Abstract
Nck is a widely expressed SH2/SH3 adaptor protein containing one SH2 and three SH3 domains. Although Nck is assumed to mediate the formation of protein-protein complexes during signaling, little is currently known about its specific function. We have constructed a series of Nck SH3 and SH2 domain mutants, expressed them in Xenopus laevis embryos, and monitored injected embryos for developmental abnormalities. This approach allows correlation of developmental phenotypes with the presence or absence of specific Nck protein-binding domains. We show that microinjection of RNA-encoding Nck with an inactivating mutation in the third SH3 domain (NckK229) into dorsal blastomeres of early embryos caused anterior truncation with high frequency, and membrane localization of both the first and second SH3 domains together was sufficient to induce this anterior-truncation phenotype. Molecular marker analysis of explants revealed that the expression of NckK229 ventralized dorsal mesoderm. Lineage tracing experiments demonstrated that the expression of Nck K229 in dorsal blastomeres affected the migratory properties of mesoderm cells in gastrulation and led to the adoption of a more posterior fate. These data suggest that protein(s) that bind the first and second SH3 domains of Nck can affect the response to signals that establish dorso-ventral patterning, and that protein(s) that bind the third SH3 domain antagonize the ventralizing effect of the first two SH3 domains.
Keywords: signal transduction, ventralization
The SH2/SH3 adaptor proteins consist almost entirely of Src homology 2 and 3 (SH2 and SH3) domains. SH2 domains bind specifically to tyrosine-phosphorylated proteins and are thought to mediate the association of signaling proteins in response to tyrosine phosphorylation, whereas SH3 domains bind to specific proline-rich sites on target proteins (1, 2). The Nck adaptor contains one SH2 and three SH3 domains, and its expression has been detected in most tissues and cell lines studied (3, 4).
Like other adaptor proteins such as Grb2, Nck is likely to serve to assemble signaling complexes by relocalizing SH3-binding effector proteins in response to changes in tyrosine phosphorylation. The SH2 domain of Nck binds either directly or indirectly to receptor tyrosine kinases such as the platelet-derived growth factor-β and fibroblast growth factor (FGF) receptors (5–8), the tyrosine-phosphorylated docking protein IRS-1 (9), and focal adhesion kinase (10). Several putative downstream effector molecules that bind to Nck SH3 domains also have been identified, including the cbl proto-oncogene product (11), the Ras guanine nucleotide exchange factor Sos (12), the Wiskott-Aldrich syndrome protein (13, 14), and the Pak serine/threonine kinase (15–18).
Relatively little is known about the specific biological function of Nck. In some cases, overexpression of Nck has been shown to cause morphological transformation and anchorage-independent growth of fibroblastic cells (3, 19). However, dominant–negative Nck mutants, unlike those of Grb2 and Crk, did not block the Ras-dependent activation of the mitogen-activated protein kinase Erk1 by oncogenic Abl or epidermal growth factor (20), and overexpression of wild-type Nck inhibited nerve growth factor and basic FGF-induced neurite outgrowth of PC12 cells via a mitogen-activated protein kinase-independent pathway (21), suggesting that Nck might have unique activities in vivo. The only clue to the role of Nck in a whole organism came from genetic studies in Drosophila, where an apparent homolog of Nck, termed dreadlocks, was shown to be important for photoreceptor cell axon guidance and targeting (22).
To explore the role of Nck and its binding partners in the context of vertebrate development, we have expressed in early Xenopus embryos a series of Nck mutants in which the binding activity of one or more of the SH3 and/or SH2 domains has been abolished. Such mutants exert effects by binding to endogenous SH2- or SH3-binding proteins, and they can have either dominant–negative effects (by sequestering Nck-binding proteins in nonproductive complexes), or gain-of-function phenotypes (if binding of proteins that normally attenuate signals is eliminated). The power of such an approach lies in the certainty that any phenotype induced by wild-type or mutant adaptors must be due to proteins that bind to the unmutated domains. Therefore the Xenopus proteins involved in developmental phenotypes subsequently can be identified by virtue of their ability to bind to specific SH2 or SH3 domains. Here we present evidence suggesting that membrane localization of Xenopus protein(s) that bind the first and second Nck SH3 domains promotes ventralization, whereas binding of another protein to the third SH3 domain of Nck inhibits this ventralizing signal.
MATERIALS AND METHODS
Construction of Nck Mutants and in Vitro Transcription.
