Abstract
Signature-tagged mutants of Desulfovibrio desulfuricans G20 were screened, and 97 genes crucial for sediment fitness were identified. These genes belong to functional categories including signal transduction, binding and transport, insertion elements, and others. Mutants with mutations in genes encoding proteins involved in amino acid biosynthesis, hydrogenase activity, and DNA repair were further characterized.
Sulfate-reducing bacteria play important roles in a variety of anaerobic environments and have the potential to be used for bioremediation of metals and hydrocarbons (19, 29, 31). The vast majority of past studies have focused on growth in laboratory media. Yet with our current knowledge of the importance of changes in gene expression in response to environmental factors (14) and recent developments demonstrating the utility of in situ microbial studies (15, 26), the limitations of studying microbial processes in the laboratory have become evident. Sediments provide unique habitats for microorganisms (12), and environmental bacteria must therefore contend with nutrient limitation, competition, osmotic changes, variation in redox potential, and other factors. Bacteria growing in sediments likely possess characteristics distinct from those grown in the laboratory under pure culture conditions (2). There have been limited efforts to prove that cellular functions observed in the laboratory are important for microorganisms growing in the natural environment. In order to address these issues, a modified signature-tagged mutagenesis technique was adopted and Desulfovibrio desulfuricans G20, a sulfate-reducing bacterium, and Shewanella oneidensis MR-1, an iron-reducing bacterium, were used as models for studying functions for in situ growth (9). Using S. oneidensis MR-1, 47 genes were identified that enhanced sediment fitness and it was further demonstrated that antibiotic efflux was a required process for bacteria in sediment (10). In this study, we identified D. desulfuricans G20 genes apparently necessary for fitness in aquifer sediments and further characterized several of them.
G20sediment (9), a G20 strain that had been adapted to sediment conditions, was used as a parent strain for construction of our tagged-transposon mutant library. Details on the generation of a mutant library were described in a previous study (9). Sediment fitness mutants were defined as mutants that were unable to grow in sediment or unable to compete with native microorganisms and were originally selected based on the fact that the oligonucleotide tag within the chromosome of the mutant was not recovered from untreated sediments after an 8-day incubation period. D. desulfuricans G20 increased in number roughly fivefold while growing in sediment during the 8-day incubation period (data not shown). Fitness mutants were retested and selected if cell numbers were less than 10% of the inoculum concentration after the 8-day sediment incubation. A total of 108 fitness mutants were identified. The transposon-inserted regions were sequenced, and insertion-deletion genes were identified. An overview of their recovery rates from sediment and their homologs to other Deltaproteobacteria whose genomes have been sequenced is shown in Table 1.
TABLE 1.
Attenuated D. desulfuricans G20 mutants identified by screening in sulfidogenic sediment
| Function and mutant | Locusa | Predicted product | % Sediment recovery | % Similarity tob:
|
||
|---|---|---|---|---|---|---|
| D. vulgaris Hildenborough | G. metallireducens | G. sulfurreducens PCA | ||||
| Energy metabolism | ||||||
| G7(pH10) | Dde_0043* | Iron-sulfur cluster-binding protein | 0.3 | 75 | 46 | 52 |
| G9(pC1) | Dde_0044* | Pyruvate flavodoxin/ferredoxin oxidoreductase, thiamine di-P-binding domain protein | 3.5 | 77 | 57 | 53 |
| G11(pH5) | Dde_0410L | Glycerol kinase | 0 | 84 | 65 | 63 |
| C10(pG11)c | Dde_2134* | Hydrogenase (NiFe) small subunit (HydA) | 0.5 | 77 | 37 | 38 |
| A3(pH11) | Dde_2334L | Succinyl-CoA synthetase, alpha subunit | 0 | 65 | 46 | 49 |
