Abstract
A multiplex PCR assay which combines detection of bacteria of the genus Listeria, Listeria monocytogenes serotypes 1/2a and 4b, and epidemic clones I, II, and III of L. monocytogenes was developed. The assay provides a rapid, reliable, and inexpensive method for screening and subgrouping this important food-borne pathogen.
Listeria monocytogenes is a gram-positive, facultative, intracellular food-borne pathogen that is widely distributed in foods, the environment, and human and animal hosts. It can cause listeriosis, a severe disease with a high case fatality rate (20% to 30%) in immunocompromised populations (5). Approximately 2,500 human listeriosis cases are reported annually in the United States, including 500 deaths (15).
Three genetic lineages have been identified for L. monocytogenes using various subtyping techniques, with lineage I containing serotypes 1/2b, 3b, 3c, and 4b, lineage II containing serotypes 1/2a, 1/2c, and 3a, and lineage III containing serotypes 4a and 4c. Lineage I isolates include major epidemic clones of L. monocytogenes which caused a large number of human listeriosis cases (17), lineage II isolates are isolated mostly from foods and the environment (13), and lineage III isolates are mostly found in animal hosts (13). Previous molecular subtyping studies have identified four major epidemic clones of L. monocytogenes (ECI, ECII, ECIII, and ECIV) (3, 12). Among these epidemic clones, ECI, a serotype 4b cluster, appears to be a cosmopolitan clonal group associated with several major outbreaks in different countries, including the coleslaw (Nova Scotia, 1981), soft cheese (Switzerland, 1983 to 1987, and California, 1985), and pork tongue (France, 1992) outbreaks (13). ECII was first observed in the 1998-1999 U.S. multistate outbreak associated with contaminated hot dogs and the 2002 U.S. multistate listeriosis outbreak associated with contaminated turkey deli meat (13). ECIV, another serotype 4b cluster, caused a pâté outbreak (United Kingdom, 1988) and a vegetable outbreak (Boston, 1983). ECIII isolates are serotype 1/2a isolates associated with the hot dog (United States, 1989) and the turkey deli meat (United States, 2000) outbreaks. These ECIII isolates were considered epidemiologically related because they were found in the same food processing plant and had identical subtypes as determined by different molecular subtyping methods (3, 12). Besides these well-identified outbreaks, Sauders et al. (17) suggested that many concurrent sporadic listeriosis cases were also caused by the major epidemic clones of L. monocytogenes. For example, during the period between 1998 and 2002, two large temporal clusters of sporadic case isolates throughout Michigan, Ohio, and New York shared the same ribotype as ECII. Sauders et al. (17) further suggested that listeriosis outbreaks and sporadic clusters may be more commonly linked than previously assumed. Based on these findings, it is reasonable to speculate that previously identified epidemic clones may be involved in future listeriosis cases and outbreaks (3). Therefore, identification and tracking of these epidemic clones are important for understanding the long-term transmission of L. monocytogenes and the establishment of efficient surveillance systems for this pathogen.
Detecting the routes by which L. monocytogenes bacteria are transmitted to humans to cause listeriosis is often difficult because the number of isolates subtyped is often limited due to the high costs of advanced molecular subtyping methods such as pulsed-field gel electrophoresis (PFGE), ribotyping, and sequence-based typing. Serotyping of L. monocytogenes is often performed prior to PFGE as a first-step screening method for subgrouping and rapidly identifying isolates that need to be further analyzed by PFGE. However, serotyping often does not provide sufficient discriminatory power for screening L. monocytogenes. For example, three major epidemic clones (ECI, ECII, and ECIV) belong to the same serotype, 4b. While many food isolates are of serotype 1/2a, only a small portion of these result in cases and outbreaks of listeriosis. Therefore, new methods that are rapid and more discriminatory are needed to screen isolates before further subtyping. Food-processing plants often do not perform subtyping following identification of Listeria spp., and this does not permit tracking and thus elimination of those epidemic clones of L. monocytogenes. Combining species identification with molecular subtyping would simplify isolate characterization, tracking, and control of L. monocytogenes, especially those serotypes and epidemic clones responsible for most food-borne disease. Therefore, the purpose of the present study was to develop a multiplex PCR assay which detects bacteria of the genus Listeria, L. monocytogenes, and major serotypes and epidemic clones of L. monocytogenes.
