP molecules. The method employs an engineered DnaB mini-intein from Synechocystis sp. strain PCC6803. The properties of DnaB, including its small size, robust nature, and ease of expression, have made it the intein of choice for many protein engineering experiments (5, 6, 32, 35, 36, 40).
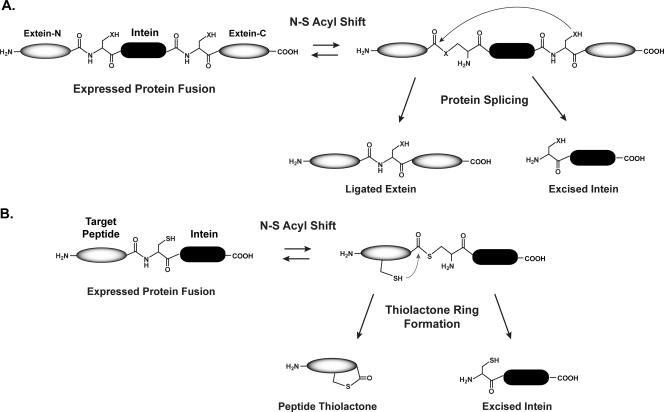
We have altered the DnaB intein function to produce the peptide thiolactone structures present in S. aureus AIP molecules. The concept is based on previous studies that demonstrated that intein splicing can be paused after the first catalytic step (Fig. 2), the conversion of a peptide bond to a thioester (often termed N-S acyl shift). By mutating the critical C-terminal asparagine residue of an intein, the splicing mechanism will stop following the N-S acyl shift (3). Additionally, without a nucleophilic (cysteine, serine, threonine) residue at the beginning of a C-terminal extein, intein-mediated splicing will be unable to occur. The ability to interrupt the intein mechanism has been utilized for various biotechnological applications, including protein purification and expressed-protein ligation (2, 22, 37), taking advantage of the activated carbonyl group made at the junction between an intein and a peptide or protein. In this report, we demonstrate that DnaB intein splicing can be interrupted to generate biologically active AIP structures.
FIG. 2.
Schematic of intein-catalyzed protein splicing and thiolactone formation. (A) Simplified version of the protein-splicing mechanism catalyzed by an intein. X, S or O from a cysteine, serine, or threonine side chain. Details of the mechanism have been reviewed elsewhere (27). (B) Interruption of intein splicing to generate peptide thiolactones. Following the N-S acyl shift, intramolecular attack from a cysteine side chain generates the thiolactone ring.
MATERIALS AND METHODS
Culture media and growth conditions.
A list of strains and plasmids used and their genotypes is provided in Table 1. Escherichia coli cultures were maintained in Luria-Bertani broth, and S. aureus strains were maintained in tryptic soy broth (TSB). E. coli antibiotic concentrations were as follows: ampicillin (Amp), 100 μg/ml; chloramphenicol (Cam), 30 μg/ml. S. aureus antibiotic concentrations were as follows: Cam, 10 μg/ml; tetracycline, 10 μg/ml. All reagents were purchased from Fisher Scientific (Pittsburg, PA) and Sigma (St. Louis, MO) unless otherwise indicated.
TABLE 1.
Strain and plasmid list
| Strain or plasmid | Genotype | Resistance | Source or reference |
|---|---|---|---|
| Escherichia coli | |||
| DH5α-E | Cloning strain | None | Invitrogen |
| ER2566 | Overexpression strain | None | New England Biolabs |
| AH394 | ER2566/ΔgshA::Cam | Cam | This work |
| AH425 | AH394/pMUT-AIPI | Amp | This work |
| AH426 | AH394/pDnaB8-AIPI | Amp | This work |
| AH495 | AH394/pDnaB8-AIPII | Amp | This work |
| AH496 | AH394/pDnaB8-AIPIII | Amp | This work |
| Staphylococcus aureus | |||
| ATCC 25923 | Agr-III | None | D. Bartels |
| FRI1169 | Agr-I | None | D. Bartels |
| RN4220 | None | G. O'Toole | |
| SH1000 | Agr-I | None | 10 |
| SH1001 | SH1000/Δagr::tet | Teta | 10 |
| SA502A | Agr-II | None | D. Bartels |
| AH429 | FRI1169/pDB59 | Cam | D. Bartels |
| AH430 | SA502A/pDB59 | Cam | D. Bartels |
| AH431 | ATCC 25923/pDB59 | Cam | D. Bartels |
| AH462 | SH1000/pDB59 | Cam | This work |
| Plasmids | |||
| pARCBD-p | SICLOPPS plasmid | Cam | 31 |
| pDNAB8 | DnaB mini-intein plasmid | Amp | This work |
| pDNAB8-AIPI | AIP-I intein plasmid | Amp | This work |
| pDNAB8-AIPII | AIP-II intein plasmid | Amp | This work |
| pDNAB8-AIPIII | AIP-III intein plasmid | Amp | This work |
| pDB59 | P3-GFP reporter | Amp, Cam | 41 |
| pEPSA5 | Expression vector | Amp, Cam | 8 |
| pET22-bsDHFR | DHFR expression vector | Amp | This work |
| pMUT-AIPI | AIP-I mutated intein plasmid | Amp | This work |
| pSU20 | Cloning vector | Cam | 1 |
| pTYB1 | Expression vector | Amp | New England Biolabs |
Tet, tetracycline.
