Abstract
The objectives of this study were to characterize Fibrobacter succinogenes glycoside hydrolases from different glycoside hydrolase families and to study their synergistic interactions. The gene encoding a major endoglucanase (endoglucanase 1) of F. succinogenes S85 was identified as cel9B from the genome sequence by reference to internal amino acid sequences of the purified native enzyme. Cel9B and two other glucanases from different families, Cel5H and Cel8B, were cloned and overexpressed, and the proteins were purified and characterized. These proteins in conjunction with two predominant cellulases, Cel10A, a chloride-stimulated cellobiosidase, and Cel51A, formerly known as endoglucanase 2 (or CelF), were assayed in various combinations to assess their synergistic interactions using ball-milled cellulose. The degree of synergism ranged from 0.6 to 3.7. The two predominant endoglucanases produced by F. succinogenes, Cel9B and Cel51A, were shown to have a synergistic effect of up to 1.67. Cel10A showed little synergy in combination with Cel9B and Cel51A. Mixtures containing all the enzymes gave a higher degree of synergism than those containing two or three enzymes, which reflected the complementarity in their modes of action as well as substrate specificities.
The degradation of complex plant cell wall polysaccharides, such as cellulose and hemicellulose, by cellulolytic microorganisms is one of the major steps of the global carbon cycle. Cellulolytic organisms typically produce multiple glycoside hydrolases that act in concert, releasing mono- and oligosaccharides from the polysaccharides. The collective activity of glycoside hydrolases is often greater than the sum of the individual enzyme activities, as a result of their synergistic interactions. Synergistic effects among endoglucanases, exoglucanases, and β-glucosidases have been reported, and the exoglucanases have a central role in cellulose degradation (25).
Fibrobacter succinogenes has a major role in biodegradation of plant cell wall polymers in the rumen, based on its predominance in the rumens of animals ingesting a forage diet (30, 37, 41) and its capacity to digest plant cell walls during growth (8). Because of these features, the cellulase system of F. succinogenes subsp. succinogenes S85 has been extensively studied (11, 17, 19). Previously, two of the major cellulolytic enzymes of F. succinogenes, EG1 and EG2 (28), which account for 32% and 10% of endoglucanase activity produced, respectively, and a chloride-stimulated cellobiosidase (ClCBase) (14) were purified and characterized. However, no further studies were conducted with these enzymes, except for the cloning and characterization of EG2 (27, 31). The sequence of the ClCBase gene was recently identified, and its product was designated Cel10A (18), but the gene encoding EG1 remained unknown. In a separate study, several glycoside hydrolases, including the family 8 cellulase Cel8B (FSU2303) and the family 5 cellulase Cel5H (FSU2914), were shown to be produced by F. succinogenes S85, as indicated by proteomic analysis (M. Morrison et al., unpublished data).
Including the cellulases cloned and/or characterized in the past, a total of 33 glycoside hydrolases that may have a role in cellulose degradation have been identified as being encoded by the genome of F. succinogenes (32). However, no glycoside hydrolase family 6 or family 48 genes, which often code for exoglucanases, were found. To explore the mechanism of cellulase biodegradation, we have undertaken a systematic study to assess the synergistic interaction of enzymes produced by F. succinogenes. In this study, the gene encoding the predominant cellulase EG1 was identified as cel9B. The synergistic interaction of this enzyme with four other cellulases from different families, EG2 (Cel51A [27]), Cel5H, Cel8B, and Cel10A, was explored to capture the diversity of hydrolase activities of these proteins produced by F. succinogenes.
MATERIALS AND METHODS
Bacterial strains, plasmids and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. F. succinogenes strain S85 (ATCC 19169) was grown in medium CDM (35) with either glucose (0.5%, wt/vol) or Avicel cellulose PH105 (0.3%, wt/vol) as the carbohydrate source. The inoculum was 3% (vol/vol), and the cultures were incubated at 37°C with shaking at 100 rpm and harvested in the late exponential phase of growth from glucose medium after 12 h and from cellulose medium after 36 h.
TABLE 1.
Strains, plasmids, and PCR primers used in this study
| Strain, plasmid, or primer | Descriptiona | Source, reference, or ATCC no. |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 | Invitrogen |
| BL21(DE3) | F−dcm ompT hsdS (rB− mB−) gal | Novagen |
| Rosetta gami(DE3) | Δara-leu7697 ΔlacX74 ΔphoAPvuII phoR araD139 ahpC galE galK rpsL F′[lac + (lacIq)pro] gor522::Tn10 (Tcr) trxB::Kan (DE3) pRARE6 (Cmr) | Novagen |
| Fibrobacter | ||
| F. succinogenes S85 | ATCC 19169 | |
| F. succinogenes A3C | ATCC 51219 | |
| F. intestinalis DR7 | ATCC 43855 | |
| F. intestinalis NR9 | ATCC 43854 | |
| Plasmids | ||
| pQE80L | Cloning and expression vector, Apr | QIAGEN |
| pET30a | Cloning and expression vector, Kmr | Novagen |
| pQE-ClCBase | Apr, cel10A gene from F. succinogenes S85 | This study |
| pQE-Cel8B | Apr, cel8B gene from F. succinogenes S85 | This study |
| pET30a-Cel9B | Kmr, cel9B gene from F. succinogenes S85 | This study |
| pET30a-Cel9BΔBTD | Kmr, cel9B gene from F. succinogenes S85, basic C-terminal domain removed | This study |
| pET30a-Cel5H | Kmr, cel5H gene from F. succinogenes S85 | This study |
| pLE69B | Apr, cel51A gene from F. succinogenes S85 | 26 |
| Primers | ||
| ClCBase-1 | TTAATGCATGCCAGTTGAACAATGGTGATATG (SphI) | This study |
| ClCBase-2C | TATTTGTCGACTTACTTCACGGTAATCTGG (SalI) | This study |
| Cel9B-P4 | ATATGGATCCGCGACTGCCTATATCAAC (BamHI) | This study |
| Cel9B-P3C | TATCTTCTCGAGTTACTTCAGATGGTTG (XhoI) | This study |
| Cel9BΔBTD-P5C | TATCTTCTCGAGTTACTTCGCAACACCCGGTG (XhoI) | This study |
| Cel8B-1 | TTAATGAGCTCGGGCGATTCCCGTTCCCGC (SacI) | This study |
| Cel8B-2C | TATTTCCCGGGTTACTTCACGTTGACGCGG (XmaI) | This study |
| Cel5H-1 | CTCTGTGGATCCGTGACTGCTAGCCGTGTTGG (BamHI) | This study |
| Cel5H-2C | ACGCTGCTCGAGTTACTTGATGGAGATACG (XhoI) | This study |
In sequences, the restriction sites for the endonucleases in parentheses are in boldface.