The genetic marker iap, previously identified by Bubert et al. (2), was incorporated into this multiplex PCR assay to detect bacteria of the genus Listeria. Different Listeria genus markers reported in the literature have been tested; however, we found that the specificities of some markers were questionable because they generated positive results in other genera, such as Escherichia and Salmonella (data not shown). Also, some other markers, such as prs (5), did not yield an amplicon that could be well separated from other amplicons in the multiplex PCR assay. Zhang et al. (18) identified a genomic region containing part of lmo2234 and part of its upstream sequence that is a specific molecular marker for L. monocytogenes identification. Genomic sequences of this region were compared between different isolates of L. monocytogenes and Listeria spp., and a primer pair that can specifically amplify L. monocytogenes was developed (Fig. 1).
FIG. 1.
Multiple-sequence comparison of the genomic region used for development of the primer set for L. monocytogenes identification. Sequences and isolate identification numbers were obtained from the Broad Institute of Harvard and MIT (http://www.broad.mit.edu). The boxes indicate the priming regions of forward and reverse primers. Serotype designations are included in parentheses following the isolate identification numbers.
Many whole-genome sequences of Listeria spp. have been completed, including those of Listeria innocua (9), Listeria welshimeri (10), L. monocytogenes EGDe (9), F2365, H7858, and F6854 (16), and more recently, 16 newly sequenced strains, including J0161, 10403S, J1-194, R2-503, J2818, HPB2262, N1-017, J2-071, N3-165, F6900, LO28, J2-003, J1-175, F2-515, J2-064, and J1-208 (1). Some genetic markers of epidemic clone I and epidemic clone II have been previously investigated (6, 9). Nelson et al. (16) compared the whole-genome sequences of five Listeria strains and identified strain-specific genomic regions. Among them, F2365, H7858, and F6854 are representative isolates of ECI, ECII, and ECIII, respectively. In the present study, these strain-specific whole-genome regions identified by Nelson et al. (16) were compared with the above-mentioned 16 newly sequenced L. monocytogenes whole genomes by web-based BLAST analysis using default settings, and genetic markers for each epidemic clone were identified (Table 1). It is interesting that many of the epidemic clone markers belong to a few gene clusters. Determination of the functions of these gene clusters may help clarify why these epidemic clones have caused multiple major listeriosis outbreaks.
TABLE 1.
Epidemic clonal-specific genetic markers identified by genome comparison
| Epidemic clone | Isolate(s) |
|---|---|
| ECI | LMOf2365_2798 to LMOf2365_2800 |
| LMOf2365_2701 to LMOf2365_2707 | |
| LMOf2365_2347 to LMOf2365_2348 | |
| LMOf2365_0687 | |
| ECII | LMOh7858_0479.5 to LMOh7858_0479.7 |
| LMOh7858_0487.6, LMOh7858_0487.8 | |
| LMOh7858_0864.1 to LMOh7858_0867 | |
| LMOh7858_1161 to LMOh7858_1170.4 | |
| LMOh7858_2409.3 to LMOh7858_2409.4 | |
| LMOh7858_2444.2 | |
| LMOh7858_2455.2 | |
| LMOh7858_2462 to LMOh7858_2465 | |
| LMOh7858_2753 to LMOh7858_2764 | |
| LMOh7858_1164.1 to LMOh7858_1167 | |
| ECIII | LMOf6854_0077 |
| LMOf6854_1150 | |
| LMOf6854_2463.4 to LMOf6854_2470.2 | |
| LMOf6854_2651.1 | |
| LMOf6854_2653.2 | |
| LMOf6854_2674.3 | |
| LMOf6854_2678.1 | |
| LMOf6854_2682.11 | |
| LMOf6854_2772 |
Among the 13 serotypes of L. monocytogenes, isolates from serotypes 1/2a and 4b account for most major listeriosis outbreaks, and all identified epidemic clones of L. monocytogenes belong to these two serotypes. Therefore, serotype 1/2a- and 4b-specific markers (lmo0737 and ORF2110, respectively) identified by a previous microarray study (5) were incorporated into this multiplex PCR assay. Different Listeria spp. can share serotypes due to their similar surface antigen structures; however, lmo0737 and ORF2110 do not encode proteins which contribute to the surface antigen structures of L. monocytogenes (6). The specificities of these markers were further tested using a total of 200 L. monocytogenes isolates from 10 serotypes and 22 Listeria isolates from 5 serotypes (5). Combining the identification of these two serotypes into the multiplex PCR assay also allowed more accurate identification of epidemic clones of L. monocytogenes. For example, in the early stages of this study, genomic region 85 M, which was identified as an ECI marker by Herd and Kocks (11), was found in six L. monocytogenes isolates of other serotypes (1/2a and 4a) (data not shown). However, combining the serotype 4b marker (present study) and 85 M still allowed accurate identification of ECI. Nonetheless, to simplify the interpretation of the multiplex PCR results, this marker was replaced by 17B because 17B alone could specifically identify ECI isolates.