Recombinant DNA techniques.
Restriction and modification enzymes were purchased from New England Biolabs (Beverly, MA) and were used according to the manufacturer's instructions. All DNA manipulations were performed using E. coli DH5α-E (Invitrogen, Carlsbad, CA). All oligonucleotides were synthesized at Integrated DNA Technologies (Coralville, IA). Plasmids were transformed into E. coli by CaCl2 heat shock as described previously (11). Nonradioactive sequencing was performed at the DNA sequencing facility at the University of Iowa.
Construction of strains.
The gshA gene was deleted from E. coli strain ER2566 using the Wanner method (4). Plasmid DNA was prepared from E. coli and transformed by electroporation into S. aureus RN4220 as described previously (29). Plasmids were moved from RN4220 into other S. aureus strains by transduction with bacteriophage α80 as described previously (25).
Construction of plasmid pDnaB8.
The portion of the gene encoding the DnaB N-terminal fragment was PCR amplified from Synechocystis sp. strain PCC6803 genomic DNA (forward oligonucleotide, 5′-GTTGTTCATATGGAATTCACTAGTGGCTCTTCCTGCATCAGTGGAGATAGTTTG-3′; reverse oligonucleotide, 5′-CAATTGTAAAGAGGAGCTTTCTAG-3′) and cloned into pGEM5-T (Promega Corporation, Madison, WI) by following the manufacturer's instructions. Similarly, portion of the gene encoding the DnaB C-terminal fragment was PCR amplified (forward oligonucleotide, 5′-CTAGAAAGCTCCTCTTTACAATTGTCACCAGAAATAGAAAAGTTGTCT-3′; reverse oligonucleotide, 5′-GTTGTTCTGCAGTTATCCGCGGCCGCCCGCATGGACAATGATGTCATTGG-3′) and cloned into pGEM5-T. The DnaB gene fragments were verified by DNA sequencing, PCR amplified from the pGEM5-T plasmid clones, and fused together with overlap extension PCR (34). The fused PCR fragment was digested with NdeI and PstI enzymes and cloned into plasmid pTYB1 (New England Biolabs). The resulting plasmid was called pDnaB4 and was verified by restriction analysis and DNA sequencing. The chitin-binding domain (CBD) was PCR amplified from pARCBD-p (forward oligonucleotide, 5′-TTATTATGCGGCCGCGGTGGCCTGACCGGTCTGAAC-3′; reverse oligonucleotide, 5′-GTTGTTCTGCAGTTATTGAAGCTGCCACAAGGCAGG-3′) and cloned into pGEM5-T. The CBD was removed from pGEM5-T with PstI and NotI enzymes and cloned downstream of the DnaB mini-intein on pDnaB4 using the same enzymes. The finished plasmid was called pDnaB8 and was verified by restriction analysis and DNA sequencing.
Construction of intein plasmids for producing intein-generated AIPs (iAIPs).
For production of iAIP-I (to distinguish intein generated from the native S. aureus AIP-I, the intein samples are referred to as iAIP-I; similarly, intein-generated AIP-II and AIP-III are referred to as iAIP-II and iAIP-III), oligonucleotides (coding, 5′-TATGTACAGCACCTGCGACTTCATCATG-3; noncoding, 5′-GCACATGATGAAGTCGCAGGTGCTGTACA-3) were hybridized and ligated into pDnaB8 digested with NdeI and SapI and the plasmid was verified and saved as pDnaB8-AIPI. A similar strategy was taken to construct the pDnaB8-AIPII (coding, 5′-TATGGGTGTTACCGCTTGCTCTTCTCTGTTC-3′; noncoding, 5′-GCAGAACAGAGAAGAGCAAGCGGTAACACCCA) and pDnaB8-AIPIII (coding, 5′-TATGATCAACTGCGACTTCCTGCTG-3′; noncoding, 5′-CGACAGCAGGAAGTCGCAGTTGATCA-3′) plasmids. For generating the cysteine mutant of DnaB, three-primer PCR (21) was performed on pDnaB8-AIPI with the following internal oligonucleotide to construct the mutation: 5′phosphorylated-GCGACTTCATCATGGCGATCAGTGGAGATAG-3′. All plasmid constructs were verified by DNA sequencing.
Preparation of iAIPs with DnaB intein.
An overnight preculture of expression strain AH426, AH495, or AH496 was prepared and inoculated into 100 ml of Luria-Bertani broth with Amp. The culture was grown at 37°C with shaking until an optical density at 600 nm of 0.5 was reached, and IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a 0.5 mM final concentration. The culture was grown with shaking at 30°C for 3 h, and the cell pellets were stored at −70°C. Cell pellets were resuspended in 20 ml chitin binding buffer consisting of 100 mM phosphate buffer, pH 7.0, with 500 mM NaCl, 1 mM EDTA, 150 μl protease inhibitor cocktail (Sigma; catalog number P8465), and 0.5 mM phenylmethylsulfonyl fluoride. The cell suspension was lysed through two passes in a French press, and insoluble material was removed by centrifugation at 19,000 rpm for 30 min at 4°C in a Beckman JA-20 rotor. The supernatant was removed, 4 ml equilibrated 50% chitin beads (New England Biolabs) was added, and the resin suspension was mixed gently at room temperature for 30 min. The chitin resin was removed by centrifugation at 500 × g for 5 min. The supernatant was removed, and the resin was washed three times for 5 min with 25 volumes of chitin binding buffer. The resin suspension was poured into a 10-ml column and allowed to settle by gravity (∼2-ml final resin volume), and the resin was equilibrated with three column volumes of elution buffer [100 mM phosphate, pH 7, 50 mM NaCl, 1 mM EDTA, 1 mM tris(2-carboxyethyl)phosphine (TCEP)]. Gravity flow from the column was stopped, and the resin was left sealed at room temperature for ∼15 h. Following incubation, fractions were eluted and assayed for activity or saved at −20°C.