To produce Nck mutants, the conserved “FLVRES” arginine of the SH2 domain was changed to lysine, or the first (absolutely conserved) tryptophan of the characteristic tryptophan doublet of the SH3 domains was changed to lysine as described (20). The same changes in the SH2 and SH3 domains of other adaptors result in dominant–negative activity in vivo (20). All mutants were cloned into the pGHXP plasmid vector (a derivative of pGEM-HE, a gift from L. Zon, Children’s Hospital, Boston) and sequenced in regions of primer binding. In all cases two independent PCR clones were characterized and had identical properties. Capped synthetic mRNAs were generated as described (23) by in vitro transcription of linearized pGHXP vectors using T7 RNA polymerase. β-Galactosidase mRNA containing a nuclear localization signal was produced from the SP64nuc-βgal plasmid (a gift from M. Mercola, Harvard Medical School).
Embryos, Microinjections, and Explant Culture.
Fertilized embryos were prepared as described (24). Staging was according to Nieuwkoop and Faber (25). RNA was microinjected following published procedures (24). Ectodermal explants (animal caps) were excised at stage 8 and cultured in 0.6× Marc’s Modified Ringer’s (MMR) containing 0.5 mg/ml BSA with or without addition of human recombinant activin A (Ajinomoto Corp., Kawasaki, Japan) for 2 h. Dorsal marginal zone (DMZ) explants consisted of a 60° wedge of the marginal zone, including the outer layer of the embryo. Stage 10+ explants (when the dorsal blastopore lip is first visible) were taken from the region above the plane of the dorsal blastopore lip. For lineage tracing experiments, β-galactosidase activity was detected by standard procedures using the chromogenic substrate 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal).
RNA Isolation and Reverse Transcriptase–PCR (RT–PCR).
Total RNA was prepared from 10 animal cap explants or 7 DMZ with Trizol (GIBCO/BRL) and examined by RT–PCR following standard procedures. PCR reactions were performed in a 25 μl reaction volume in the presence of trace amounts of [α-32P]dCTP using an annealing temperature of 55°C and 20 cycles for EF1α and muscle actin, and 25 cycles for others. The sequences of primer pairs used for RT–PCR will be gladly provided on request.
Northern Blot Analysis.
Total RNA was extracted from staged, pooled embryos with Trizol, and mRNA was selected by oligo(dT)-cellulose chromatography. Full-length 32P-labeled Nck cDNA probe was prepared by the random priming method. Antisense RNA probe to ornithine decarboxylase (gift of M. Whitman, Harvard Medical School) was prepared by in vitro transcription using T7 RNA polymerase.
Histological Analysis.
Embryos were fixed in 0.1 M Mops/2 mM ethylene glycol bis(β-aminoethyl ether)-N,N,N′,N′,-tetraacetic acid/1 mM MgSO4/3.7% formaldehyde for 1 h, embedded in plastic according to the manufacturer’s instructions (JB-4, Polysciences), and sectioned at 5–10 μm. Sectioned tissue was stained with hematoxylin and eosin.
Western Blot Analysis.
Protein extracts from three whole embryos at stage 12 were boiled in sample buffer for 3 min and separated by electrophoresis through a 10% SDS/PAGE gel. Anti-Nck blots were performed by standard procedures using monoclonal antibody against Nck (Transduction Laboratories, Lexington, KY).
RESULTS AND DISCUSSION
Expression of Nck K229 Causes Anterior Truncations.
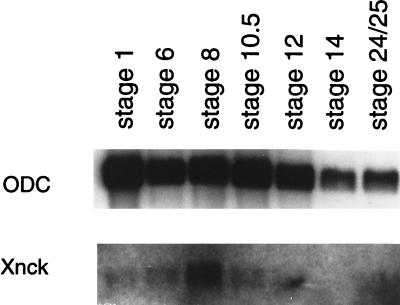
To examine Nck expression during Xenopus laevis embryogenesis, we cloned the cDNA encoding the Xenopus Nck homolog from a tadpole cDNA library. The full-length cDNA (GenBank accession number U85781U85781) encoded 378 deduced amino acids with 87% identity to human Nck. Xenopus Nck mRNA was present at low levels in fertilized one-cell stage embryos, was strongly induced in the blastula with the onset of zygotic transcription (stage 8), and decreased sharply from late gastrula (stage 12) until the tadpole stage (Fig. 1). In contrast, expression of messages for two other adaptors, Crk and Grb2, was more constant throughout Xenopus development (W.L. and B.J.M., unpublished observation). This expression pattern suggests that Nck might play a unique and important role during early development.