| D5(pB3) | Dde_2334L | Succinyl-CoA synthetase, alpha subunit | 1.1 | 65 | 46 | 49 |
| C10(pB6)c | Dde_3282* | Formate C-acetyltransferase | 0.3 | 35 | 28 | 33 |
| A8(pF3) | Dde_3709* | Ferredoxin, 4Fe-4S, putative | 7.2 | 81 | 24 | 23 |
| C8(pE11)c | Dde_0081* | Fe-only hydrogenase | 0.3 | 73 | 27 | 28 |
| Amino acid biosynthesis | ||||||
| A2(pE11)c | Dde_0079 | Tryptophan synthase, beta subunit | 0.5 | 81 | 62 | 32 |
| G12(pD6)c | Dde_1081L | Arginine biosynthesis bifunctional protein ArgJ | 0.3 | 65 | 50 | 48 |
| C10(pF5) | Dde_3111 | l-Serine dehydratase | 1.6 | 42 | / | / |
| A5(pA9) | Dde_3130* | Acetolactate synthase, small subunit | 0.3 | 64 | 34 | 33 |
| E2(pG11) | Dde_0104 | Glutamine synthetase | 0 | 85 | 51 | 49 |
| B12(pE4)c | Dde_3487* | Chorismate mutase/prephenate dehydratase | 0 | 72 | 43 | 41 |
| C6(pF6) | Dde_3487* | Chorismate mutase/prephenate dehydratase | 0 | 72 | 43 | 41 |
| A5(pB6) | Dde_3635L | Glutamate synthase (NADPH), homotetrameric | 0 | 75 | 33 | 53 |
| A9(pC7) | Dde_3635L | Glutamate synthase (NADPH), homotetrameric | 0.5 | 75 | 33 | 53 |
| Nucleotide metabolism | ||||||
| A1(pA10) | Dde_0113* | Ribonucleoside-diphosphate reductase | 0.3 | 79 | 54 | 54 |
| A4(pE7) | Dde_3016* | Anaerobic ribonucleoside-triphosphate reductase, putative | 0 | 83 | / | / |
| C12(pB6) | Dde_3016* | Anaerobic ribonucleoside-triphosphate reductase, putative | 0 | 83 | / | / |
| H7(pG4) | Dde_3016* | Anaerobic ribonucleoside-triphosphate reductase, putative | 0 | 83 | / | / |
| Carbohydrate metabolism | ||||||
| C10(pE3) | Dde_0415L | Alpha amylase domain protein | 3.5 | / | 44 | / |
| E1(pF11) | Dde_1178 | Glycerone kinase | 0.8 | 86 | / | / |
| H8(pG2) | Dde_1424 | Alpha-glucan phosphorylase | 1.3 | 73 | 49 | 49 |
| Coenzyme metabolism | ||||||
| G2(pA11) | Dde_1379 | Thiamine-phosphate pyrophosphorylase | 0.4 | 62 | 45 | 45 |
| H8(pF5) | Dde_2713L | Lipoate-protein ligase B | 1.3 | 69 | 41 | 40 |
| H10(pG10) | Dde_3495* | Cobinamide kinase and cobinamide phosphate guanylyltransferase | 0.3 | 30 | 33 | 32 |
| Lipid metabolism | ||||||
| G9(pB10) | Dde_2001 | Phospholipid/glycerol acyltransferase | 0.3 | 58 | 27 | 28 |
| H4(pD3) | Dde_2001 | Phospholipid/glycerol acyltransferase | 0 | 58 | 27 | 28 |
| Cell envelope | ||||||
| A1(pG11) | Dde_0428L | Glycosyltransferase-like | 0.3 | 49 | / | / |
| A9(pC4) | Dde_0438* | UDP-N-acetylglucosamine pyrophosphorylase-related protein | 0 | / | / | / |
| A6(pA2) | Dde_1370* | N-Acetylmuramoyl-l-alanine amidase | 5.6 | 41 | 38 | 37 |
| D8(pA12)d | Dde_1437* | Carboxyl-terminal protease | 0.1 | 63 | 46 | 45 |
| B2(pF1) | Dde_1551 | d-Alanyl-d-alanine dipeptidase | 3.7 | / | / | / |
| D2(pE11) | Dde_1551 | d-Alanyl-d-alanine dipeptidase | 0.8 | / | / | / |
| A3(pF6) | Dde_1682 | Uncharacterized protein involved in outer membrane biogenesis-like | 0 | 33 | / | 21 |
| B1(pF7) | Dde_1682 | Uncharacterized protein involved in outer membrane biogenesis-like | 0.8 | 33 | / | 21 |
| H6(pH11) | Dde_3043* | d-Alanine-d-alanine ligase and related ATP-grasp enzyme-like | 1.1 | / | 26 | / |
| B8(pC6) | Dde_3694* | Glucose-1-phosphate cytidylyl-transferase | 0 | 76 | 27 | 27 |
| E2(pG5) | Dde_3138* | Membrane protein | 0 | / | / | / |
| A4(pH9) | Dde_3102 | Membrane protein, putative | 0.3 | 60 | / | / |
| Transport and binding protein | ||||||
| G5(pB9) | Dde_0495* | Heavy metal-translocating P-type ATPase | 0.5 | 45 | 36 | 37 |
| E11(pD9) | Dde_3504 | Chloride channel family protein | 0.3 | 71 | 29 | 30 |
| C10(pG3) | Dde_3725* | Phosphonate uptake transporter | 0.3 | 83 | / | / |
| A9(pG2) | Dde_0208 | Conserved hypothetical protein (domain Na+/H+ antiporter) | 1.6 | 28 | / | / |
| D8(pH9) | Dde_2476 | Na+/H+ antiporter family protein | 0 | 63 | / | / |
| E2(pB11) | Dde_0371* | Dipeptide ABC transporter substrate-binding protein | 1.0 | 26 | 27 | 26 |
| G2(pG2) | Dde_0396L | Amino acid ABC transporter, permease protein, 3-TM region, His/Glu/Gln/Arg/opine | 7.2 | 42 | / | 39 |