Determining iAIP concentration.
A Sep-Pak Plus cartridge (Waters, Milford, MA) was conditioned according to the manufacturer's instructions. To remove TCEP, an AIP sample from an intein purification was bound to the cartridge, washed with 20 ml of water with 0.1% trifluoroacetic acid (TFA), and eluted with 2 ml of 60% acetonitrile with 0.1% TFA. The concentration of the iAIP was determined using assays with 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB), also called Ellman's reagent (Pierce, Rockford, IL). The thiolactone ring was opened with 1 M (final concentration) NaOH and neutralized with HCl, and DTNB assays were performed before and after base treatment. For the assays, a 1-ml reaction mixture was prepared with 100 mM Tris-HCl, pH 8, and 0.1 mM DTNB (prepared fresh) and different amounts of untreated and base-treated AIP were added. The reaction mixtures were incubated for 10 min at room temperature, and the absorbance was measure at 412 nm. The concentrations were determined with an extinction coefficient of 13,600 M−1 cm−1, and the prebase reading was subtracted to get the final iAIP concentration.
AIP inhibition assays.
For monitoring inhibition of the Agr system, an overnight culture of the appropriate reporter strain was inoculated into TSB with Cam and grown to an optical density at 600 nm of 0.05 (0.25 for AH431). In triplicate, 475 μl of reporter culture was aliquoted into test tubes (13 by 100 mm), and 25 μl of spent media or intein-generated AIP was added to each tube. As controls, separate tubes were prepared with the addition of either 25 μl of TSB or chitin elution buffer. The tubes were shaken at 250 rpm and 37°C and assayed at the following times unless otherwise indicated: AH429, 12 h; AH430, 3 h; AH431, 4 h. Both cell density (optical density at 595 nm) and green fluorescent protein (GFP) fluorescence (excitation at 485 nm, emission at 535 nm) were measured in a Tecan GENios (Research Triangle Park, NC) microtiter plate reader by removing 100 μl from each tube and assaying in a microtiter plate (3606 plates; Corning). Fluorescence is reported as an average of the three samples. Control AIP samples were prepared from the appropriate S. aureus producing strains. Each strain was grown in TSB until an optical density at 600 nm of ∼2.0 to 2.5, the cells were pelleted, and the supernatant was filtered through a 0.2-μm syringe filter. The filtered supernatants were stored at 4°C and used within 48 h.
AIP activation assays.
Overnight cultures of S. aureus reporter strains were grown in TSB and subcultured 1:50 into TSB plus 0.2% glucose supplemented with AIP (final volume, 300 μl; AIP concentration, 50 nM). Cultures were grown in test tubes (13 by 100 mm) at 37°C with shaking at 250 rpm. Cell density and fluorescence were monitored using a Tecan GENios microtiter plate reader at 4, 6, 8, and 10 h after inoculation. Optimal induction was observed at 6 h for reporter strains AH462 and AH430 and at 8 h for reporter strain AH431 (data not shown).
RESULTS
Construction of a DnaB mini-intein plasmid.
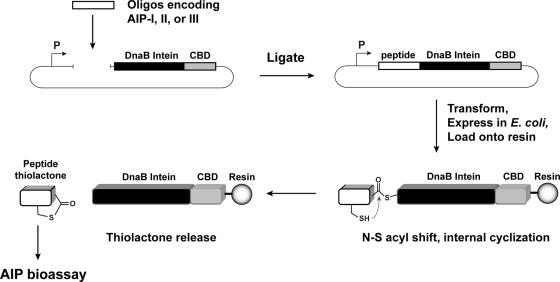
The molecular design of the mini-intein plasmid was based on the gene deletion studies performed by Liu and colleagues (32, 40), who determined that the 429-amino-acid DnaB intein in Synechocystis sp. strain PCC6803 could be reduced to a 154-amino-acid active protein. To construct the mini-intein, the gene fragments encoding the two domains of DnaB were PCR amplified from chromosomal DNA, fused together to create the mini-intein, and ligated into an IPTG-inducible expression vector. Additionally, to inactivate splicing without affecting the N-S acyl shift (3), the C-terminal asparagine residue was mutated to an alanine. For cloning and protein purification, restriction sites were added to the 5′ end of the intein and a CBD was fused to the 3′ end. The resulting plasmid, called pDnaB8, allows cloning and expression of any peptide or protein with a C-terminal intein-CBD fusion (Fig. 3).
FIG. 3.
Schematic of the method for generating the S. aureus AIP signals using the DnaB mini-intein. First, an oligonucleotide encoding the AIP peptide is ligated at the 5′ end of the DnaB intein in plasmid pDnaB8. The construct is then expressed in E. coli, cells are lysed, and the fusion protein is purified on resin. The intein performs the N-S acyl shift, creating the thioester, allowing internal attack from a cysteine side chain to release the thiolactone-containing peptide. Elution fractions are then tested for biological activity with S. aureus reporter strains.