Figure 1.
Xenopus Nck mRNA expression during embryonic development. Equal amounts of poly(A)+ RNA extracted from embryos at stage indicated above were loaded in each lane and blot was hybridized to full-length Xenopus Nck cDNA probe. Hybridization with ornithine decarboxylase (ODC) riboprobe (Top) is a control for RNA loading.
To explore the potential role of Nck and Nck-binding proteins in embryonic development, in vitro-synthesized capped mRNAs encoding wild-type or mutated forms of Nck were microinjected into both dorsal blastomeres of Xenopus embryos at the four-cell stage. Mutant mRNAs (diagrammed in Fig. 2) encoded site-directed point mutations in one or more of the SH2 and/or SH3 domains of Nck designed to abolish detectable ligand-binding activity to the mutated domains without affecting the binding activity of unmutated domains. A Nck mutant in which all SH3 and SH2 domains are mutated, NckKALL, serves as a negative control for nonspecific effects of microinjection and protein expression.
Figure 2.
Ventralization induced by expression of Nck constructs. The structure of proteins encoded by microinjected RNAs is represented. Synthetic mRNA was injected into marginal zone region of the two dorsal blastomeres at four-cell stage. Phenotypes were scored morphologically at stage 36 according to the dorso-anterior index described previously (26). SH2 and SH3 domains are represented by shaded boxes; mutant domains in which ligand binding activity was ablated are indicated by large “X” on diagram. Amino acid number of residues substituted by lysine (K) is indicated on left. For truncation mutants (Bottom), number in shaded box indicates first, second, or third SH3 domain. Wavy line, myristoylation signal. ∗∗, 10 embryos (10 of 19) also had “split tail” resulting from incomplete closure of the blastopore. ∗, All embryos had “split tail.”
The only whole-embryo phenotype observed at high frequency was a truncation of the anterior structures of the embryo (Fig. 3A). Anterior truncation is characteristic of many known factors that induce ventralization (causing ventral tissues to be overrepresented in the embryo at the expense of more dorsal tissues), including UV light (27), BMP-4 (28), Xvent-1 (29), and Mix.1 (30). The extent of ventralization can be represented using the dorso-anterior index scale in which DAI 5 is normal and DAI 0 is completely ventralized (26).
Figure 3.
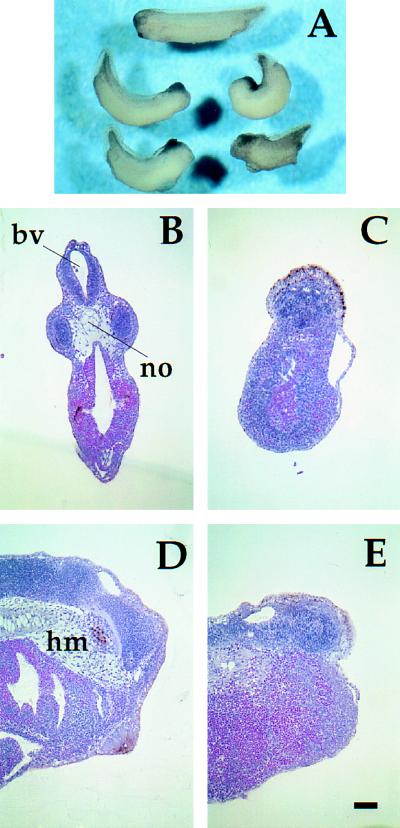
Nck K229 interferes with head and anterior notochord development. (A) Phenotype of embryos microinjected with KALL control mRNA (top embryo) or K229 mRNA (bottom four embryos) as in Fig. 2 legend. (B–E) Histological analysis of KALL-injected control embryos (B and D) and K229-injected embryos (C and E). Embryos were fixed at stage 36, sectioned, and stained with hematoxylin/eosin. (B and C) Transverse sections at the level of the eye in the control embryo. Note absence of notochord (no), head mesenchyme and brain ventricle (bv) in C. (D and E) Sagittal (midline) sections. Note absence of head mesenchyme (hm) and anterior notochord in E. (Bar = 100 μm.)