| G9(pB1) | Dde_1386 | Amino acid ABC transporter, permease protein, 3-TM region, His/Glu/Gln/Arg/opine | 4.0 | 70 | / | 37 |
| C7(pE7) | Dde_0339 | Outer membrane protein, OMPP1/FadL/TodX family | 0.3 | 54 | / | / |
| G11(pG11) | Dde_0339 | Outer membrane protein, OMPP1/FadL/TodX family | 0 | 54 | / | / |
| A3(pE2) | Dde_0132 | Permease, putative | 9.1 | 53 | 22 | 22 |
| A6(pH4) | Dde_0630* | Sodium/solute symporter family protein | 0.3 | 34 | 34 | 32 |
| G9(pB4) | Dde_1335L | ABC transporter, permease protein | 0 | 58 | 28 | 25 |
| Signal transduction | ||||||
| H9(pF8) | Dde_0602* | Multisensor signal transduction histidine kinase | 0 | 28 | 38 | 40 |
| G5(pA9) | Dde_1945 | Putative PAS/PAC sensor protein | 0.3 | 29 | 35 | 38 |
| D12(pB8) | Dde_3715L | Multisensor signal transduction histidine kinase | 0 | 69 | 27 | 39 |
| C8(pC10)d | Dde_1569L | Metal-dependent phosphohydrolase | 0 | 71 | 58 | 52 |
| B12(pG11) | Dde_3047L | Serine phosphatase | 0.3 | 31 | 26 | 33 |
| B8(pB6) | Dde_3047L | Serine phosphatase | 0 | 31 | 26 | 33 |
| G9(pD12) | Dde_3096* | Putative PAS/PAC sensor protein | 1.5 | 28 | 25 | 25 |
| A4(pG11) | Dde_2734 | Methyl-accepting chemotaxis sensory transducer | 0 | 41 | 28 | 32 |
| D5(pD2) | Dde_0458 | Methyl-accepting chemotaxis sensory transducer | 3.7 | 41 | 42 | 44 |
| D12(pC8) | Dde_1755 | Methyl-accepting chemotaxis sensory transducer | 0.3 | 35 | 31 | 27 |
| B2(pG1) | Dde_3212L | CheD, stimulates methylation of monocyte chemoattractant proteins | 2.9 | 52 | 44 | 47 |
| DNA replication, recombination, and repair | ||||||
| E12(pC12) | Dde_0534 | Putative transposase protein | 0.5 | / | / | / |
| B4(pB9) | Dde_0618 | ISxcd1 transposase | 0 | 50 | 36 | 24 |
| G9(pG3) | Dde_0618 | ISxcd1 transposase | 1.6 | 50 | 36 | 24 |
| D5(pC2) | Dde_3364L | ISxcd1 transposase | 2.4 | 50 | 36 | 24 |
| A8(pF7)c | Dde_2322* | Holliday junction DNA helicase RuvB | 0.3 | 84 | 62 | 63 |
| A12(pF10) | Dde_2872 | Transposase-like | 0.3 | / | / | 30 |
| B12(pF11)c | Dde_2973L | DNA-directed DNA polymerase UmuC | 0.5 | 54 | 26 | 25 |
| D12(pF8) | Dde_3099 | Methylated-DNA-protein-cysteine methyltransferase | 0 | / | / | / |
| Ribosomal structure and biogenesis | ||||||
| C6(pB7) | Dde_0389* | Dihydrouridine synthase family protein | 0.3 | 65 | 41 | 41 |
| G9(pH3) | Dde_1432L | Ribosomal protein L11 methyltransferase, putative | 3.2 | 63 | 37 | 37 |
| Transcription regulatory functions | ||||||
| C6(pB5) | Dde_0247* | DNA-directed RNA polymerase, omega subunit | 0 | 84 | 61 | 59 |
| D12(pE9) | Dde_1614* | Regulatory protein GntR, helix turn helix (HTH) | 0.3 | 58 | / | 49 |
| G2(pC4) | Dde_3684* | Hypothetical protein (domain, HTH-ARSR) | 0.5 | / | / | / |
| A1(pD3) | Dde_0289 | Putative transcriptional regulator, Fis family | 0 | 45 | 42 | 39 |
| Posttranslational modification | ||||||
| B10(pF9) | Dde_0278* | Radical sterile alpha motif (SAM) domain protein | 0 | 58 | / | / |
| G6(pH7) | Dde_1002L | Peptide methionine sulfoxide reductase | 0.8 | 41 | 61 | 42 |
| B8(pF6) | Dde_1203* | Thioredoxin reductase | 0 | 56 | 34 | 36 |
| B10(pF3) | Dde_2313 | Thiol peroxidase | 0.8 | 82 | 66 | 63 |
| H8(pE2) | Dde_3318* | Hypothetical protein (domain, trans-aconitate methyltransferase) | 7.2 | / | 29 | / |
| Others | ||||||
| C8(pA7) | Dde_0151* | Metallo-beta-lactamase family protein | 0.3 | 73 | / | / |
| D11(pC10) | Dde_0266 | Acetyltransferase-like | 0.3 | / | / | / |
| A9(pB11) | Dde_0966 | Metal-dependent phosphohydrolase | 0.8 | / | / | / |
| B11(pC2)d | Dde_1652* | Metal-dependent phosphohydrolase | 0 | 55 | 35 | 34 |
| C6(pG11) | Dde_3164* | DHH family protein | 0.5 | 53 | 36 | 34 |
| A4(pH11) | Dde_3451* | Conserved hypothetical protein (domain, predicted SAM-dependent methyltransferases) | 0.3 | 60 | 43 | 44 |
| B11(pF2)d | Dde_1729* | Protein of unknown function DUF34 | 0 | 46 | 30 | 28 |
| G9(pF3) | Dde_2698* | ATP synthase protein I | 1.3 | 57 | / | 38 |
| D11(pF11) | Dde_3374* | Phage putative head morphogenesis protein, SPP1 gp7 | 0.3 | 56 | / | / |
| C12(pG2) | Dde_3386* | Phage tail tape measure protein TP901, core region | 1.3 | 30 | / | / |