Preliminary testing of the DnaB mini-intein plasmid.
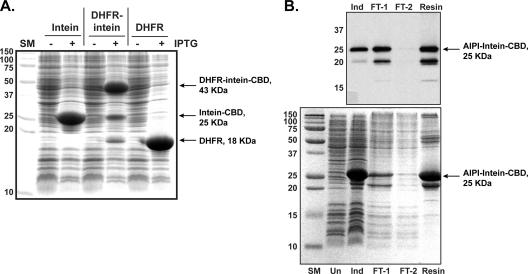
To gauge the activity of the intein, the Bacillus subtilis dihydrofolate reductase (DHFR) gene was cloned into pDnaB8. If the intein is active, the peptide bond at the DHFR-intein junction will be changed to a thioester, creating a labile bond that can be cleaved by nucleophiles and visualized on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. DHFR was chosen as an enzyme that is small (18 kDa), straightforward to express in E. coli, and amenable to C-terminal fusions (12). The overexpression of the DHFR-intein-CBD fusion was performed in E. coli strain AH394, which has a deletion of the glutathione synthetase gene (gshA). Following overexpression, three distinct bands could be identified by SDS-PAGE analysis (Fig. 4A), indicating that some intracellular cleavage occurs. One band corresponded to the DHFR-intein-CBD fusion (43 kDa), the second band was the intein-CBD fusion alone (25 kDa), and the last band was native DHFR (18 kDa). Western blotting with chitin domain antibody confirmed the presence of CBD in the two larger protein bands (data not shown). The same experiments were performed with a gshA+ strain, which resulted in higher levels of intracellular cleavage (data not shown), indicating that blocking glutathione biosynthesis increased the pools of the unprocessed DHFR-intein-CBD fusion. Overall, these observations indicate that the DnaB mini-intein is active and can efficiently create thioester bonds in this molecular arrangement.
FIG. 4.
DnaB intein activity and AIP-I purification. (A) A DHFR protein fusion was used to test DnaB activity. Plasmids pDnaB8, pDnaB8-DHFR, and pET22-bsDHFR were expressed in strain AH394 with or without IPTG induction as indicated. Overexpressed bands corresponding to intein-CBD, DHFR-intein-CBD, and DHFR are shown. (B) Samples of an iAIP-I purification were separated by SDS-PAGE and probed with CBD antibody (shown on top). Gel lanes are as follows: SM, size marker; Un, uninduced; Ind, induced; FT-1, early flowthrough sample; FT-2, late flowthrough sample; resin, chitin resin.
Generating and testing biological activity of iAIP-I.
Two plasmids were constructed to test the DnaB production of iAIP-I. The first plasmid, pDnaB8-AIPI (Fig. 3), has linear AIP-I fused to the DnaB mini-intein, with an additional methionine residue added for translation initiation (amino acid sequence MYSTCDFIM). The second plasmid, pDnaB8-AIPmut, is a control with the cysteine nucleophilic residue of DnaB mutated to an alanine. Without the cysteine residue, the DnaB mini-intein is unable to perform the N-S acyl shift.
The two intein fusions were overexpressed in E. coli and purified on chitin resin (Fig. 4B). Both protein fusions looked equivalent by SDS-PAGE and Western analysis with chitin domain antibody (data not shown). The intein fusions were kept on resin in the presence of buffer with TCEP, a non-thiol-reducing agent, added to maintain DnaB activity and keep the AIP cysteine reduced. Following incubation, the resin buffer was eluted and tested for biological activity. The bioassay is based on the observation that there are three inhibition classes among S. aureus Agr systems (Fig. 1). S. aureus Agr-II reporter strain AH430 has a plasmid with the RNAIII transcript promoter driving GFP expression. Culture supernatants of an S. aureus Agr-I strain competitively inhibit the quorum-sensing response in a type II strain, creating a convenient and sensitive bioassay for AIP-I activity (17). Gratifyingly, the pDnaB8-AIPI fractions inhibited the quorum-sensing response in strain AH430 in a dose-dependent manner, indicating the presence of iAIP-I in the sample. Through dilutions and bioassay tests, optimal iAIP-I activity was observed in the first eluted column volume (data not shown), and these fractions were subsequently used in structure verification and inhibition profiling. The resin elutions prepared from the pDnaB8-AIPmut control plasmid did not inhibit GFP expression in AH430 (Fig. 5), demonstrating that DnaB activity is essential for generating iAIP-I. In a typical purification, a 100-ml culture of E. coli overexpressing the intein fusion yielded ∼400 nmol of iAIP-I.
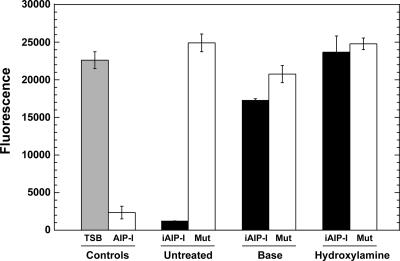
FIG. 5.