The frequency and severity of anterior truncation caused by each Nck construct is summarized in Fig. 2. Nck K229, which has a mutation in the third SH3 domain, caused the highest frequency of anterior truncation with over 70% of embryos affected. Among other Nck constructs, microinjection of RNA encoding any Nck clone (including wild-type Nck) in which both the first and second SH3 domains were intact induced a similar head defect phenotype, but at lower frequency than K229. By contrast, embryos microinjected with mRNA encoding any Nck clone with a mutation in either the first or second SH3 domain developed normally. Taken together these results strongly implicate proteins that bind the first two SH3 domains of Nck in ventralization. Furthermore, the observation that the K229 mutant has a stronger effect than wild-type Nck suggests that an intact third SH3 domain antagonizes ventralization, whereas the observation that K229 had a stronger effect than the K229,308 mutant suggests that a functional SH2 domain facilitates the ventralizing activity.
The SH2 domain is likely to function to relocalize Nck and its associated SH3-binding effector proteins in response to changes in tyrosine phosphorylation. Because the SH2 promotes ventralization by Nck mutants, we were interested in whether constitutive localization of the Nck SH3 domains to the membrane (where many SH2-binding sites, such as autophosphorylated growth factor receptors, reside) would ventralize. We therefore constructed Nck deletion mutants consisting of SH3 domains with or without addition of an N-terminal myristoylation signal derived from Src (Fig. 2). The Src myristoylation signal has been shown to direct the localization of heterologous proteins to membranes (31, 32).
Dorsal injection of a myristoylated Nck clone containing the first and second SH3 domains (Fig. 2) resulted in essentially the same head defect phenotype illustrated in Fig. 3, and its effect was more potent than K229 per ng RNA injected. In contrast, myristoylated clones encoding individual SH3 domains or lacking a myristoylation signal did not induce anterior truncation. Furthermore, coinjection of two RNAs, one encoding a myristoylated first SH3 domain and one encoding a myristoylated second SH3 domain, did not result in head defects. These data show that localization of SH3–1 and SH3–2 (linked in cis) to membranes is sufficient for Nck-induced ventralization. This suggests that an endogenous protein or proteins binds simultaneously to both the first and second Nck SH3 domains, and that membrane localization of this complex results in ventralization. Finally, a myristoylated mutant containing all three SH3 domains of Nck had much weaker ventralizing activity than the construct encoding only SH3–1 and SH3–2 (Fig. 2), confirming that the presence of SH3–3 antagonizes ventralization, consistent with a model in which a protein that binds the third SH3 domain negatively regulates the ventralizing signal.
Fig. 3A shows the phenotype of embryos observed when K229 mRNA was injected. Although there was considerable embryo-to-embryo variation in morphology, K229-injected embryos typically exhibited loss of eyes, cement gland, and forehead; heart development was normal. Histological analysis revealed a small mass of brain tissue anterior to the otic vesicle of K229-injected embryos, whereas the brain ventricle, eyes, and notochord were absent (Fig. 3 B and C). The most severe cases lacked evidence of any anterior brain tissue. Head mesenchyme was also reduced or absent (Fig. 3 D and E). There was no apparent difference, however, in the structure of the more posterior spinal cord and notochord between K229-injected and control embryos (data not shown).
The anterior truncation phenotype induced by K229 was dose-dependent in both frequency and severity (Fig. 2 and data not shown). In mild cases caused by the microinjection of lower amounts of RNA (0.5 ng per embryo), embryos showed tiny eyes or a fused single eye accompanied with reduced forehead, while increasing the RNA above 2.5 ng per embryo resulted in failure of the blastopore to close after gastrulation, leading to a “split tail” along with anterior truncation and in severe cases to gastrulation arrest. Injection of K229 mRNA into the ventral blastomeres (as opposed to dorsal injection as in the experiments above) at the 4- or 8-cell stage had no apparent effect on embryogenesis. A Xenopus Nck K229 mutant caused the same embryonic phenotypes as expression of human Nck K229 (not shown, but see Fig. 4 C and D), demonstrating that ventralization was not specific to the human Nck gene used for most experiments.
Figure 4.