| E2(pA10) | Dde_0833* | ATPase | 0.3 | 54 | 49 | 45 |
| C10(pF3) | Dde_2869* | Type I restriction-modification system, S subunit | 7.7 | / | / | / |
| Hypothetical and conserved hypothetical proteins | ||||||
| B9(pF11) | Dde_0728L | Conserved hypothetical protein | 0.3 | / | / | / |
| C6(pF11) | Dde_0222* | Conserved hypothetical protein | 0 | 61 | / | / |
| B8(pA10) | Dde_0229* | Conserved hypothetical protein | 0.3 | 65 | 49 | 49 |
| G11(pA11) | Dde_0983* | Hypothetical protein | 0.3 | 65 | 39 | 38 |
| B12(pC10) | Dde_1127* | Hypothetical protein | 0.3 | 67 | 51 | 50 |
| D5(pF5) | Dde_2572 | Hypothetical protein | 6.1 | 32 | / | / |
| B8(pG6) | Dde_2586 | Hypothetical protein | 0.8 | / | / | / |
| G9(pF5) | Dde_3771 | Hypothetical protein | 2.4 | / | / | / |
| H8(pC4) | Dde_3760 | Hypothetical protein | 0 | / | / | / |
| G7(pE11) | Dde_0940 | Hypothetical protein | 0.5 | / | / | / |
*, locus is the gene within an operon; L, locus is the last gene of an operon.
/, homologs to D. desulfuricans G20 proteins were not found.
Mutant was further characterized in this work.
Mutant had slower growth in LS medium than the parent strain.
These mutants represent transposition events into 97 open reading frames (ORFs) whose predicted products fall into a wide variety of functional categories (Table 1). The positions of ORFs with transposon insertions were identified, and their locations on the chromosome were not biased, as indicated by the gene locus number. It is noted that these genes were distributed throughout the chromosome, and this result is consistent with previous studies with pathogens for growth and viability in vivo (22, 25). Nine genes were identified twice, and one gene was identified three times by different tagged transposons in different locations within the gene (Table 1). Bioinformatics analysis indicated that proteins from all of the functional category groups (based on analyses of clusters of orthologous groups [COG]) were identified, except for cell division genes, which are likely to be essential not only for in situ growth but for growth in general. Such mutants would not be present in the library, as they would have been eliminated during the initial selection on lactate-sulfate (LS) antibiotic plates (17, 21).
All sediment fitness mutants were able to grow in LS medium. The majority of mutants had growth rates identical to strain G20sediment, with only four growing more slowly than G20sediment (Table 1). During the pooled incubation in sediment, it is possible that these slow-growing mutants were unable to grow or compete with native bacteria or other mutants in the sediment due to a lower level of inoculum (caused by slower growth in the inoculum tubes). However, these mutants were then grown and inoculated into sediment individually and shown to be unable to survive, confirming that the functions of these interrupted genes were also needed for sediment survival.
Growth of mutants for amino acid biosynthesis genes.
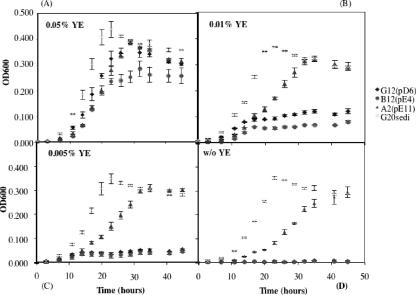
Desulfovibrio desulfuricans G20 was grown in LS medium prepared as described by Groh et al. (9). A mineral medium, prepared as described by Castaneda-Carrion (3) with a few modifications, was also used. The mineral medium contained 10 mM sodium sulfate, 25 mM sodium lactate, 0.05% yeast extract, and vitamins and minerals. N2-CO2 (4:1) was used for the headspace. Prior to autoclaving, the pH was adjusted to 7.2, and after autoclaving, 8 mM NaHCO3 and 1.6 mM Na2S were added from anaerobic stock solutions. Three mutants [G12(pD6), B12(pE4), and A2(pE11)] with mutations in genes involved in amino acid biosynthesis (Table 1) were individually cultured in LS medium and then transferred (0.1 ml) to lactate mineral medium. A further transfer was then made of log-phase cultures (optical density at 600 nm [OD600] of 0.4) into mineral media containing 0, 0.05%, 0.01%, and 0.005% yeast extract. OD600s were recorded from duplicate tubes for each mutant with strain G20sediment as the control.