Verification of the iAIP-I structure. Strain AH430 (Agr-II) served as the reporter for all the tests, and GFP readings were taken 12 h after sample addition. For testing, each sample was diluted 20-fold into the AH430 culture at the beginning of logarithmic phase. As controls, TSB and supernatant from SH1000 (AIP-I) were added to AH430. To test the DnaB intein method, iAIP-I was purified from strains AH426 (shown as iAIP-I) and AH425 (cysteine mutant; shown as Mut). As indicated, the samples were left untreated or, to check for the thiolactone ring, were treated with base or hydroxylamine.
Confirming the AIP-I structure.
To check the mass of iAIP-I, matrix-assisted laser desorption ionization (MALDI) analysis was performed and yielded a major peak at m/z 1,092.3. This peak matched the expected molecular mass of 1,092 Da for S. aureus AIP-I with an additional methionine at the N terminus. All iAIP-I peptides identified by MALDI had this extra residue (data not shown). In the DnaB mutant control, no iAIP-I was detected by MALDI analysis.
Two additional approaches were taken to confirm the thiolactone structure. These approaches are based on the principle that linear iAIP-I will not function as a quorum-sensing inhibitor (15). As a control, synthetic linear AIP-I with an extra methionine residue was tested and did not inhibit the Agr response (data not shown). For the structure verification tests, iAIP-I was treated with sodium hydroxide base to open the thiolactone ring, neutralized with acid, and assayed for activity. In an additional test, iAIP-I was treated with hydroxylamine, which will react with thioesters to form a peptide hydroxymate, again opening the thiolactone ring. Both the base- and hydroxylamine-treated samples were tested for Agr-II inhibition, and neither sample inhibited the Agr response, while untreated iAIP-I did inhibit activity (Fig. 5). The slightly reduced GFP level with the base treatment was due to the higher salt concentrations in these samples (data not shown). Altogether, these experiments demonstrated that the correct iAIP-I modification is generated by the DnaB mini-intein system and that, most importantly, iAIP-I has biological activity.
Agr reporter strains.
To test the iAIPs, reporter strains had to be developed for each of the Agr-I, Agr-II, and Agr-III systems. Since many of the available S. aureus isolates are uncharacterized, the agrD gene was sequenced in several strains to type the Agr system, and the quorum-sensing response was tested in each strain with plasmid pDB59 (plasmid with RNAIII promoter driving GFP expression). For Agr-I, strain FRI1169 gives a strong, reproducible Agr response with the pDB59 plasmid. The resulting strain, AH429, was used as the Agr-I reporter for testing AIP samples, and quorum-sensing in this strain was inhibited by supernatants from Agr-II and -III strains (Fig. 6). Other Agr-I strains, such as SH1000, were suitable alternatives to FRI1169 in all conditions tested (data not shown). As described above, an Agr-II reporter, strain AH430, was already developed, and this reporter is inhibited by culture supernatants from Agr-I and Agr-III strains (Fig. 6). After several strains were screened for a suitable Agr-III reporter, it was found that S. aureus ATCC 25923 gave the strongest, most reproducible Agr response with the pDB59 plasmid. The resulting strain, AH431, was used as the Agr-III reporter for testing AIP samples, and quorum sensing was inhibited in AH431 by supernatants from Agr-I and -II strains.
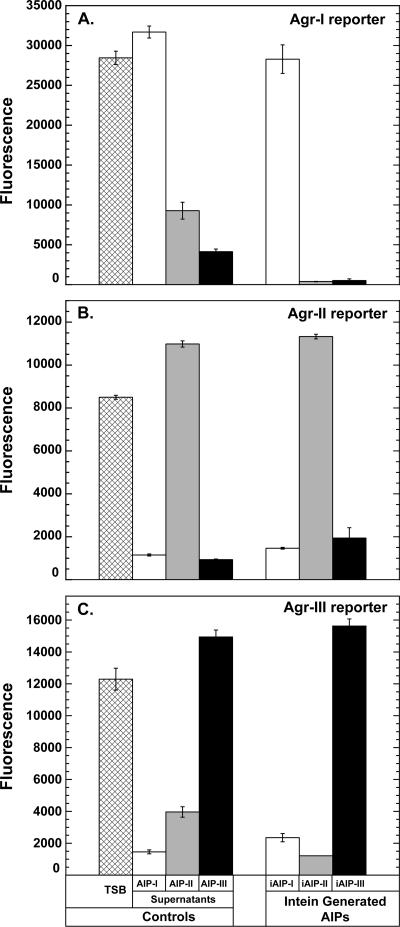
FIG. 6.
Inhibition profiling with the iAIPs. For testing, each sample was diluted 20-fold into the appropriate reporter strain at the beginning of log phase. GFP fluorescence was monitored over time and compared to that of control samples of TSB and filtered supernatants from AIP-I-, AIP-II-, and AIP-III-producing strains. (A) Strain AH429 (Agr-I reporter). (B) Strain AH430 (Agr-II reporter). (C) Strain AH431 (Agr-III reporter).
Inhibition profiling of iAIP-I.
The biological activity of iAIP-I was tested against the developed Agr-I, Agr-II, and Agr-III reporter strains. With an Agr-I strain, the iAIP-I sample did not inhibit quorum sensing, as evidenced by the negligible change in GFP levels. However, quorum sensing in Agr-II and -III strains was inhibited with the iAIP-I sample (Fig. 6), and the observed effects were consistent with multiple other iAIP-I purifications. Each reporter strain behaved as expected with control S. aureus culture supernatants. In these experiments, SA502A produced low levels of AIP-II, and testing of these supernatants resulted in weak inhibition of the Agr-I and Agr-III reporters.