Expression of K229 ventralizes dorsal mesoderm. (A and B) Animal caps explanted from stage 8 embryos injected at one-cell stage with 1 ng of control KALL mRNA (A) or K229 mRNA (B) were treated with 30 ng/ml activin. Photographs were taken after 8 h of culture. (C and D) Induction of mesodermal marker genes by activin in animal caps expressing various Nck clones. mRNA indicated above lanes was injected as in A and B, and animal caps explanted at stage 8. In XK229(0.25) and K229(0.25) lanes, 0.25 ng of RNA was injected, all others 1 ng. XK229 indicates K229 mutant of Xenopus Nck gene; all other constructs were derived from human gene. K229(−), XK229(−), unij(−): K229- or XK229-injected or uninjected control caps without activin treatment. WE, whole embryo. EF1α is control for template levels in RT–PCR. Results shown are representative of three experiments. (C) After 2-h incubation with activin at 100 ng/ml, animal caps were harvested for RT–PCR at stage 11. (Bottom) Injected embryos were cultured until stage 12 for examination of Nck protein expression levels by immunoblot. (D) Animal caps treated with 5 ng/ml activin for 2 h and harvested for RT–PCR at stage 36. (E and F) Induction of mesodermal marker genes in DMZ explants. mRNA indicated above lanes was injected into dorsal blastomeres of 4-cell embryos (0.5 ng/blastomere). DMZ explants from early gastrula embryos were cultured until harvested for RT–PCR at stage 12 (E) or stage 19 (F). KALL, RT(−); KALL-injected control reaction lacking RT.
Nck K229 Interferes with Mesodermal Patterning.
We next performed explant experiments to address whether anterior truncation was due to a direct effect on the dorso-ventral patterning of mesoderm. When Xenopus blastula ectodermal explants (animal caps) are cultured in saline solution they differentiate into ciliated epidermis. In contrast, when such explants are treated with activin or other factors mesoderm is induced; the caps differentiate into a variety of mesodermal and neural tissues and undergo morphogenetic movements resulting in elongation (33).
We found that elongation of animal caps in response to activin was strongly inhibited by expression of K229 (Fig. 4B), but not by control KALL (Fig. 4A). RT–PCR analysis revealed that expression of K229 in activin-treated animal caps reduced the induction of the dorsal mesoderm markers goosecoid (gsc) and muscle actin, had no effect on the induction of general mesoderm marker brachyury (Xbra), and increased the expression of the ventral mesoderm marker genes Xwnt-8 and α-globin relative to KALL-injected controls (Fig. 4 C and D). In contrast, animal caps derived from embryos injected with constructs that induced lower frequencies of anterior truncation (wild-type Nck, K308, and K229,308) did not differ significantly from control caps by RT–PCR analysis. No mesodermal markers were induced in animal caps derived from K229-injected embryos without activin treatment (Fig. 4 C and D). In all cases, exogenous Nck protein expression levels were shown to be equivalent by immunoblotting (Fig. 4C). Taken together, these results demonstrate that expression of K229 can respecify dorsal mesoderm to adopt a more ventral fate, but cannot block mesoderm induction itself or induce mesoderm in the absence of exogenous signals.
To confirm that K229 alters dorso-ventral patterning in the context of the whole embryo, DMZ explants from embryos that had been microinjected with Nck mutant mRNAs were also assayed by RT–PCR for expression of various mesodermal marker genes. In normal embryos, the DMZ is destined to form dorsal mesodermal tissues and corresponds to the Spemann organizer, a special region of the embryo that is responsible for induction of muscle and neural tissue and for coordinating cell movements during gastrulation (34). The dorsal mesoderm marker gsc was repressed by K229 in DMZ explants cultured to the late gastrula stage (stage 12; Fig. 4E) and both gsc and another dorsal marker, noggin, were repressed in explants cultured to the tailbud stage (stage 19; Fig. 4F). In contrast, expression of K229 up-regulated the ventral mesoderm marker Xwnt-8 (Fig. 4E). As in animal cap explants, the wild type and K308 had little, if any, effect on the expression of mesoderm marker genes in the DMZ.
Taken together, these data demonstrate that K229 affects dorso-ventral patterning. Its expression alters the manner in which induced mesoderm responds to exogenous or endogenous signals, thereby causing cells that normally would adopt dorsal mesodermal fates to adopt more ventral fates. The anterior truncation phenotype caused by mutants other than K229 might be due to a weaker ventralizing activity that cannot be detected under these experimental conditions, or it is possible that these mutants exert their effects through different mechanisms. None of the Nck mutants could dorsalize induced mesoderm (Fig. 4 C and D), and ventral injection of K229 had no phenotypic effect in whole embryos (not shown), suggesting that Nck is only capable of respecifying presumptive dorsal mesoderm.
Expression of Nck K229 Affects the Migration of Mesoderm Cell.