Identification of the gene coding for EG1.
EG1 was purified from the extracellular culture fluid of F. succinogenes S85 grown on cellulose as described by McGavin and Forsberg (28). The purified enzyme was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to in-gel trypsin digestion, matrix-assisted laser desorption ionization-time of flight mass spectrometry (MS) (19) and tandem MS at the Advanced Protein Technology Centre of the Hospital for Sick Children in Toronto (www.sickkids.on.ca/APTC) to obtain peptide masses and internal amino acid sequences that were used to identify the gene in the F. succinogenes genome (www.tigr.org).
Cloning, overexpression, and purification of Cel9B (FSU2361; EG1), Cel10A (FSU0257; ClCBase), Cel5H (FSU2914), and Cel8B (FSU2303).
The genes coding for each of the individual proteins were amplified by PCR using genomic DNA of F. succinogenes S85 as the template and primers listed in Table 1. Restriction endonuclease sites were introduced by the primers to facilitate subsequent cloning. The PCR amplicons were digested by appropriate restriction endonucleases and cloned into the corresponding restriction sites of expression plasmid pQE80L or pET30a. All constructs were fusion proteins with an N-terminal six-histidine tag. Cel9B, Cel9BΔBTD, Cel5H, and Cel51A were produced in E. coli BL21(DE3). The cells were cultured in Luria-Bertani (LB) medium containing either kanamycin (34 μg/ml; for the expression of Cel9B, Cel9BΔBTD, and Cel5H) or ampicillin (100 μg/ml; for the expression of Cel51A) at 18°C until an optical density at 600 nm of ≅0.7 was reached, when expression was induced with a final concentration of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 24 h. Cel10A and Cel8B were produced in E. coli Rosetta gami(DE3) in LB medium supplemented with 2.5 mM betaine and 0.5 M d-sorbitol (5) and the antibiotics chloramphenicol (34 μg/ml), kanamycin (34 μg/ml), tetracycline (12.5 μg/ml), and ampicillin (100 μg/ml) at 18°C, and expression was induced by IPTG as described above. The cells were collected by centrifugation (9,000 × g, 15 min, 4°C), resuspended in binding buffer (20 mM sodium phosphate, 0.5 M NaCl, and 30 mM imidazole, pH 7.5), and disrupted by three passes through a French press (Thermo Electron Corporation, Burlington, Ontario, Canada) at a pressure of 8,300 kPa, and cell debris, including inclusion bodies, was removed by centrifugation (15,000 × g, 30 min, 4°C).
The fractions of Cel5H, Cel8B, Cel9B, and Cel10A not sedimentable at 15,000 × g for 30 min were subjected to immobilized metal affinity chromatography (IMAC) as described in the instruction manual (instruction 11-0008-87 AB; http://www.chromatography.amersham-biosciences.com). Cel51A was purified as described before (27).
For further purification of Cel9B, 2 ml of eluted sample containing approximately 10 mg of protein from IMAC was diluted in 200 ml of 0.05 M potassium phosphate buffer (pH 6.5) and concentrated to 5 ml using a PM-10 membrane with a 10-kDa cutoff. The concentrate was applied to a 2.5- by 10-cm column of DEAE-Sepharose CL-6B equilibrated in starting buffer (50 mM potassium phosphate buffer, pH 6.5, 0.01% [wt/vol] sodium azide), followed by application of a 500-ml linear gradient from 0 to 1 M NaCl in 50 mM potassium phosphate, pH 6.5. The flow rate was 50 ml/h, and 5-ml fractions were collected. The enzyme was eluted from the column with 0.08 to 0.12 M of NaCl, as determined by measuring the conductivity of the fractions. The fractions containing carboxymethyl cellulase activity were combined and concentrated, and the purity was checked using SDS-PAGE.
For further purification of Cel10A, a portion of the eluted sample from IMAC was diluted 10-fold with 0.02 M potassium phosphate buffer (pH 6.5) and loaded onto a hydroxylapatite column (1.0 by 30 cm) preequilibrated with 0.02 M potassium phosphate buffer (pH 6.5). After the sample was applied, the column was washed with 30 ml of 0.05 M potassium phosphate buffer (pH 6.5), and the proteins were eluted with a linear potassium phosphate gradient (0.05 to 0.6 M). The enzyme was eluted from the column with 0.4 to 0.5 M of potassium phosphate, as determined by measuring the conductivity of the fractions. The fractions containing recombinant ClCBase (rcClCBase) activity were combined, and the purity was checked using SDS-PAGE. Prior to enzyme assays, the purified rcClCBase and native ClCBase were dialyzed against 10 mM imidazole buffer (pH 6.5) to remove chloride.
Enzyme assays and protein estimation.
Cellobiosidase activity was assayed as described previously (14), and protein concentration was determined using the method of Bradford (6) except for solutions containing urea where the bicinchoninic acid method was used (36). The protein standard was bovine serum albumin (Sigma).