The mutation in G12(pD6) is in the gene encoding N-acetylglutamate synthase (ArgJ), which catalyzes two activities in the cyclic version of arginine biosynthesis: the synthesis of acetylglutamate from glutamate and acetyl coenzyme A (acetyl-CoA) and that of ornithine by transacetylation between acetylornithine and glutamate (27). This protein has 37% similarity to the Escherichia coli protein ArgJ. B12(pE4) is mutated in the gene encoding chorismate mutase/prephenate dehydratase, a cytoplasmic protein with 31% similarity to the E. coli enzyme. Chorismate mutase catalyzes the conversion of chorismate to prephenate in the tyrosine and phenylalanine biosynthesis pathways (39). A2(pE11) is mutated in the gene encoding the tryptophan synthase (TrpB), beta subunit, responsible for the final step of l-tryptophan biosynthesis. It has very high homology to TrpB in other bacteria, including E. coli (55%) and D. vulgaris (81.15%). This protein also has a paralog in G20, with 57% identity.
In order to be certain that the loss in sediment fitness for these mutants was due to the loss of the ability to synthesize amino acids, growth was assessed with different concentrations of yeast extract, which provides trace amounts of amino acids (34). Figure 1 shows growth curves in decreasing yeast extract concentrations. Without yeast extract in the medium, all of these mutants had impaired growth. These results also indicate that D. desulfuricans G20 has the ability to synthesize all of the necessary amino acids when growing in mineral medium. Two of the mutants lost the ability to grow without added amino acids, a function apparently needed for growth in sediments. The slow growth of mutant A2(pE11) suggests to us that the paralog of TrpB may be functional.
FIG. 1.
Growth of D. desulfuricans G20sediment and the mutants G12(pD6), B12(pE4), and A2(pE11) in mineral medium with or without (w/o) yeast extract (YE) at concentrations of 0.05% (A), 0.01% (B), 0.005% (C), and 0% (D). The data shown represent the average of duplicate cultures. Error bars represent standard deviations.
Growth experiments with mutants G12(pD6) and B12(pE4) suggest that they have lost the ability to synthesize arginine or phenylalanine for growth. Given that free amino acids are likely present at very low levels in sediments, the inability to generate all of the needed amino acids likely influenced the ability to survive. Pathogens and commensal microorganisms, on the other hand, do not likely have similar constraints (11). Others have shown that amino acid biosynthesis can be elevated in response to nutrient limitation, stress, or amino acid restriction (37), perhaps having an indirect effect on fitness under those conditions.
Growth of strains with UV treatment.
Mutants with mutations in the umuC and ruvB genes were individually cultured in LS medium to an OD600 of 0.5 to 0.7. Cells (2 ml) were added to 18 ml of LS medium in a petri dish and then exposed to UV light at 254 nm at 10 cm for 10, 30, 60, 180, and 300 s. After UV exposure, serial dilutions were made into 2-ml 96-well plates (Beckman Instruments, Inc.; no. BK609681) for 3-well most probable number counts. Growth was recorded after a 2-day incubation at 37°C. UV and many chemicals cause mutagenesis by a process of translesion synthesis that requires DNA polymerase III and the SOS-regulated proteins UmuD, UmuC, and RecA. This machinery allows replication to continue through DNA lesions, therefore avoiding lethal interruption of DNA replication after DNA damage (30). UmuC is a well-conserved protein in prokaryotes and is present in all kingdoms of life (33). UmuC in G20 has an ortholog in E. coli with 41.67% identity, and it is also conserved within the Deltaproteobacteria (Table 1).
RuvB is part of the RuvABC complex of proteins, which are involved in Holliday junction resolution. During DNA replication, recombination, and repair processes, Holliday junctions are formed (5). RuvA forms a helicase complex with RuvB, mediating the Holliday junction migration by localized denaturation and reannealing (6).
To verify that the fitness mutants were deficient in survival after DNA damage, we compared rates of mutant survival after exposing cells to UV light (254-nm wavelength). The survival rate of each mutant is shown in Table 2. Both mutants had at least 10-fold-lower survival rates than the parent strain after exposure to UV light, providing strong evidence for a role of the interrupted genes in the response to mutagens.
TABLE 2.
Comparison of the survival rates after UV treatment based on most probable number counts
| Mutant | Survival ratio after UV exposure fora:
|
||||
|---|---|---|---|---|---|
| 10 s | 30 s | 1 min | 3 min | 5 min | |
| A8(pF7) | 2.3 × 10−2 | 1.3 × 10−3 | 1.0 × 10−5 | 4.2 × 10−8 | 1.79 × 10−8 |
| B12(pF11) | 0.6 × 10−2 | 1.0 × 10−3 | 2.77 × 10−7 | 5.5 × 10−8 | 2.29 × 10−8 |
| G20sediment | 1.05 × 10−1 | 1.2 × 10−2 | 7.69 × 10−4 | 6.5 × 10−7 | 6.5 × 10−7 |
The survival ratio was calculated from the number of cells that survived after UV exposure divided by the original number of cells.