Migration of involuted mesoderm cells over the surface of the blastocoel is known to be important for gastrulation and subsequent anterior-posterior neural patterning of the embryo (35). We therefore examined the migration and ultimate fate of mesodermal cells in K229-injected embryos by lineage tracing with the histochemical marker β-galactosidase (36). Embryos were microinjected with β-galactosidase mRNA plus either Nck K229 or control KALL mRNA in the C1 blastomeres at the 32-cell stage. Embryos were later fixed, stained with X-Gal, and examined as whole mounts or sections. We selected the C1 blastomeres for injection because these cells give rise to the migratory deep marginal zone cells of the gastrula, which migrate toward the most anterior region of the embryo and ultimately populate the anterior structures most affected by Nck mutant expression (37, 38). The whole-embryo phenotype of embryos microinjected with K229 RNA into C1 blastomeres at the 32-cell stage was apparently normal, presumably because induced changes of cell fate were compensated for by other (uninjected) cells.
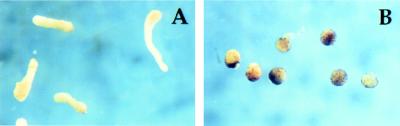
At the midgastrula stage, cells expressing the β-galactosidase lineage tracer in control embryos comprised migratory deep zone cells (leading endomesoderm cells), which crawl along the blastocoel roof as shown in Fig. 5A. In K229-injected embryos, however, fewer marked cells were in the leading endomesoderm, and most were found instead in the involuting marginal zone (Fig. 5B), suggesting that their migration over the blastocoel roof was delayed. In control tadpoles, descendants of C1 blastomeres ultimately populated head mesenchyme and head endoderm (Fig. 5C), whereas in embryos injected with K229 mRNA, far less staining was seen in the corresponding sites but was prominent in somites (Fig. 5D). These results show that K229 expression retards the migration of presumptive anterior mesoderm cells and respecifies their ultimate fate.
Figure 5.
Nck K229 disrupts migration of mesoderm cells. mRNA (0.1 ng) encoding nuclear-localized β-galactosidase plus 0.2 ng of either KALL control (A and C) or K229 (B and D) mRNA was injected into C1 blastomeres of 32-cell embryos. Embryos were fixed and stained with X-Gal at stage 12 (A and B) or stage 30 (C and D). (A and B) Embryos are positioned with animal pole at top and dorsal side to the right. (Bar = 100 μm.)
Implications.
In Xenopus embryos the dorso-ventral patterning of mesodermal tissues is determined by antagonizing dorsalizing and ventralizing signals (39–41). Members of the transforming growth factor-β, FGF, and Wnt families all have been shown to affect mesoderm patterning, and it will be of considerable interest to determine the specific pathways affected by expression of Nck mutants. The anterior defects we observe, including those in ectodermal derivatives such as the eyes and neural tissue, are all likely to be secondary effects of mesoderm respecification. However, at this time we cannot rule out direct effects on the adhesive or migratory properties of mesoderm in addition to effects on dorso-anterior patterning. For example, the expression of K229 inhibited spreading of mesoderm cells of both DMZ and activin-treated animal caps on a fibronectin substrate (not shown).
A number of genes can indirectly affect mesoderm cell migration in Xenopus gastrulation; among these, gsc and Xvent-1 are reported to mediate both mesoderm patterning and cell migration. Ectopic expression or overexpression of gsc dorsalizes ventral mesoderm and leads to accelerated involution toward the anterior of the embryo, probably by converting more cells into prospective head mesoderm (42). Xvent-1 expression can be induced by BMP-4, and overexpression of Xvent-1 ventralizes dorsal mesoderm and leads to respecification of notochord and head mesoderm to a more posterior fate due to a retardation of involution movements at gastrulation (29). The ventralizing activity and retardation of the mesoderm cell migration of K229-injected embryos is similar to the effects of Xvent-1 overexpression. Consistent with this, K229 expression induces endogenous Xvent-1 expression approximately 5-fold relative to controls in activin-treated animal caps (not shown).
The observation that constitutive membrane localization of Nck SH3 domains is sufficient for the anterior truncation phenotype, as is overexpression of wild-type Nck in some cases, suggests that ventralization might be due to an exaggerated or inappropriate form of a normal Nck-mediated signal. Among the tyrosine-phosphorylated molecules known to associate with Nck, FGF receptor and platelet-derived growth factor receptor are reported to affect cell movements during Xenopus gastrulation. However, data from the literature are inconsistent with these molecules being upstream of Nck in a ventralization pathway. For example, unlike K229, eFGF overexpression was shown to autoinduce ventral mesoderm in animal cap explants (43), and dominant–negative platelet-derived growth factor receptor-α did not affect mesoderm formation or patterning induced by FGF or activin (36). Focal adhesion kinase also can affect mesoderm formation and is highly phosphorylated during early development in mice (44). However, Focal adhesion kinase-deficient mice display a general defect of mesoderm development, and the cells from these embryos have reduced mobility in vitro (45, 46), unlike the ventralizing activity seen with the K229 mutant. Epistasis experiments will definitively establish whether Nck might function to relay signals from these proteins.