Glycoside hydrolase activities on polysaccharides were assayed by incubating appropriately diluted enzymes in an assay mixture containing 1% (wt/vol) low-viscosity carboxymethyl cellulose (CMC; with a degree of derivatization of 0.7), 1% (wt/vol) medium-viscosity CMC, 1% (wt/vol) Avicel PH105, 1% (wt/vol) hydroxyethyl cellulose, 1% (wt/vol) barley β-glucan, 1% (wt/vol) lichenin, 0.5% (wt/vol) ball-milled Avicel cellulose, or 1% (wt/vol) barley β-glucan in 0.05 M sodium phosphate buffer (pH 6.5) at 37°C for 10 min to 2 h with end-over-end rotation (8 rpm). The reducing sugar was detected by the p-hydroxybenzoic acid hydrazide methods (22). To measure the time course in synergism experiments with a mixture of enzymes, 800 μl substrate and enzyme mixture was incubated at 37°C with end-over-end rotation and 50-μl samples were taken. The assay mixture contained 50 mM HEPES buffer (pH 6.5), 1 mM of MgCl2, 200 mM NaCl, 0.01% (wt/vol) sodium azide, and appropriately diluted enzymes. All assays were done in triplicate and repeated at least twice.
Ball-milled cellulose and barley straw were prepared by mixing 200 ml of a 6% (wt/vol) aqueous suspension of Avicel cellulose PH105 or barley straw with flint balls in an 800-ml Mill jar (Norton Chemical Process Products Division, OH) at 70 rpm for 24 h at 4°C. Acid-swollen cellulose (amorphous) was prepared as described previously (28).
Binding assay.
For qualitative analysis of binding, the purified proteins (100 μg) were mixed with 12 mg of Avicel cellulose PH105 or ball-milled barley straw in a final volume of 0.5 ml of 20 mM Tris, pH 7.5. After incubation at 4°C for 1 h with end-over-end rotation, the mixtures were centrifuged at 10,000 × g for 5 min to sediment the substrates and bound proteins. The cellulose substrates were washed twice, once with 20 mM Tris (pH 7.5) and then once with 20 mM Tris (pH 7.5) containing 1 M NaCl buffer, each time sedimenting the cellulose by centrifugation at 10,000 × g for 5 min. To assess qualitative binding of enzymes to cellulose, the cellulose with the bound proteins was mixed with 50 μl of SDS sample buffer by heating at 90°C for 3 min and the supernatant was analyzed by 10% SDS-PAGE.
For quantitative analysis of binding, the purified Cel10A proteins (5 to 150 μg) were mixed with 1 mg Avicel cellulose PH105, ball-milled barley straw, or 50 μl of a 20% (wt/vol) bacterial microcrystalline cellulose (BMCC) prepared as described by Väljamäe et al. (38) in a final volume of 0.5 ml of a 20 mM Tris buffer (pH 7.5). The mixtures were incubated at 4°C for 1 h with end-over-end rotation, followed by centrifugation at 13,000 × g for 5 min. The supernatant containing unbound proteins was collected and the protein concentration measured at 280 nm by reference to a known concentration of Cel10A. All assays were conducted in duplicate. The following equation of Sakoda and Hiromi (34) was used to obtain affinity parameters: [PC] = [P][PC]max/(Kd + [P]), where [PC] is the amount of bound protein, [P] is the amount of unbound protein expressed in μM, Kd (μM) is the equilibrium dissociation constant, and [PC]max (μmol per g cellulose) is the maximum amount of protein bound, respectively.
Computational analysis.
Peptide sequence data were used as query sequences in tBLASTx searches of the F. succinogenes S85 genome sequence data available at the TIGR unfinished genome website (http://www.tigr.org). The open reading frames (ORF) including the query sequences within the genomic DNA sequence were identified using ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Each ORF was analyzed to determine the protein family and domain organization using the Pfam search server (http://www.sanger.ac.uk/Software/Pfam/) and NCBI BLAST server (http://www.ncbi.nlm.nih.gov/BLAST). The signal peptide cleavage site was predicted by using SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/). Multiple sequence alignments were performed by ClustalW (http://www.ebi.ac.uk/clustalw). Statistical analysis was completed by using the General Linear Model procedure in SAS (version 9.1, 2003; SAS Institute, Inc., Cary, NC).
Nucleotide sequence accession numbers.
The nucleotide sequences of the genes cel9B, cel8B, and cel5H from F. succinogenes S85 have been deposited in the GenBank database under accession numbers EU055604, EU055605, and EU055606, respectively.
RESULTS
Identification of the gene coding for EG1 purified from F. succinogenes S85 and sequence analysis of EG1, Cel8B, and Cel5H.
Endoglucanase 1 (EG1) was purified previously (28). Matrix-assisted laser desorption ionization-time of flight MS analysis of tryptic peptides of the EG1 protein enabled identification of a cellulase gene annotated as cel9B (FSU2361) from the genome of F. succinogenes S85. The tryptic peptides identified accounted for 46.5% of the mature protein sequence, and the sequences were dispersed throughout the entire protein except for the C-terminal 88-amino-acid region (see Fig. S1 in the supplemental material). Among peptides detected by MS, five were sequenced by tandem MS (see Fig. S1 in the supplemental material). Cel9B contained a short signal peptide, an N-terminal immunoglobulin (Ig)-like structure, a family 9 glycoside hydrolase catalytic domain, and a 43-amino-acid basic C-terminal domain (BTD) with a pI of 12.0 (Fig. 1; see Fig. S2 in the supplemental material). The gene encoding the enzyme was found to be the celE gene cloned by Malburg et al. (26), except that the cel9B sequence encoding amino-terminal residues 1 to 149 was missing from celE. The amino-terminal region contained the entire N-terminal Ig-like domain as well as the N-terminal 36 amino acids of the catalytic domain. Truncation of the N-terminal region apparently had inactivated the enzyme. Consequently, CelE was not further characterized by Malburg et al. (26). The celE gene, now cel9B, encodes a protein with 70% amino acid sequence identity to that encoded by the celD (present designation: cel9C) gene, characterized by Malburg et al. (26).
FIG. 1.
Cellulases used in this study and their domain structures. GH, glycoside hydrolase; pentagons, His6 tags from vectors. BTDs were divided into two groups (I and II) based on their sequence similarity.