Generating and testing iAIP-II and iAIP-III.
Oligonucleotides encoding the AIP-II and AIP-III linear peptides were cloned into plasmid pDnaB8 with an additional methionine residue at the N terminus for translation initiation. By the intein method, iAIP-II and iAIP-III were generated, and initial tests with elution fractions indicated the presence of biologically active samples. Purifications of iAIP-II and iAIP-III had yields similar to those of the AIP-I preparations, approximately 300 to 400 nmol per 100 ml of E. coli culture. To check the structures of both iAIP samples, MALDI analysis was performed. The iAIP-II sample had an m/z of 879.4, matching the expected molecular mass of 879 Da for complete removal of the N-terminal methionine. No iAIP-II with the initiator methionine was detected by MALDI. For iAIP-III, the sample had an m/z of 950.4, matching the expected molecular mass of 950 Da for AIP-III with an additional methionine. The presence of processed iAIP-III, with an expected molecular mass of 819 Da, was not detected.
Samples of iAIP-II and iAIP-III were tested against all three Agr reporter strains. As anticipated, iAIP-II inhibited quorum sensing in the Agr-I and Agr-III reporter strains but not the Agr-II strain. In a parallel test, iAIP-III inhibited the Agr-I and Agr-II reporter strains but not the Agr-III reporter strain (Fig. 6). All control culture supernatants behaved as expected in both sets of experiments. Additional purifications of iAIP-II and iAIP-III fractions yielded similar results, demonstrating that both peptide signals displayed the correct inhibition profile.
Agr activation with iAIP-I, iAIP-II, and iAIP-III.
While all the iAIP samples have the expected Agr inhibition profiles, it is important to test Agr activation, a more stringent indication of the correct AIP structure (18). When S. aureus is grown in TSB, RNAIII transcription is high, as observed with our GFP reporter strains (Fig. 6). When the cognate AIP from culture supernatant or an intein purification is added to a reporter strain, there is little change in the RNAIII response, presumably because the RNAIII response is already near maximum levels.
In published reports, the addition of AIP samples results in a significant increase in the RNAIII levels (15). We reasoned that the S. aureus medium conditions limit the Agr response, allowing detection of AIP activation. Glucose is a common medium component, and growth on glucose is known to repress RNAIII transcription (28). When our reporter strains were grown in TSB with 0.2% glucose, we observed an approximately 70% drop in GFP expression (data not shown). The addition of AIP to the glucose-treated strains restores the maximal levels of RNAIII, creating a simple test for AIP activation.
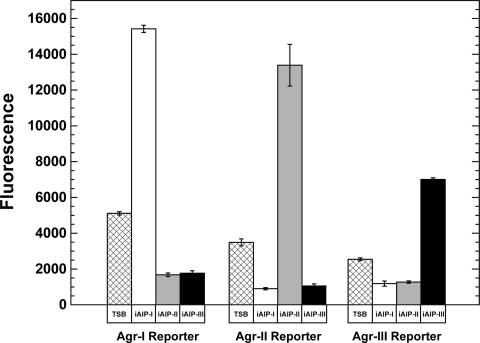
Using the activation assay, we tested the iAIP-I, iAIP-II, and iAIP-III samples with the S. aureus reporter strains. A method was developed to determine iAIP concentrations using Ellman's reagent (see Materials and Methods), and the iAIPs were added to each reporter at a 50 nM final concentration. Gratifyingly, each iAIP sample activated only its cognate reporter strain (Fig. 7), and the activation levels for iAIP-I, iAIP-II, and iAIP-III over the TSB control were 3.0-, 3.8-, and 2.8-fold, respectively. Overall, these results demonstrate that the iAIPs are functional quorum-sensing signals.
FIG. 7.
Agr activation with the iAIPs. S. aureus Agr-I (AH462), Agr-II (AH430), Agr-III (AH431) reporter strains were grown in TSB with 0.2% glucose, and 50 nM iAIP was added at the beginning of logarithmic phase. Over time, GFP fluorescence was monitored and compared to controls without additions (TSB) or with competing iAIP signals. Activation or inhibition results with control S. aureus supernatants are not shown, but all yielded the same pattern as observed with the iAIPs.
DISCUSSION
In this report, we present a new intein-based method for generating the S. aureus AIP signals. The method is simple, robust, and cost-effective, presenting an attractive alternative to synthetically based approaches. We demonstrate that AIP-I, AIP-II, and AIP-III can be generated with the intein system and that each of these iAIPs displayed the expected biological activity against S. aureus reporter strains.
The method in this report uses the Synechocystis DnaB mini-intein, and it is likely that other engineered inteins, such as the Saccharomyces cerevisiae Vma intein and the Mycobacterium tuberculosis RecA intein, could perform a similar function. The properties of the DnaB mini-intein, such as its small size and high level of expression in E. coli, made it an attractive choice for these experiments. As evidenced with the DHFR studies, DnaB is active in the E. coli cytoplasm, which may lead to lower yields of purified iAIPs. In an effort to minimize in vivo cleavage, E. coli strains with a more oxidizing cytoplasm were tested, such as the Origami (Novagen) series of strains and a glutathione synthetase (gshA) mutant. Due to poor expression of the DnaB intein in the Origami strains, these strains were not used further (data not shown). Only the gshA mutant helped reduce the level of in vivo cleavage, presumably due to lower levels of intracellular free thiols. Thus, some linear AIP may be lost from the intein fusion in the E. coli cells prior to resin purification. With the scalable nature of the method, it should be possible to generate sufficient concentrations of iAIPs for most experimental designs.