Among the ECI-specific genes, LMOf2365_2798 corresponds to fragment 17B, which was previously identified as an ECI marker by microarray analysis (11). Therefore, LMOf2365_2798 was chosen as the genetic maker for ECI in the multiplex PCR assay. Evans et al. (7) analyzed a special genomic region of L. monocytogenes between wap and inlA and identified some negative genetic markers for ECII. Analysis of this region between an ECI strain (F2365) and an ECII strain (H7858) allowed us to identify some positive markers for ECII. These positive markers were then confirmed by comparison of the newly finished 16 whole-genome sequences (Table 1). The 16 newly sequenced genomes include those of three ECIII strains and four other serotype 1/2a strains, which allowed identification of genetic markers specific to ECIII. One of these markers, gene LMOF6854_2463.4, was chosen as the ECIII marker in the multiplex PCR assay. Genomic properties and genome sequences of ECIV have not been reported, and therefore, ECIV markers were not identified and incorporated into the multiplex assay in this study. The ECIV isolates show patterns identical to those for other 4b isolates that do not belong to ECI or ECII.
The primer sequences used in this study, along with the putative functions of the selected genetic markers and the lengths of PCR products, are listed in Table 2. The specificity and sensitivity of the PCR method were evaluated with a total of 89 representative Listeria isolates obtained from the U.S. Centers for Disease Control and Prevention and the Food Safety Laboratory at Cornell University (Table 3) and 11 non-Listeria isolates from the American Type Culture Collection. All L. monocytogenes isolates were previously characterized by serotyping and ribotyping (3). L. monocytogenes isolates were grown overnight on trypticase soy yeast extract agar (Difco Laboratories, Becton Dickinson, Sparks, MD), and genomic DNA was extracted using an UltraClean Microbial DNA extraction kit (Mo Bio Laboratories, Solana Beach, CA) and stored at −20°C before use. One microliter of genomic DNA was added as a template. The seven primer sets were added at the following final concentrations: 50 nM for the ECI marker, 90 nM for the ECII marker, 30 nM for the ECIII marker, 100 nM for the serotype 4b marker, 30 nM for the serotype 1/2a marker, 30 nM for the L. monocytogenes marker, and 35 nM for the Listeria genus marker (Table 1). PCR was performed using QIAGEN multiplex PCR kits (QIAGEN Inc., Valencia, CA) and a Mastercycler thermocycler (Eppendorf Scientific, Hamburg, Germany), with an initial activation step for 15 min at 95°C prior to 15 cycles of 1 min at 94°C, 1 min with a touchdown from 55°C to 51°C (3 cycles per temperature), and 1 min at 72°C, followed by 15 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C and 1 final cycle for 8 min at 72°C. The 10-μl reaction mixture was mixed with 3 μl of loading buffer and separated on a 1.5% agarose gel in a 0.5× Tris-borate-EDTA buffer with a 1-h run time. The PCR product was visualized by ethidium bromide staining. The accuracy parameters (sensitivity and specificity) of this multiplex PCR were subsequently calculated. The sensitivity, sometimes referred as inclusivity, is defined as the percentage of positive samples that yield positive PCR signals (4, 14). The specificity, sometimes referred to as exclusivity, is defined as the percentage of negative samples that yield negative PCR signals (4, 14). To evaluate the reproducibility, the multiplex PCR was performed on each epidemic clonal isolate at least four times and on all other isolates at least three times.