Two other novel cellulases cloned in this study included family 8 cellulase Cel8B (FSU2303) and family 5 cellulase Cel5H (FSU2914), both of which were known to be produced by F. succinogenes S85 (Morrison et al., unpublished). Cel8B contained an N-terminal glycoside hydrolase family 8 catalytic domain followed by an internal domain of unknown function (Fig. 1; see Fig. S2 in the supplemental material). There was a basic terminal domain (Fig. 1; pI = 10.3) at the C terminus, but no sequence similarity was found between this domain and BTDcel9B. The N-terminal catalytic domain was homologous to those of endoglucanase C and CelA from two strains of Clostridium spp. (1, 9), while no protein sequence was found to have similarity to the internal domain. Cel5H (Fig. 1; see Fig. S2 in the supplemental material) contained an N-terminal GH5 catalytic domain followed by two repeat domains, which were homologues to the putative CBM11 domain of endoglucanase 3 from F. succinogenes (29). There was also a basic terminal domain (pI = 10.8) which had low but significant similarity to BTDcel8B. Cel5H showed 95% sequence identity to an endoglucanase from the anaerobic fungus Orpinomyces joyonii, which suggests a recent horizontal gene transfer occurred between the two organisms (24).
The above-mentioned cellulases as well as Cel10A (ClCbase/FSU0257; Fig. 1; see Fig. S2 in the supplemental material) and Cel51A (CelF/FSU0382) were purified as described in Material and Methods, and the purified proteins were used in the subsequent studies (see Fig. S3 in the supplemental material for an SDS-PAGE analysis of the purified proteins).
Substrate specificity.
Table 2 shows the activities of the four enzymes on polymeric substrates. All four enzymes exhibited activity on CMC, with Cel9B the highest (25.0 U/mg) and Cel5H the lowest (0.091 U/mg).
TABLE 2.
Substrate specificities of rcCel9B, rcCel10A, rcCel8B, and rcCel5H on different polysaccharides
| Substrate | Substrate specificitya (U/mg) ± SD of:
|
|||
|---|---|---|---|---|
| rcCel9B | rcCel10A | rcCel8B | rcCel5H | |
| CMC | 25.00 ± 5.13 (100) | 2.22 ± 0.34 (100) | 0.83 ± 0.17 (100) | 0.09 ± 0.05 (100) |
| Hydroxyethyl cellulose | 9.75 ± 4.13 (39) | 1.11 ± 0.40 (50) | 0.13 ± 0.04 (16) | 0.03 ± 0.00 (32) |
| Ball-milled cellulose | 0.17 ± 0.03 (1) | 0.04 ± 0.00 (2) | 0.08 ± 0.01 (9) | 0.01 ± 0.00 (12) |
| Avicel PH105 | 0.05 ± 0.02 (<1) | 0 | 0 | 0 |
| Sigmacel 100 | 0.07 ± 0.01 (<1) | 0.05 ± 0.00 (2) | 0.02 ± 0.00 (2) | 0 |
| Lichenin | 51.83 ± 5.36 (207) | 0 | 0.30 ± 0.08 (37) | 0.03 ± 0.00 (30) |
| Barley β-glucan | 47.78 ± 1.96 (191) | 6.09 ± 0.53 (274) | 0.56 ± 0.01 (67) | 0.04 ± 0.02 (45) |
| Oat-spelled xylan | 0.15 ± 0.05 (<1) | 1.35 ± 0.34 (61) | 0.10 ± 0.05 (12) | 0.02 ± 0.00 (21) |
| Mannan | 0 | 0.13 ± 0.04 (6) | 0.02 ± 0.00 (3) | 0 (0) |
One unit of enzyme activity is the amount that produces 1 μmol of product per min. Values in the parentheses are relative activities (percent) compared to respective activities on CMC.
The specific activities of Cel9B on barley β-glucan and lichenin were similar and twice as much as on CMC. It was reported that the native EG1 had similar activities on the three substrates (28). Cel10A had the highest activity on the barley β-glucan but no detectable activity on lichenin. Cel8B and Cel5H had highest activity on CMC. Cel10A had higher activity on oat-spelled xylan than the other enzymes. All four enzymes had either low or no activity on Avicel, Sigmacel, and mannan.
Biochemical characteristics of individual enzymes.
The kinetic parameters and other biochemical properties of the purified enzymes are presented in Table 3. All enzyme assays were carried out with CMC as the substrate, except for those of the native ClCBase and rcClCBase, where p-nitrophenyl cellobioside was used. rcCel9B and rcCel9BΔBTD (reCel9B with the BTD removed) showed higher Km and lower Vmax values (Table 3) than those previously reported for the native enzyme (28). However, rcCel9BΔBTD had a lower Km and higher Vmax than the intact rcCel9B. Divalent metal ions Ca2+ and Mg2+ at 1 mM stimulated enzyme activity of both rcCel9B and rcCel9BΔBTD by 33% and 48%, respectively, while 5 mM EDTA abolished enzyme activity completely. Both rcCel9B and rcCel9BΔBTD had pH optima of 6.0 and retained over 80% of activity at pH values from 5.5 to 8.0. The temperature optima of both constructs were identical at 37°C. Both pH and temperature optima were similar to those of the native EG1. When 100 μg of Cel9B was incubated with a 0.8-mg Whatman no. 1 filter paper disk at 37°C for 16 h, 1.7 μg of reducing sugar (glucose equivalent) was produced. The soluble reducing sugar accounted for 89.2% of all the reducing sugar produced, while 10.8% of the reducing end was associated with the filter paper.
TABLE 3.
Catalytic propertiesa and other characteristics of EG1, rcCel9B, rcCel9BΔBTD, ClCBase, rcCel8B, and rcCel5H
| Enzyme (reference) | Km | Vmax (U/mg) | kcat (s−1) | kcat/Km (ml/s · mg) | Binding to cellulose | Temp optimum (oC) | pH optimum | Divalent cation requirement |
|---|---|---|---|---|---|---|---|---|
| EG1 (28) | 3.6 mg/ml | 84 | 91 | 25.3 | No | 39 | 6.5 | Yes |
| rcCel9B | 9.20 ± 0.83 mg/ml | 36.9 ± 1.46 | 39.79 ± 1.57 | 4.24 | No | 37 | 6.5 | Yes |
| rcCel9BΔBTD | 5.93 ± 0.78 mg/ml | 47.8 ± 3.00 | 46.29 ± 2.91 | 7.80 | No | 37 | 6.5 | Yes |
| rcCel5H | 16.2 ± 1.91 mg/ml | 0.31 ± 0.02 | 0.51 ± 0.03 | 0.031 | Yes | 37 | 6.5 | No |
| rcCel8B | 10.9 ± 1.25 mg/ml | 1.08 ± 0.06 | 1.47 ± 0.08 | 0.14 | No | 42 | 7.0 | No |
| ClCBase (14) | 0.1 mM | 14.2 | 15.9 | 159 | Yes | 45/37b | 6.5 | No |
| rcCel10A | 0.36 ± 0.02 mM | 11.6 ± 1.02 | 12.1 ± 1.06 | 33.6 | Yes | 37/37b | 6.5 | No |
Substrates used: p-nitrophenyl cellobioside for native ClCBase and rcCel10A and CMC for the enzymes EG1, rcCel9B, rcCel9BΔBTD, rcCel5H, and rcCel8B.