With protein expression in E. coli, the initiator methionine is not always removed by aminopeptidase (9), which may introduce some variability at the N terminus of the AIP-intein fusion proteins. Removal of the initiator methionine is dependent on the structure of the second amino acid, the penultimate residue, as aromatic and charged residues block N-terminal processing (9). In the intein-generated iAIP-I, tyrosine is the penultimate residue and completely blocks aminopeptidase activity in this position; not surprisingly, only a structure with the extra methionine is detectable by MALDI. Considering that experiments with chemically synthesized AIP-I indicated that N-terminal additions had little effect on biological activity (19), it is not surprising that iAIP-I was an effective activator of the Agr-I reporter. In the case of iAIP-II, the N-terminal methionine is completely removed, as expected with an adjacent glycine residue. However, the methionine was not removed from iAIP-III, presumably due to poor processing with the isoleucine in the penultimate position. Even with the N-terminal addition, iAIP-III induced the Agr-III reporter 2.8-fold (Fig. 7), which was similar to the Agr activation with other iAIPs. This result contrasted with a report that synthetic AIP-III with an N-terminal alanine extension could not activate an Agr reporter (19). The deleterious effects of additional residues have also been observed with AIP pheromones in Staphylococcus epidermidis (26). On the basis of the approaches reported herein, the additional N-terminal methionine on iAIP-III does not appear to limit Agr-III activation, but further studies would be necessary to confirm this result.
Assays for AIP activation have been used since the pioneering studies on the elucidation of the AIP structure (15). Many laboratories grow S. aureus in a medium called CYGP, which contains 0.5% glucose (25). It is known that growth of S. aureus in the presence of glucose lowers the pH, and the acidity reduces the levels of RNAIII expression (28). In tests with CYGP, the activation assay was similar to our experiments with TSB containing 0.2% glucose, suggesting that our observations are comparable to those previously reported (data not shown). While the assay allowed convenient monitoring of AIP, the mechanism for Agr activation is not clear. In our experimental analysis, the low pH appeared to attenuate the Agr system, presumably decreasing AIP production, allowing the Agr response to “jump-start” in the presence of externally provided AIP. A strong activation occurred only early in the growth phase (data not shown), supporting this proposal. However, further experimental analysis is necessary to confirm the glucose effects on the Agr system.
While proof-of-principle experiments were performed with the S. aureus AIP signals, the intein-based approach to generating peptide thiolactones may have broader implications. There are many AIP-like signaling molecules produced by other Staphylococcus spp. (24), and the DNA encoding features of the intein system would allow straightforward production of these structures. Moreover, by simply swapping a serine side chain for the cysteine, it may be possible to generate peptide lactone structures for various studies. There are numerous peptide lactones that function as bacterial signaling molecules, such as the GBAP signal of Enterococcus faecalis and the AIP signal of Staphylococcus intermedius (16, 23). The DNA-encoding benefits of inteins should also allow library generation, a strategy that has been exploited to make genetically encoded libraries of cyclic peptides (30, 31). These libraries could enable screening for novel bioactive peptide structures.
Acknowledgments
We thank Doug Bartels for providing plasmids, strains, and experimental advice on the AIP bioassays. We thank George O'Toole for providing plasmids and strains.
B. R. Boles was supported by NIH training grant no. T32 AI07511. This work was supported by a Cystic Fibrosis Foundation pilot grant and a Roy J. Carver Charitable Trust medical research initiative grant.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 2.Chong, S., F. B. Mersha, D. G. Comb, M. E. Scott, D. Landry, L. M. Vence, F. B. Perler, J. Benner, R. B. Kucera, C. A. Hirvonen, J. J. Pelletier, H. Paulus, and M. Q. Xu. 1997. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192:271-281. [DOI] [PubMed] [Google Scholar]
- 3.Chong, S., Y. Shao, H. Paulus, J. Benner, F. B. Perler, and M. Q. Xu. 1996. Protein splicing involving the Saccharomyces cerevisiae VMA intein. The steps in the splicing pathway, side reactions leading to protein cleavage, and establishment of an in vitro splicing system. J. Biol. Chem. 271:22159-22168. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding, Y., M. Q. Xu, I. Ghosh, X. Chen, S. Ferrandon, G. Lesage, and Z. Rao. 2003. Crystal structure of a mini-intein reveals a conserved catalytic module involved in side chain cyclization of asparagine during protein splicing. J. Biol. Chem. 278:39133-39142. [DOI] [PubMed] [Google Scholar]
- 6.Evans, T. C., Jr., J. Benner, and M. Q. Xu. 1999. The cyclization and polymerization of bacterially expressed proteins using modified self-splicing inteins. J. Biol. Chem. 274:18359-18363. [DOI] [PubMed] [Google Scholar]