TABLE 2.
Gene targets, primer sequences, and PCR amplicon sizes in the multiplex PCR assay
| Gene target | Isolate group | Primer sequencea | Amplicon size (bp) | Protein encoded | Source or reference |
|---|---|---|---|---|---|
| LMOf2365_2798 | ECI | (F) AATAGAAATAAGCGGAAGTGT | 303 | Unknown function | This study |
| (R) TTATTTCCTGTCGGCTTAG | |||||
| LMOh7858_0487.8 to inlA | ECII | (F) ATTATGCCAAGTGGTTACGGA | 889 | Unknown function | This study |
| (R) ATCTGTTTGCGAGACCGTGTC | |||||
| LMOF6854_2463.4 | ECIII | (F) TTGCTAATTCTGATGCGTTGG | 497 | Putative helicase-like | This study |
| (R) GCGCTAGGGAATAGTAAAGG | |||||
| ORF2110 | Serotype 4b | (F) AGTGGACAATTGATTGGTGAA | 597 | Putative secreted | 5 |
| (R) CATCCATCCCTTACTTTGGAC | |||||
| lmo0737 | Serotype 1/2a | (F) GAGTAATTATGGCGCAACATC | 724 | Unknown function | This study |
| (R) CCAATCGCGTGAATATCGG | |||||
| lmo2234 | Listeria monocytogenes | (F) TGTCCAGTTCCATTTTTAACT | 420 | Unknown function | This study |
| (R) TTGTTGTTCTGCTGTACGA | |||||
| iap | Listeria spp. | (F) ATGAATATGAAAAAAGCAAC | 1,450-1,600 | P60 | 2 |
| (R) TTATACGCGACCGAAGCCAAC |
(F), forward primer; (R), reverse primer.
TABLE 3.
Evaluation of multiplex PCR using representative isolates
| Species | Serotype | No. of isolates | Multiplex PCR result for:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| ECI | ECII | ECIII | 1/2a | 4b | L. monocytogenes | Listeria genus | |||
| L. monocytogenes | 4b (ECI) | 11 | + | − | − | − | + | + | + |
| 4b (ECII) | 20 | − | + | − | − | + | + | + | |
| 4b (non-ECs) | 24a | − | − | − | − | + | + | + | |
| 1/2a (ECIII) | 4 | − | − | + | + | − | + | + | |
| 1/2a (non-ECs) | 10 | − | − | − | + | − | + | + | |
| Non-1/2a, 4b | 12 | − | − | − | − | − | + | + | |
| L. innocua | NAb | 2 | − | − | − | − | − | − | + |
| Listeria ivanovii | NA | 2 | − | − | − | − | − | − | + |
| L. welshimeri | NA | 2 | − | − | − | − | − | − | + |
| Listeria seeligeri | NA | 2 | − | − | − | − | − | − | + |
| Salmonella enterica | Enteritidis | 2 | − | − | − | − | − | − | − |
| Escherichia coli | O157 | 1 | − | − | − | − | − | − | − |
| NA | 1 | − | − | − | − | − | − | − | |
| Bacillus cereus | 2 | − | − | − | − | − | − | − | |
| Bacillus subtilis | 2 | − | − | − | − | − | − | − | |
| Bacillus licheniformis | 1 | − | − | − | − | − | − | − | |
| Staphylococcus aureus | NA | 2 | − | − | − | − | − | − | − |
Two 4b isolates that do not belong to any of the epidemic clones have the ECII marker.
NA, not available.