With/without 0.2 M NaCl.
The affinity of Cel10A for p-nitrophenyl cellobioside was not affected in the absence or presence of 0.02 M of chloride, with a Km value of 0.16 mM, while the Km value was significantly increased to 0.36 mM in the presence of 0.05 and 0.1 M of chloride. The Vmax values were increased from 3.3 to 11.6 U/mg when the concentration of chloride was increased from 0 to 100 mM. We confirmed that rcClCBase is not glycosylated, while the native ClCBase is glycosylated (data not shown). The effects of chloride ion concentration on enzyme activity of glycosylated and nonglycosylated forms of ClCBase were compared (Fig. 2). A 100-fold increase in chloride concentration was necessary for the recombinant enzyme to exhibit the same level of activity as the native enzyme.
FIG. 2.
Effect of chloride on cellobiosidase activity of the native ClCBase (open triangles) and rcClCBase (filled triangles). The enzyme assays were performed with p-nitrophenyl cellobioside as the substrate in 10 mM imidazole buffer (pH 6.5) at various concentrations of chloride ions. The bars represent the standard errors (n = 3). Values for 100% activity of the native ClCBase and rcClCBase were 13.8 and 12.0 U/mg, respectively.
Cel8B and Cel5H had Km values similar to that of Cel9B, while the Vmax values were much lower. Divalent metal ions did not stimulate the activities of Cel8B and Cel5H, and 5 mM EDTA did not inhibit their activities. pH and temperature optima of Cel5H were similar to those of the other enzymes. Cel8B retained over 80% of activity between pH 5 and 8, with a pH optimum of 7.0. The temperature optimum of Cel8B was 42°C.
To test and confirm the mode of action of Cel8B and Cel51A, hydrolysis products of acid-swollen cellulose were analyzed by high-pressure liquid chromatography. Cel8B produced (on a molar basis) cellobiose (38%), cellotriose (41%), and cellotetraose (20%) (see Fig. S4A in the supplemental material). No glucose was detected. Cel51A produced cello-oligosaccharide with a degree of polymerization ranging from 2 to 5, with cellotetraose as the major product (33%) (see Fig. S4B in the supplemental material). Glucose and cellobiose accounted for 2.9% and 31%, respectively, of the products on a molar basis.
Binding to cellulose.
The ability of Cel5H, Cel8B, Cel9B, and Cel10A to bind to Avicel was tested. Cel9B did not bind to Avicel under our experimental conditions, which was consistent with previously reported data for the native EG1 (28). Cel8B also did not bind to cellulose, which ruled out a possible substrate binding role for the internal unknown domain. Cel5H bound to cellulose, which presumably was due to the presence of the two CBM11 domains. Both rcCel10A and native Cel10A bound to Avicel. The affinity of rcCel10A for cellulose was assessed using Avicel cellulose, BMCC, and ball-milled barley straw as the substrates (Table 4). Cel10A exhibited the highest affinity for BMCC but bound maximally to ball-milled barley straw.
TABLE 4.
Kd and the maximum amount of rcCel10A bound for each substratea
| Substrate | Kd (μM) | [PC]max (μmol/g substrate) |
|---|---|---|
| Avicel cellulose PH105 | 4.76 | 3.36 |
| BMCC | 1.75 | 4.33 |
| Ball-milled barley straw | 2.86 | 5.99 |
The binding assay was performed in duplicate, and the Kd and [PC]max values were calculated by using a double-reciprocal plot.
Synergistic interaction between five cellulases in F. succinogenes.
Since all five enzymes showed very low activity on Avicel cellulose but moderate activity against ball-milled cellulose, the latter was used as the substrate for testing synergy.
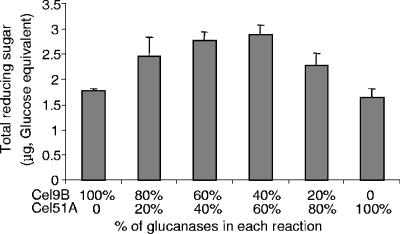
The initial experiments examining synergy between Cel9B and Cel51A (CelF) were conducted with a substrate concentration of 0.5% ball-milled cellulose and an incubation time of 2 h (Fig. 3). Purified rcCel9B and rcCel51A were each diluted to equal ball-milled cellulase activities and combined at different ratios in the assay mixture to maintain a constant sum of individual activities. Cel9B and Cel51A were tested individually as controls. Activities of all mixtures were significantly higher than that of either enzyme assayed alone, indicating a synergistic interaction. Maximum synergy (1.67) was observed with a ratio of Cel9B and Cel51A of 3:2 to 2:3 (Fig. 3) by activity, which corresponded to a molar ratio of 1:2.6 to 1:5.9.
FIG. 3.
Reducing sugar produced (glucose equivalents) from ball-milled cellulose by binary mixtures of Cel9B and Cel51A after 2 h of incubation at 37°C. Purified rcCel9B and rcCel51A were each diluted to equal ball-milled cellulase activities and combined at different ratios, as indicated, in the assay mixture to maintain a constant sum of individual activities. The mixture that showed the highest degree of synergism (Cel9B/Cel51A = 40:60) contained 2.9 pmol of Cel9B and 17.1 pmol of Cel51A in a 100-μl reaction mixture.