Published analyses of sediments have clearly demonstrated the presence of mutagens, which can threaten the viability of aquatic biota (4). DNA-damaging agents range from UV light, to fungal metabolites, to reactive oxygen species (33). Although the roles of specific DNA repair pathways have not been studied in natural systems, both error-free (RuvABC) (28) and error-prone (UmuDC) (30) pathways are universally present in environmental bacteria (1, 7, 38). Previous studies have shown that DNA repair mechanisms (specifically RecA) are induced upon exposure of pure cultures living in natural environments to UV light (1) or chemical mutagens (7). The results presented here showing the importance of umuC and ruvB genes in sediment survival clearly demonstrate a role for DNA repair systems in sediment-dwelling bacteria in dealing with in situ concentrations of mutagens.
Growth of strains with mutations in energy production genes.
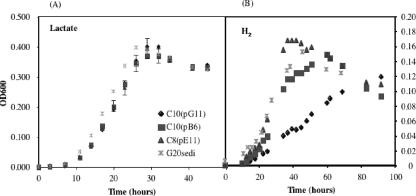
With H2 as an electron donor, lactate was omitted, and 10 mM sodium acetate-H2 (10 ml) was added to the mineral medium. Three mutants [C10(pG11), C10(pB6), and C8(pE11)] with mutations in genes annotated to be involved in energy metabolism (Table 1) were cultured in LS medium to test their abilities to grow with H2. Lactate-grown cultures (OD600 of about 0.4) were used to inoculate (0.1 ml) cultures incubated with H2 (shaken) or lactate as the electron donor. Duplicate tubes were used for each mutant with strains G20wildtype and G20sediment as controls. C10(pB6)had a mutation in the gene encoding formate C-acetyltransferase (also known as pyruvate/formate lyase), a key enzyme of anaerobic glucose metabolism, converting CoA and pyruvate to acetyl-CoA and formate (20). Previous studies have shown that pyruvate/formate lyase was required when carbon-starved E. coli entered the stationary phase (23). The G20 enzyme has 33.5% homology to formate C-acetyltransferase in E. coli and is conserved in Streptococcus species. Finding the ortholog for G20 during the sediment selection suggests that G20 may be similarly experiencing carbon limitation. The similar growth relative to the parent strain in both LS medium (66 mM lactate) and mineral medium (25 mM lactate) (Fig. 2) indicated that with adequate carbon in the form of lactate, the mutant was able to grow as well as the parent strain.
FIG. 2.
Growth of the D. desulfuricans G20sediment (G20sedi) strain and the mutants C10(pG11), C10(pB6), and C8(pE11) in mineral medium using lactate (A) and H2 (B) as electron donors. Cultures were incubated at 37°C with shaking for H2 tubes and without shaking for lactate tubes. The data shown represent the average of duplicate cultures. Error bars represent standard deviations.
C10(pG11) had a mutation in the gene encoding the NiFe hydrogenase small subunit (HydA), a periplasmic protein believed to be involved in H2 uptake (36). The small subunit in G20 has 77.0% homology to hydrogenase (HydA) in Desulfovibrio vulgaris and 37.1% homology to hydrogenase in Geobacter metallireducens. This enzyme has two paralogs (HynB-1 and HynB-2) in G20. The deletion mutant of its ortholog in D. vulgaris was found to grow similarly during the exponential phase and quickly die during the stationary growth phase (8).
C8(pE11) has a mutation in the gene encoding the Fe-only hydrogenase, a periplasmic protein which contains 4Fe-4S clusters. Cytochrome c3 is likely the physiological electron carrier for the enzyme. However, the role of the Fe-only hydrogenase as an uptake or production (24, 35) hydrogenase is still being debated.
Both the parent strain and mutants grew similarly in lactate mineral medium (Fig. 2). However, only the NiFe hydrogenase mutant C10(pG11) grew more slowly with H2 than the G20sediment strain (Fig. 2), confirming a role of the NiFe hydrogenase in H2 uptake.
H2 is a common intermediate in natural environments, and sulfate-reducing bacteria are capable of using it as an energy source (18). The interruption of the NiFe hydrogenase gene, coding for a well-studied protein thought to be involved in uptake of H2 during growth (36), decreased its H2-dependent growth rate. Our experiments with another mutant C8(pE11) containing the gene encoding the Fe-only hydrogenase showed no growth effect with H2 as the electron donor, suggesting an alternative role for this protein during sediment growth. H2 is a key intermediate in aquatic sediments and in anoxic sediments (13), and H2 partial pressures are strictly maintained at low, steady-state levels by H2-consuming organisms (18). As H2 levels drop below 10 nM, sulfate-reducing bacteria are known to outcompete methanogens and acetogens for H2 (13, 16). The selection of the uptake hydrogenase in the assay for loss of sediment fitness provides direct evidence for a role in sediment H2 uptake by Desulfovibrio.
Identical growth characteristics in both LS medium and lactate mineral medium for the selected mutants and the parent strain provide strong evidence that these genes are specifically involved in sediment fitness. Although the cellular functions of genes from one species cannot always be determined based on database searches, similar functions of proteins, originally identified through homolog analysis, have been subsequently proven (10, 32). Thus, genes that have been identified as critical for G20 sediment fitness might have similar functions to their homologs in other microorganisms. It is important to note that transposon insertions located in an operon would likely influence expression of downstream genes and the observed phenotype may be attributed to this (polar) effect. Our results only identify the transposon-inserted gene and whether the gene is located within an operon or as the terminal ORF of an operon. The latter type of insertion is less likely to have a polar effect.