Figure 2 shows some typical examples of multiplex PCR results for representative isolates of Listeria spp. and other species, and Table 3 summarizes the results for all isolates. Gel electrophoresis and sequencing of amplicons confirmed that all primer pairs specifically amplified the desired PCR products. The maximum number of amplicons generated per isolate was four, and the different amplicons were always well separated. The multiplex PCR specifically identified bacteria of the genus Listeria, L. monocytogenes serotypes 1/2a and 4b, and all three epidemic clones, with the exception that two serotype 4b isolates that are not part of ECII generated ECII amplicons. Based on the results from our limited number of isolates, the sensitivity is 100% for all seven pairs of primers, and the specificity is 100% for all primers, except that the specificity of the ECII primer is 97.5%. The intralaboratory reproducibility of this assay is 100%. The seven different targets (amplicons) in this multiplex PCR assay had sharp and uniform intensities among all isolates except the Listeria genus marker iap. The primer pair for iap amplification was designed based on the regions in iap conserved among Listeria spp. It is possible that there are one or two nucleotide mutations in the priming regions that caused the differing amplicon intensities between isolates. One L. innocua isolate yielded a 490-bp amplicon with a size similar to that of the ECIII marker; however, this isolate did not yield an L. monocytogenes marker and therefore was correctly identified as a member of the Listeria genus. A 1,000-bp nonspecific amplification was observed in one L. monocytogenes serotype 4a isolate and one serotype 4c isolate; however, this did not affect the unambiguous characterization of these two isolates.
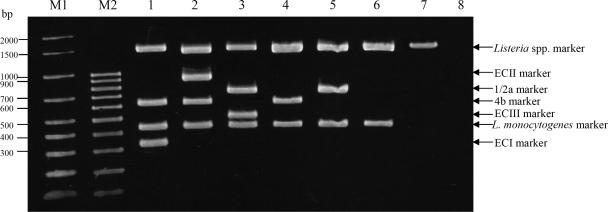
FIG. 2.
Agarose gel electrophoresis results for the multiplex PCR assay with representative isolates of Listeria species. Lanes 1 to 6, L. monocytogenes reference isolates of ECI (F2365), ECII (H7858), ECIII (F6854), 4b but non-EC (J1-129), 1/2a but non-ECIII (F2-663), and non-4b and -1/2a (R2-503), respectively; lane 7, Listeria ivanovii (C2-011); lane 8, Bacillus cereus; lanes M1 and M2, Bio-Rad molecular weight markers (Bio-Rad, Hercules, CA). Genetic markers corresponding to the amplified fragments are indicated on the right. Molecular sizes are given (in base pairs) at the left. Isolates were obtained from the CDC and Cornell University, and original isolate identification numbers are used.
The multiplex PCR assay that was developed did not differentiate serotype 1/2a from 3a or serotype 4b from 4d or 4e. However, serotypes 3a, 4e, 4d, and 7 are rarely found in food and patients. Farber and Peterkin (8) analyzed the serotype distribution of isolates of worldwide listeriosis cases and found that 95% of isolates in listeriosis cases belong to serotypes 1/2a, 1/2b, and 4b. The data from the National Reference Center in France showed that over 98% of the food and clinical isolates of L. monocytogenes between 2001 and 2004 belonged to serotypes 1/2a, 1/2b, 1/2c, and 4b (5). Therefore, the inability to differentiate rare serotypes would not decrease the utility of this multiplex PCR assay for the long-term surveillance and epidemiology of L. monocytogenes.
Combining the seven genetic markers allowed simultaneous species identification and detection of major serotypes and epidemic clones associated with human listeriosis. By using a single multiplex PCR, one can quickly tell if an isolate belongs to the genus Listeria or L. monocytogenes serotype 1/2a or 4b and/or epidemic clone I, II, or III. Therefore, this single assay provides critical information about the subtypes of test isolates. This multiplex PCR assay can be used for screening and subgrouping L. monocytogenes after pure cultures are obtained and significantly reduces the number of isolates that need to be subtyped by more expensive and discriminatory molecular methods, like PFGE and sequence-based typing. This assay can also be adopted by small public health laboratories, food-testing laboratories, and food industries which cannot afford more expensive methods and equipment or trained technicians.
Acknowledgments
This study was supported by a U.S. Department of Agriculture Special Milk Safety grant to The Pennsylvania State University.
We thank Martin Wiedmann, Bala Swaminathan, and Stephanie Doores for providing some of the bacterial strains used in this research. We also thank Wei Zhang for his advice on genomic sequence analysis.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Broad Institute of Harvard and MIT. 27 September 2006. Listeria monocytogenes sequencing project. http://www.broad.mit.edu.