We then determined the catalytic interactions with different combinations of Cel9B, Cel10A, and Cel51A (Table 5). Binary mixtures containing combinations of two enzymes were assayed, and reaction mixtures containing individual enzymes were also included as controls, with ball-milled cellulose as the substrate. In all combinations tested, the binary mixture containing 20 pmol of Cel9B and 20 pmol of Cel51A but no Cel10A gave the highest activity and produced 10.4 μg of reducing sugar in 4 h. The inclusion of Cel10A decreased the activities of the mixtures, although ternary combinations which contained more than 4 pmol of Cel9B, 16 pmol of Cel51A, and less than 8 pmol of Cel10A retained over 80% of the highest activity. Cel10A showed a little synergistic interaction with Cel51A. The binary mixture of Cel10A and Cel9B had a synergism degree less than 1, indicating the existence of either inhibition or competition. The highest synergy was observed with 12 pmol of Cel9B, 16 pmol of Cel10A, and 16 pmol of Cel51A, and the degree of synergism remained nearly this high when the mixture contained more than 4 pmol of Cel9B, 16 pmol of Cel51A, and less than 8 pmol of Cel10A (Table 5). Data in Table 5 were fit to a generalized linear model by the SAS procedure “Proc GLM,” and multivariate regression equations were developed. The relative activity could be calculated as 0.76 · [Cel10A] + 3.22 · [Cel51A] + 2.62 · [Cel9B] − 22.2 (R2 = 0.761; coefficient of variation = 17.1), while the degree of synergism may be calculated as 0.007 · [Cel10A] + 0.035 · [Cel51A] + 0.016 · [Cel9B] + 0.57 (R2 = 0.523; coefficient of variation = 14.9), where [Cel10A] stands for the amount (in pmol) of Cel10A enzyme used. In both formulas, Cel10A had a lower weight than the other two enzymes.
TABLE 5.
Relative activity and synergism degree of enzyme mixtures containing recombinant Cel51A, Cel9B, and Cel10A catalyzing hydrolysis of ball-milled cellulose
| Amt (pmol) of: | Relative activitya (%) | Degree of synergism | ||
|---|---|---|---|---|
| rcCel10A | rcCel51A | rcCel9B | ||
| 0 | 20 | 20 | 100.0 | 1.63 |
| 8 | 16 | 20 | 91.8 | 1.53 |
| 8 | 20 | 16 | 93.3 | 1.60 |
| 16 | 12 | 20 | 76.8 | 1.31 |
| 16 | 16 | 16 | 84.8 | 1.49 |
| 16 | 20 | 12 | 94.9 | 1.72 |
| 24 | 8 | 20 | 55.0 | 0.96 |
| 24 | 12 | 16 | 74.4 | 1.34 |
| 24 | 16 | 12 | 87.9 | 1.64 |
| 24 | 20 | 8 | 70.7 | 1.36 |
| 32 | 4 | 20 | 64.9 | 1.16 |
| 32 | 8 | 16 | 77.8 | 1.44 |
| 32 | 12 | 12 | 75.9 | 1.45 |
| 32 | 16 | 8 | 84.0 | 1.66 |
| 32 | 20 | 4 | 83.0 | 1.70 |
| 40 | 0 | 20 | 34.0 | 0.63 |
| 40 | 4 | 16 | 76.1 | 1.45 |
| 40 | 8 | 12 | 69.8 | 1.37 |
| 40 | 12 | 8 | 71.0 | 1.45 |
| 40 | 16 | 4 | 75.1 | 1.59 |
| 40 | 20 | 0 | 52.2 | 1.14 |
| 40 | 0 | 0 | 19.3 | 1.00 |
| 0 | 20 | 0 | 26.3 | 1.00 |
| 0 | 0 | 20 | 35.1 | 1.00 |
One hundred percent activity equals 10.4 μg reducing sugar produced in 4 h.
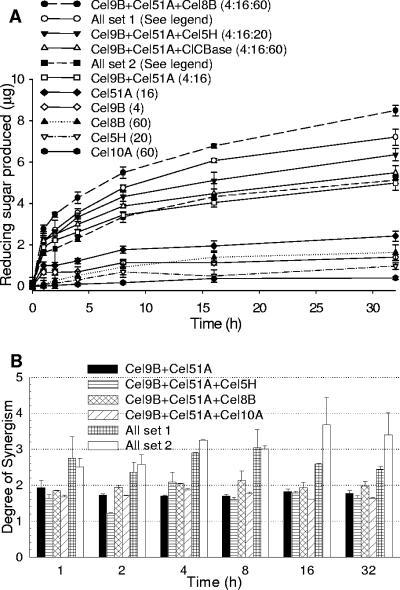
The specific and total activities of the mixture of the five enzymes were determined. Enzyme mixtures were incubated with 0.5% ball-milled cellulose for 32 h in 800-μl reaction mixtures. Samples (100 μl) were taken at 1, 2, 4, 8, 16, and 32 h. The combination Cel9B/Cel51A/Cel8B gave the highest activity at all time points (Fig. 4A). The greatest extent of ball-milled cellulose hydrolysis achieved after 32 h was 1.6%. The greatest degree of synergism was attained in the presence of all five enzymes (Fig. 4B).
FIG. 4.
Reducing sugar produced (glucose equivalents) (A) and degree of synergism (B) during the hydrolysis of ball-milled cellulose (0.5%) by different combinations of cellulases. The compositions of the mixtures (in pmol/100 μl) are indicated in parentheses. The cellulase mixture “all set 1” contains 2 pmol Cel9B, 8 pmol Cel51A, 30 pmol ClCBase, 30pmol Cel8B, and 10 pmol Cel5H (per 100 μl). “All set 2” contains 1 pmol Cel9B, 4 pmol Cel51A, 15 pmol ClCBase, 15 pmol Cel8B, and 5 pmol Cel5H (per 100 μl).
DISCUSSION
In the present study, we identified and characterized four recombinant β-glucanases and reported for the first time the synergistic interactions among five cellulases from different glycoside hydrolase families of F. succinogenes.