Although much work remains for understanding specific roles for identified genes during sediment growth, our limited studies demonstrated several functions needed by G20 during growth in sediment. Identification of all 97 genes important for growth/fitness gives us an idea of the variety of proteins required by environmental microbes to adapt to their niches. Based on the fact that more than 70% of the identified gene products have homologs in D. vulgaris Hildenborough, G. metallireducens, and Geobacter sulfurreducens PCA, it is likely that many of these genes have universally required properties.
Acknowledgments
We thank Judy Wall (University of Missouri—Columbia) for critical reading of the manuscript.
This work was supported by the U.S. Department of Energy, Environmental Remediation Science Program (ERSP) of the Office of Biological and Environmental Research of the Office of Science.
Footnotes
Published ahead of print on 17 August 2007.
REFERENCES
- 1.Booth, M. G., W. H. Jeffrey, and R. V. Miller. 2001. RecA expression in response to solar UVR in the marine bacterium Vibrio natriegens. Microb. Ecol. 42:531-539. [DOI] [PubMed] [Google Scholar]
- 2.Brune, A., P. Frenzel, and H. Cypionka. 2000. Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol. Rev. 24:691-710. [DOI] [PubMed] [Google Scholar]
- 3.Castaneda-Carrion, I. N. 2001. Isolation and genetic characterization of subsurface microorganisms. M.S. thesis. University of Oklahoma, Norman, OK.
- 4.Chen, G., and P. A. White. 2004. The mutagenic hazards of aquatic sediments: a review. Mutat. Res. 567:151-225. [DOI] [PubMed] [Google Scholar]
- 5.Dickman, M. J., S. M. Ingleston, S. E. Sedelnikova, J. B. Rafferty, R. G. Lloyd, J. A. Grasby, and D. P. Hornby. 2002. The RuvABC resolvasome. Eur. J. Biochem. 269:5492-5501. [DOI] [PubMed] [Google Scholar]
- 6.Donaldson, J. R., C. T. Courcelle, and J. Courcelle. 2006. RuvABC is required to resolve Holliday junctions that accumulate following replication on damaged templates in Escherichia coli. J. Biol. Chem. 281:28811-28821. [DOI] [PubMed] [Google Scholar]
- 7.Elasri, M. O., T. Reid, S. Hutchens, and R. V. Miller. 2000. Response of a Pseudomonas aeruginosa biofilm community to DNA-damaging chemical agents. FEMS Microbiol. Ecol. 33:21-25. [DOI] [PubMed] [Google Scholar]
- 8.Goenka, A., J. K. Voordouw, W. Lubitz, W. Gartner, and G. Voordouw. 2005. Construction of a [NiFe]-hydrogenase deletion mutant of Desulfovibrio vulgaris Hildenborough. Biochem. Soc. Trans. 33:59-60. [DOI] [PubMed] [Google Scholar]
- 9.Groh, J. L., Q. Luo, J. D. Ballard, and L. R. Krumholz. 2005. A method adapting microarray technology for signature-tagged mutagenesis of Desulfovibrio desulfuricans G20 and Shewanella oneidensis MR-1 in anaerobic sediment survival experiments. Appl. Environ. Microbiol. 71:7064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groh, J. L., Q. Luo, J. D. Ballard, and L. R. Krumholz. 2007. Genes that enhance the ecological fitness of Shewanella oneidensis MR-1 in sediments reveal the value of antibiotic resistance. Appl. Environ. Microbiol. 73:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 12.Hewson, I., G. A. Vargo, and J. A. Fuhrman. 2003. Bacterial diversity in shallow oligotrophic marine benthos and overlying waters: effects of virus infection, containment, and nutrient enrichment. Microb. Ecol. 46:322-336. [DOI] [PubMed] [Google Scholar]
- 13.Hoehler, T. M., D. B. Albert, M. J. Alperin, B. M. Bebout, C. S. Martens, and D. J. Des Marais. 2002. Comparative ecology of H2 cycling in sedimentary and phototrophic ecosystems. Antonie Leeuwenhoek 81:575-585. [DOI] [PubMed] [Google Scholar]
- 14.Holmes, D. E., K. P. Nevin, and D. R. Lovley. 2004. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl. Environ. Microbiol. 70:7251-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krumholz, L. R., D. A. Elias, and J. M. Suflita. 2003. Immobilization of cobalt by sulfate-reducing bacteria in subsurface sediments. Geomicrobiol. J. 20:61-72. [Google Scholar]
- 16.Krumholz, L. R., S. H. Harris, S. T. Tay, and J. M. Suflita. 1999. Characterization of two subsurface H2-utilizing bacteria, Desulfomicrobium hypogeium sp. nov. and Acetobacterium psammolithicum sp. nov., and their ecological roles. Appl. Environ. Microbiol. 65:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehoux, D. E., and R. C. Levesque. 2002. Polymerase chain reaction-based signature-tagged mutagenesis. Methods Mol. Biol. 182:127-137. [DOI] [PubMed] [Google Scholar]
- 18.Lovley, D. R., and S. Goodwin. 1988. Hydrogen concentrations as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments. Geochim. Cosmochim. Acta 52:2993-3003. [Google Scholar]