- 7.Fleming, V., E. Feil, A. K. Sewell, N. Day, A. Buckling, and R. C. Massey. 2006. Agr interference between clinical Staphylococcus aureus strains in an insect model of virulence. J. Bacteriol. 188:7686-7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, G. C. Kedar, P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu Zy, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 9.Hirel, P. H., M. J. Schmitter, P. Dessen, G. Fayat, and S. Blanquet. 1989. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. USA 86:8247-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 12.Iwakura, M., and T. Tanaka. 1992. Dihydrofolate reductase from Bacillus subtilis and its artificial derivatives: expression, purification, and characterization. J. Biochem. (Tokyo) 111:638-642. [DOI] [PubMed] [Google Scholar]
- 13.Jarraud, S., G. J. Lyon, A. M. Figueiredo, L. Gerard, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 16.Ji, G., W. Pei, L. Zhang, R. Qiu, J. Lin, Y. Benito, G. Lina, and R. P. Novick. 2005. Staphylococcus intermedius produces a functional agr autoinducing peptide containing a cyclic lactone. J. Bacteriol. 187:3139-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kavanaugh, J. S., M. Thoendel, and A. R. Horswill. 2007. A role for type I signal peptidase in Staphylococcus aureus quorum-sensing. Mol. Microbiol. 65:780-798. [DOI] [PubMed] [Google Scholar]
- 18.Lyon, G. J., and R. P. Novick. 2004. Peptide signaling in Staphylococcus aureus and other gram-positive bacteria. Peptides 25:1389-1403. [DOI] [PubMed] [Google Scholar]
- 19.Lyon, G. J., J. S. Wright, T. W. Muir, and R. P. Novick. 2002. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry 41:10095-10104. [DOI] [PubMed] [Google Scholar]
- 20.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 96:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michael, S. F. 1994. Mutagenesis by incorporation of a phosphorylated oligo during PCR amplification. BioTechniques 16:410-412. [PubMed] [Google Scholar]
- 22.Muir, T. W. 2003. Semisynthesis of proteins by expressed protein ligation. Annu. Rev. Biochem. 72:249-289. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. Akkermans, W. M. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 24.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 25.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 26.Otto, M., R. Sussmuth, G. Jung, and F. Gotz. 1998. Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett. 424:89-94. [DOI] [PubMed] [Google Scholar]
- 27.Perler, F. B. 2006. Protein splicing mechanisms and applications. IUBMB Life 58:63. [DOI] [PubMed] [Google Scholar]
- 28.Regassa, L. B., R. P. Novick, and M. J. Betley. 1992. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 60:3381-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 30.Scott, C. P., E. Abel-Santos, A. D. Jones, and S. J. Benkovic. 2001. Structural requirements for the biosynthesis of backbone cyclic peptide libraries. Chem. Biol. 8:801-815. [DOI] [PubMed] [Google Scholar]
- 31.Scott, C. P., E. Abel-Santos, M. Wall, D. C. Wahnon, and S. J. Benkovic. 1999. Production of cyclic peptides and proteins in vivo. Proc. Natl. Acad. Sci. USA 96:13638-13643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, W., J. Yang, and X. Q. Liu. 2004. Synthetic two-piece and three-piece split inteins for protein trans-splicing. J. Biol. Chem. 279:35281-35286. [DOI] [PubMed] [Google Scholar]
- 33.Sung, J. M., P. D. Chantler, and D. H. Lloyd. 2006. Accessory gene regulator locus of Staphylococcus intermedius. Infect. Immun. 74:2947-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urban, A., S. Neukirchen, and K. E. Jaeger. 1997. A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res. 25:2227-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams, N. K., E. Liepinsh, S. J. Watt, P. Prosselkov, J. M. Matthews, P. Attard, J. L. Beck, N. E. Dixon, and G. Otting. 2005. Stabilization of native protein fold by intein-mediated covalent cyclization. J. Mol. Biol. 346:1095-1108. [DOI] [PubMed] [Google Scholar]
- 36.Williams, N. K., P. Prosselkov, E. Liepinsh, I. Line, A. Sharipo, D. R. Littler, P. M. Curmi, G. Otting, and N. E. Dixon. 2002. In vivo protein cyclization promoted by a circularly permuted Synechocystis sp. PCC6803 DnaB mini-intein. J. Biol. Chem. 277:7790-7798. [DOI] [PubMed] [Google Scholar]
- 37.Wood, D. W., W. Wu, G. Belfort, V. Derbyshire, and M. Belfort. 1999. A genetic system yields self-cleaving inteins for bioseparations. Nat. Biotechnol. 17:889-892. [DOI] [PubMed] [Google Scholar]
- 38.Wright, J. S., III, R. Jin, and R. P. Novick. 2005. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. USA 102:1691-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright, J. S., III, G. J. Lyon, E. A. George, T. W. Muir, and R. P. Novick. 2004. Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphylococcal quorum sensing. Proc. Natl. Acad. Sci. USA 101:16168-16173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, H., M. Q. Xu, and X. Q. Liu. 1998. Protein trans-splicing and functional mini-inteins of a cyanobacterial dnaB intein. Biochim. Biophys. Acta 1387:422-432. [DOI] [PubMed] [Google Scholar]
- 41.Yarwood, J. M., D. J. Bartels, E. M. Volper, and E. P. Greenberg. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, L., and G. Ji. 2004. Identification of a staphylococcal AgrB segment(s) responsible for group-specific processing of AgrD by gene swapping. J. Bacteriol. 186:6706-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]