Genes coding for the enzymes characterized in this study may be present in both F. succinogenes and Fibrobacter intestinalis, as documented by Western immunoblotting, Southern hybridization (M. Qi, unpublished data) and suppressive subtractive hybridization (33). Several other glycoside hydrolases were also reported to be conserved among strains in different Fibrobacter species, including endoglucanase 3 (Cel5G) (23), Cel51A/EG2, a cellodextrinase (Cel5C), and a lichenase (Lic16C), as well as xylanase C (Xyn11C) (4).
Cel9B is a major cellulase secreted by F. succinogenes S85 and accounted for approximately 32% of the total endoglucanase activity present in the nonsedimentable fraction (28). Cel9B has 86% identity to the endoglucanase C protein from F. succinogenes BL2 (2). A partial gene identified from F. intestinalis DR7 (33) showed higher similarity (37% identity) to cel9B than to any other GH9 enzyme gene, indicating that this gene might be widespread among the species in the genus Fibrobacter.
Cel9B contained an N-terminal Ig-like structure, a family 9 glycoside hydrolase catalytic domain, and a BTD. It did not contain a carbohydrate binding module (CBM), which is in accordance to the fact the Cel9B did not bind to Avicel cellulose in contrast to Cel51A (28), which contains N-terminal CBMs (27). Truncation of the N-terminal Ig-like module as well as the N-terminal part of the catalytic domain probably explains the lack of activity of CelE previously reported (26). The N-terminal Ig-like domain exists in many family 9 glycoside hydrolases of Clostridium thermocellum, and it is believed to interact with the catalytic domain in the CbhA through a large number of hydrogen bonds and hydrophobic interactions (20). Deletion of this module from the Ig-GH9 construct of CbhA resulted in complete loss of activity of the GH9 module (20).
Both Cel9B and Cel9C (CelD [26]) have C-terminal domains with high pI values that have been called BTDs (26). The function of the domain is unclear, but it apparently does not have a role in cellulose binding and cell surface adherence since the intact protein did not bind to cellulose and was released as a monomeric protein into the extracellular culture fluid during growth (28). In the present study, the BTD-truncated Cel9B had a higher specific activity than the intact enzyme, indicating that this domain somehow modulated the catalytic property of the enzyme. In contrast, Bera-Maillet et al. (3) reportedly removed the C-terminal domain of endoglucanase from F. succinogenes BL2 and mentioned that deletion of this domain abolished the activity of endoglucanase. However, no details on the number of C-terminal amino acid residues removed was provided. Consequently a precise comparison cannot be made at the present time.
Cel9B is known as an endoglucanase with a high activity on CMC. When acting on filter paper, the insoluble reducing sugar accounted for 10.8% of all the reducing sugar produced. A previous study showed that exoglucanases usually produced less than 10% insoluble reducing groups, endoglucanases generate over 30% insoluble reducing groups, and processive endoglucanases generate 10% to 30% insoluble reducing groups (15). Thus, Cel9D may be classified as a processive endoglucanase enzyme. However, detailed kinetic study will be needed to elucidate the processive manner of Cel9B.
The CMCase activity of native EG1, Cel9B, and Cel9BΔBTD was increased by the presence of 1 mM Ca2+ or Mg2+. These cations seem to stimulate several GH9 glucanases (3, 7). The activities of Cel9B and Cel9BΔBTD were lower than that of the native enzyme reported previously (28). A reasonable possibility for this difference is that, because the EG1 protein was purified from culture supernatant, traces of other cellulase proteins not removed may have increased the activity due to a synergistic effect as previously reported for purified glucanases of Thermomonospora fusca (40). However, we cannot preclude the possibility that the native EG1 was glycosylated or otherwise modified, without a significant change in the mass, which could have enhanced its catalytic activity.
rcCel10A was shown to retain the general properties of the native form of the ClCBase from F. succinogenes S85, including specific activity, stimulation of enzyme activity by anions, and binding to cellulose. However, it was different from the native Cel10A in terms of a higher Km and a higher concentration of chloride for maximum catalytic activity. The difference may be attributed to the absence of glycosylation present of the native enzyme (14), which could influence the tertiary structure. Differences in properties between native and recombinant enzymes are common. For example, two recombinant xylanases were documented as showing significant differences in catalytic properties and hydrolysis end products from the native forms of the proteins (12, 21).
Over 100 carbohydrate active enzymes were identified from the genome of F. succinogenes. However, the expression patterns of many of these enzymes are not documented. The two major enzymes produced by F. succinogenes, Cel9B and Cel51A (28), were shown to have the highest synergistic effect on ball-milled cellulose. Cel51A is a typical endoglucanase with two carbohydrate binding modules. It degrades acid-swollen cellulose (a type of amorphous cellulose) to cello-oligosaccharides with a degree of polymerization from 2 to 5, with cellotetraose as the major product (see Fig. S4B in the supplemental material) (28). In contrast, EG1 produced cellobiose and cellotriose as major products (28). Therefore EG1 is able to digest some of the cello-oligosaccharides produced by Cel51A. The ternary mixture containing Cel10A, Cel51A, and Cel9B had similar or overall lower activity on the ball-milled cellulose (Table 5) compared with the binary mixture containing Cel51A and Cel9B. This may be attributed to the fact that Cel10A has catalytic properties similar to those of Cel9B, cleaving cello-oligosaccharides to primarily cellobiose (14).
It was shown that a major portion of Cel10A produced by F. succinogenes was cell associated and located in the periplasmic fraction (18). Therefore it is likely that Cel10A is involved in the degradation of soluble oligo- or polysaccharides that are transported into the cell. However, F. succinogenes is known to have an outer membrane that is easily detached from the peptidoglycan layer and to form vesicles (10). Thus, periplasm enzymes may have access to the insoluble substrates as well. If Cel10A has no opportunity to contact cellulose, the high affinity for cellulose of the N-terminal CBM4 cannot be explained. Indeed a significant amount of Cel10A was identified in the outer membrane released into the extracellular culture fluid during growth on cellulose (18). Therefore, the role of Cel10A in cellulose digestion is equivocal. Another cellodextrinase identified from periplasmic contents, Cel5C (FSU2070), was previously identified in F. succinogenes (13, 16). Cel5C lacked a CBM and has limited activity on cellulose; it was not considered to be an important enzyme for crystalline cellulose degradation (13, 16). Therefore this enzyme was not included in this study.