- 19.Lovley, D. R., P. K. Widman, J. C. Woodward, and E. J. P. Phillips. 1993. Reduction of uranium by cytochrome c3 of Desulfovibrio vulgaris. Appl. Environ. Microbiol. 59:3572-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matheron, C., A. M. Delort, G. Gaudet, and E. Forano. 1997. Re-investigation of glucose metabolism in Fibrobacter succinogenes, using NMR spectroscopy and enzymatic assays. Evidence for pentose phosphates phosphoketolase and pyruvate formate lyase activities. Biochim. Biophys. Acta 1355:50-60. [DOI] [PubMed] [Google Scholar]
- 21.Mecsas, J. 2002. Use of signature-tagged mutagenesis in pathogenesis studies. Curr. Opin. Microbiol. 5:33-37. [DOI] [PubMed] [Google Scholar]
- 22.Merrell, D. S., D. L. Hava, and A. Camilli. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43:1471-1491. [DOI] [PubMed] [Google Scholar]
- 23.Nystrom, T. 2004. Stationary-phase physiology. Annu. Rev. Microbiol. 58:161-181. [DOI] [PubMed] [Google Scholar]
- 24.Pohorelic, B. K. J., J. K. Voordouw, E. Lojou, A. Dolla, J. Harder, and G. Voordouw. 2002. Effects of deletion of genes encoding Fe-only hydrogenase of Desulfovibrio vulgaris Hildenborough on hydrogen and lactate metabolism. J. Bacteriol. 184:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potvin, E., D. E. Lehoux, I. Kukavica-Ibrulj, K. L. Richard, F. Sanschagrin, G. W. Lau, and R. C. Levesque. 2003. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ. Microbiol. 5:1294-1308. [DOI] [PubMed] [Google Scholar]
- 26.Purdy, K. J., T. M. Embley, and D. B. Nedwell. 2002. The distribution and activity of sulphate reducing bacteria in estuarine and coastal marine sediments. Antonie Leeuwenhoek 81:181-187. [DOI] [PubMed] [Google Scholar]
- 27.Sakanyan, V., D. Charlier, C. Legrain, A. Kochikyan, I. Mett, A. Pierard, and N. Glansdorff. 1993. Primary structure, partial purification and regulation of key enzymes of the acetyl cycle of arginine biosynthesis in Bacillus stearothermophilus: dual function of ornithine acetyltransferase. J. Gen. Microbiol. 139:393-402. [DOI] [PubMed] [Google Scholar]
- 28.Shinagawa, H., K. Makino, M. Amemura, S. Kimura, H. Iwasaki, and A. Nakata. 1988. Structure and regulation of the Escherichia coli ruv operon involved in DNA repair and recombination. J. Bacteriol. 170:4322-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sierra-Alvarez, R., S. Karri, S. Freeman, and J. A. Field. 2006. Biological treatment of heavy metals in acid mine drainage using sulfate reducing bioreactors. Water Sci. Technol 54:179-185. [DOI] [PubMed] [Google Scholar]
- 30.Smith, B. T., and G. C. Walker. 1998. Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics 148:1599-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.So, C. M., C. D. Phelps, and L. Y. Young. 2003. Anaerobic transformation of alkanes to fatty acids by a sulfate-reducing bacterium, strain Hxd3. Appl. Environ. Microbiol. 69:3892-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sung, H.-M., G. Yeamans, C. A. Ross, and R. E. Yasbin. 2003. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J. Bacteriol. 185:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 34.Tanner, R. S. 1997. Cultivation of bacteria and fungi, p. 52-60. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, DC.
- 35.van den Berg, W. A. M., W. M. A. M. van Dongen, and C. Veeger. 1991. Reduction of the amount of periplasmic hydrogenase in Desulfovibrio vulgaris (Hildenborough) with antisense RNA: direct evidence for an important role of this hydrogenase in lactate metabolism. J. Bacteriol. 173:3688-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voordouw, G., V. Niviere, F. G. Ferris, P. M. Fedorak, and D. W. S. Westlake. 1990. Distribution of hydrogenase genes in Desulfovibrio spp. and their use in identification of species from the oil field environment. Appl. Environ. Microbiol. 56:3748-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weerasinghe, J. P., T. Dong, M. R. Schertzberg, M. G. Kirchhof, Y. Sun, and H. E. Schellhorn. 2006. Stationary phase expression of the arginine biosynthetic operon argCBH in Escherichia coli. BMC Microbiol. 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson, C., T. M. Caton, J. A. Buchheim, M. A. Buchheim, M. A. Schneegurt, and R. V. Miller. 2004. DNA-repair potential of Halomonas spp. from the Salt Plains Microbial Observatory of Oklahoma. Microb. Ecol. 48:541-549. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, S., G. Pohnert, P. Kongsaeree, D. B. Wilson, J. Clardy, and B. Ganem. 1998. Chorismate mutase-prephenate dehydratase from Escherichia coli. Study of catalytic and regulatory domains using genetically engineered proteins. J. Biol. Chem. 273:6248-6253. [DOI] [PubMed] [Google Scholar]