The ventralizing activity, anterior truncation, and retardation of the involuting migration reported here all might result from a defect in a single signaling pathway leading to respecification of dorso-ventral fate, or be the composite of effects on several signal pathways. It is therefore essential to identify the downstream effector protein(s) of Nck that mediate the biological effects described in this report. Because these effectors must bind to the SH3 domains of the Nck mutants that give rise to the phenotype, we have a straightforward means to biochemically isolate the critical interacting proteins and a rationale for assessing the relevance of candidate Nck-binding proteins.
Acknowledgments
We thank S. Sokol, M. Mercola, M. Whitman, and L. Zon for kindly providing plasmids and primers, Y. Eto of Ajinomoto Corp. for providing human recombinant activin A, S. Sokol and K. Symes for critically reading this manuscript, and E. Heinrich and E. Marieb for excellent technical assistance. B.J.M. is an Assistant Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- FGF
fibroblast growth factor
- DMZ
dorsal marginal zone
- RT–PCR
reverse transcriptase–polymerase chain reaction
Footnotes
References
- 1.Cohen G B, Baltimore D. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 2.Pawson T. Nature (London) 1995;373:573–579. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 3.Li W, Hu E, Skolnik E Y, Ullrich A, Schlessinger J. Mol Cell Biol. 1992;12:5824–5833. doi: 10.1128/mcb.12.12.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehman J M, Riethmuller G, Johnson J P. Nucleic Acids Res. 1990;18:1048. doi: 10.1093/nar/18.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W, Nishimura R, Kashishian A, Batzer A G, Kim W J H, Cooper J A, Schlessinger J. Mol Cell Biol. 1994;14:509–517. doi: 10.1128/mcb.14.1.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meisenhelder J, Hunter T. Mol Cell Biol. 1992;12:5843–5856. doi: 10.1128/mcb.12.12.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishimura R, Li W, Kashishian A, Mondino A, Zhou M, Cooper J, Schlessinger J. Mol Cell Biol. 1993;13:6889–6896. doi: 10.1128/mcb.13.11.6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan P J, Gillespie L L. Dev Biol. 1994;166:101–111. doi: 10.1006/dbio.1994.1299. [DOI] [PubMed] [Google Scholar]
- 9.Lee C-H, Li W, Nishimura R, Zhou M, Batzer A G, Myers J, White M F, Schlessinger J, Skolnik E Y. Proc Natl Acad Sci USA. 1993;90:11713–11717. doi: 10.1073/pnas.90.24.11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlaepfer D D, Hanks S K, Hunter T, van der Geer P. Nature (London) 1994;372:786–791. doi: 10.1038/372786a0. [DOI] [PubMed] [Google Scholar]
- 11.Rivero-Lezcano O M, Sameshima J H, Marcilla A, Robbins K C. J Biol Chem. 1994;269:17363–17366. [PubMed] [Google Scholar]
- 12.Hu G, Milfay D, Williams L T. Mol Cell Biol. 1995;15:1169–1174. doi: 10.1128/mcb.15.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rivero-Lezcano O M, Marcilla A, Sameshima J H, Robbin K C. Mol Cell Biol. 1995;15:5725–5731. doi: 10.1128/mcb.15.10.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derry J M J, Ochs H D, Francke U. Cell. 1994;78:635–644. [PubMed] [Google Scholar]
- 15.Bagrodia S, Derijard B, Davis R J, Cerione R A. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 16.Bokoch G, Wang Y, Bohl B P, Sells M A, Quilliam L A, Knaus U G. J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 17.Galisteo M L, Chernoff J, Su Y C, Skolnik E Y, Schlessinger J. J Biol Chem. 1996;271:20997–21004. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 18.Lu W, Katz S, Gupta R, Mayer B J. Curr Biol. 1997;7:85–94. doi: 10.1016/s0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- 19.Chou M M, Fajardo J E, Hanafusa H. Mol Cell Biol. 1992;12:5834–5842. doi: 10.1128/mcb.12.12.5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka M, Gupta R, Mayer B J. Mol Cell Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rockow S, Tang J, Xiong W, Li W. Oncogene. 1996;12:2351–2359. [PubMed] [Google Scholar]