- 2.Bubert, A., S. Kohler, and W. Goebel. 1992. The homologous and heterologous regions within the iap gene allow genus- and species-specific identification of Listeria spp. by polymerase chain reaction. Appl. Environ. Microbiol. 58:2625-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Y., W. Zhang, and S. J. Knabel. 2007. Multi-virulence-locus sequence typing identifies single nucleotide polymorphisms which differentiate epidemic clones and outbreak strains of Listeria monocytogenes. J. Clin. Microbiol. 45:835-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Agostino, M., M. Wagner, J. A. Vazquez-Boland, T. Kuchta, R. Karpiskova, J. Hoorfar, S. Novella, M. Scortti, J. Ellison, A. Murray, I. Fernandes, M. Kuhn, J. Pazlarova, A. Heuvelink, and N. Cook. 2004. A validated PCR-based method to detect Listeria monocytogenes using raw milk as a food model—towards an international standard. J. Food Prot. 67:1646-1655. [DOI] [PubMed] [Google Scholar]
- 5.Doumith, M., C. Buchrieser, P. Glaser, C. Jacquet, and P. Martin. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doumith, M., C. Cazalet, N. Simoes, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, M. R., B. Swaminathan, L. M. Graves, E. Altermann, T. R. Klaenhammer, R. C. Fink, S. Kernodle, and S. Kathariou. 2004. Genetic markers unique to Listeria monocytogenes serotype 4b differentiate epidemic clone II (hot dog outbreak strains) from other lineages. Appl. Environ. Microbiol. 70:2383-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 10.Hain, T., C. Steinweg, C. T. Kuenne, A. Billion, R. Ghai, S. S. Chatterjee, E. Domann, U. Karst, A. Goesmann, T. Bekel, D. Bartels, O. Kaiser, F. Meyer, A. Puhler, B. Weisshaar, J. Wehland, C. Liang, T. Dandekar, R. Lampidis, J. Kreft, W. Goebel, and T. Chakraborty. 2006. Whole-genome sequence of Listeria welshimeri reveals common steps in genome reduction with Listeria innocua as compared to Listeria monocytogenes. J. Bacteriol. 188:7405-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herd, M., and C. Kocks. 2001. Gene fragments distinguishing an epidemic-associated strain from a virulent prototype strain of Listeria monocytogenes belong to a distinct functional subset of genes and partially cross-hybridize with other Listeria species. Infect. Immun. 69:3972-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kathariou, S. 2003. Foodborne outbreaks of listeriosis and epidemic-associated lineages of Listeria monocytogenes, p. 243-256. In M. E. Torrence and R. E. Isaacson (ed.), Microbial food safety in animal agriculture. Iowa State University Press, Ames, IA.
- 13.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 14.Lübeck, P. S., P. Wolffs, S. L. On, P. Ahrens, P. Rådström, and J. Hoorfar. 2003. Toward an international standard for PCR-based detection of food-borne thermotolerant Campylobacters: assay development and analytical validation. Appl. Environ. Microbiol. 69:5664-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mead, P. S., E. F. Dunne, L. Graves, M. Wiedmann, M. Patrick, S. Hunter, E. Salehi, F. Mostashari, A. Craig, P. Mshar, T. Bannerman, B. D. Sauders, P. Hayes, W. Dewitt, P. Sparling, P. Griffin, D. Morse, L. Slutsker, and B. Swaminathan. 2006. Nationwide outbreak of listeriosis due to contaminated meat. Epidemiol. Infect. 134:744-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauders, B. D., Y. Schukken, L. Kornstein, V. Reddy, T. Bannerman, E. Salehi, N. Dumas, B. J. Anderson, J. P. Massey, and M. Wiedmann. 2006. Molecular epidemiology and cluster analysis of human listeriosis cases in three U.S. states. J. Food Prot. 69:1680-1689. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, W., A. Hughes, G. Wilt, and S. J. Knabel. 2004. The BAX PCR assay for screening Listeria monocytogenes targets a partial putative gene lmo2234. J. Food Prot. 67:1507-1511. [DOI] [PubMed] [Google Scholar]