- 22.Garrity P A, Rao Y, Salecker I, McGlade J, Pawson T, Zipursky S L. Cell. 1996;85:639–650. doi: 10.1016/s0092-8674(00)81231-3. [DOI] [PubMed] [Google Scholar]
- 23.Krieg P A, Melton D A. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newport J, Kirschner M. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 25.Nieuwkoop P D, Faber J. Normal Table of Xenopus laevis. Amsterdam: North–Holland; 1967. [Google Scholar]
- 26.Kao K R, Elinson R P. Dev Biol. 1988;127:64–77. doi: 10.1016/0012-1606(88)90189-3. [DOI] [PubMed] [Google Scholar]
- 27.Malacinski G M, Brothers A J, Chung H M. Dev Biol. 1977;56:24–39. doi: 10.1016/0012-1606(77)90152-x. [DOI] [PubMed] [Google Scholar]
- 28.Jones C M, Lyons K M, Lapan P M, Wright C V E, Hogan B L M. Development (Cambridge, UK) 1992;115:639–647. doi: 10.1242/dev.115.2.639. [DOI] [PubMed] [Google Scholar]
- 29.Gawantka V, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. EMBO J. 1995;24:6268–6279. doi: 10.1002/j.1460-2075.1995.tb00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mead P E, Brivanlou I H, Kelley C M, Zon L I. Nature (London) 1996;382:357–360. doi: 10.1038/382357a0. [DOI] [PubMed] [Google Scholar]
- 31.Holsinger L J, Spencer D M, Austin D J, Schreiber S L, Crabtree G R. Proc Natl Acad Sci USA. 1995;92:9810–9814. doi: 10.1073/pnas.92.21.9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellman D, Garber E A, Cross R R, Hanafusa H. Nature (London) 1985;314:374–377. doi: 10.1038/314374a0. [DOI] [PubMed] [Google Scholar]
- 33.Smith J C. Development (Cambridge, UK) 1987;99:3–14. doi: 10.1242/dev.99.1.3. [DOI] [PubMed] [Google Scholar]
- 34.Spemann H. Embryonic Development and Induction. New York: Yale Univ. Press; 1938. [Google Scholar]
- 35.Keller R, Winklebauer R. Curr Top Dev Biol. 1992;27:39–89. doi: 10.1016/s0070-2153(08)60532-3. [DOI] [PubMed] [Google Scholar]
- 36.Ataliotis P, Symes K, Chou M M, Mercola M. Development (Cambridge, UK) 1995;121:3099–3110. doi: 10.1242/dev.121.9.3099. [DOI] [PubMed] [Google Scholar]
- 37.Dale L, Slack J M W. Development (Cambridge, UK) 1987;99:527–551. doi: 10.1242/dev.99.4.527. [DOI] [PubMed] [Google Scholar]
- 38.Moody S A. Dev Biol. 1987;122:300–319. doi: 10.1016/0012-1606(87)90296-x. [DOI] [PubMed] [Google Scholar]
- 39.Kessler D S, Melton D A. Science. 1994;266:596–604. doi: 10.1126/science.7939714. [DOI] [PubMed] [Google Scholar]
- 40.De Robertis E M, Sasai Y. Nature (London) 1996;380:37–40. doi: 10.1038/380037a0. [DOI] [PubMed] [Google Scholar]
- 41.Smith J C. Curr Opin Cell Biol. 1995;7:856–861. doi: 10.1016/0955-0674(95)80070-0. [DOI] [PubMed] [Google Scholar]
- 42.Niehrs C, Keller R, Cho K W Y, De Robertis E M. Cell. 1993;72:491–503. doi: 10.1016/0092-8674(93)90069-3. [DOI] [PubMed] [Google Scholar]
- 43.Isaacs H V, Pownall M E, Slack J M W. EMBO J. 1994;13:4469–4481. doi: 10.1002/j.1460-2075.1994.tb06769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polte T R, Naftilan A J, Hanks S K. J Cell Biochem. 1994;55:106–119. doi: 10.1002/jcb.240550113. [DOI] [PubMed] [Google Scholar]
- 45.Ilic D, Furuta Y, Kanazawa S, Takeda N, Sobue K, Nakatsuji N, Nomura S, Fujimoto J, Okada M, Yamamoto T, Aizawa S. Nature (London) 1995;377:539–544. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- 46.Furuta Y, Ilic D, Kanazawa S, Takeda N, Yamamoto T, Aizawa S. Oncogene. 1995;11:1989–1995. [PubMed] [Google Scholar]