The ternary mixtures containing Cel9B, Cel51A, and Cel8B gave a higher degree of synergism than the binary mixture of Cel9B and Cel51A (Fig. 3 and 4B). Cel8B hydrolyzed acid-swollen cellulose and produced cellobiose, cellotriose, and cellotetraose (see Fig. S4 in the supplemental material). Cellotriose and cellotetraose produced could subsequently be degraded by Cel9B. The mixtures containing five enzymes give a higher degree of synergism than those containing two or three enzymes, which may be due to different substrate specificities. Given that F. succinogenes contains 33 cellulases from different families, the synergism degrees of all the cellulases could be even higher in vivo. The mixture containing more enzyme (all set 1) had a lower degree of synergism than the one containing one-half the amount of enzymes (all set 2). This observation indicated that the ratio of substrate to enzyme had an effect on the synergistic interaction, which was also reported in the synergy study of endoglucanase I, cellobiohydrolase I, and cellobiohydrolase II from Trichoderma reesei by Woodward et al. (39).
This is the first time that the synergism of the F. succinogenes cellulases was tested. Although a synergistic effect was identified among some of the enzymes, both the total activity and the extent of ball-milled cellulose degradation were low. Therefore the apparent synergism may apply only to a very small portion of the substrate. Thus, other proteins important for cellulose degradation in F. succinogenes still remain to be identified. Moreover, differences in catalytic properties between native and recombinant enzymes are problematic because the synergistic interactions observed may not be a true reflection of the interaction of native enzymes; however, there is little choice in using native enzymes when they are produced in comparatively low concentrations. On the other hand, native enzymes may be contaminated with traces of other cellulolytic enzymes that may influence the outcome as well (40). Experiments are now in progress to identify other cellulases that may be important for cellulose degradation by F. succinogenes.
Supplementary Material
Acknowledgments
This research was supported by Initiative for Future Agriculture and Food Systems grant no. 2000-52100-9618 from USDA-CSREES to the North American Consortium for Genomics of Fibrolytic Ruminal Bacteria and by the Natural Science and Engineering Research Council of Canada. The Fibrobacter succinogenes genome project was undertaken at The Institute for Genomic Research and supported by USDA-CSREES funds.
Footnotes
Published ahead of print on 27 July 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Beguin, P., P. Cornet, and J. P. Aubert. 1985. Sequence of a cellulase gene of the thermophilic bacterium Clostridium thermocellum. J. Bacteriol. 162:102-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bera, C., V. Broussolle, E. Forano, and G. Gaudet. 1996. Gene sequence analysis and properties of EGC, a family E (9) endoglucanase from Fibrobacter succinogenes BL2. FEMS Microbiol. Lett. 136:79-84. [DOI] [PubMed] [Google Scholar]
- 3.Bera-Maillet, C., V. Broussolle, P. Pristas, J. P. Girardeau, G. Gaudet, and E. Forano. 2000. Characterisation of endoglucanases EGB and EGC from Fibrobacter succinogenes. Biochim. Biophys. Acta 1476:191-202. [DOI] [PubMed] [Google Scholar]
- 4.Bera-Maillet, C., Y. Ribot, and E. Forano. 2004. Fiber-degrading systems of different strains of the genus Fibrobacter. Appl. Environ. Microbiol. 70:2172-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell, J. R., and R. Horgan. 1991. A novel strategy for production of a highly expressed recombinant protein in an active form. FEBS Lett. 295:10-12. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Chauvaux, S., P. Beguin, J. P. Aubert, K. M. Bhat, L. A. Gow, T. M. Wood, and A. Bairoch. 1990. Calcium-binding affinity and calcium-enhanced activity of Clostridium thermocellum endoglucanase D. Biochem. J. 265:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dehority, B. A. 2003. Rumen microbiology. Nottingham University Press, Thrumpton, Nottingham, United Kingdom.
- 9.Fierobe, H. P., C. Bagnara-Tardif, C. Gaudin, F. Guerlesquin, P. Sauve, A. Belaich, and J. P. Belaich. 1993. Purification and characterization of endoglucanase C from Clostridium cellulolyticum. Catalytic comparison with endoglucanase A. Eur. J. Biochem. 217:557-565. [DOI] [PubMed] [Google Scholar]
- 10.Forsberg, C. W., T. J. Beveridge, and A. Hellstrom. 1981. Cellulase and xylanase release from Bacteroides succinogenes and its importance in the rumen environment. Appl. Environ. Microbiol. 42:886-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsberg, C. W., E. Forano, and A. Chesson. 2000. Microbial adherence to plant cell wall and enzymatic hydrolysis, p. 79-88. In P. B. Cronje (ed.), Ruminant physiology: digestion, metabolism, growth and reproduction. CABI Publishing, Wallingford, United Kingdom.
- 12.Honda, H., T. Kudo, and K. Horikoshi. 1985. Purification and partial characterization of alkaline xylanase from Escherichia coli carrying Pcx311. Agric. Biol. Chem. 49:3165-3169. [Google Scholar]
- 13.Huang, L., and C. W. Forsberg. 1987. Isolation of a cellodextrinase from Bacteroides succinogenes. Appl. Environ. Microbiol. 53:1034-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, L., C. W. Forsberg, and D. Y. Thomas. 1988. Purification and characterization of a chloride-stimulated cellobiosidase from Bacteroides succinogenes S85. J. Bacteriol. 170:2923-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irwin, D. C., M. Spezio, L. P. Walker, and D. B. Wilson. 1993. Activity studies of 8 purified cellulases—specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42:1002-1013. [DOI] [PubMed] [Google Scholar]
- 16.Iyo, A. H., and C. W. Forsberg. 1994. Features of the cellodextrinase gene from Fibrobacter succinogenes S85. Can. J. Microbiol. 40:592-596. [DOI] [PubMed] [Google Scholar]
- 17.Jun, H. S., J. K. Ha, L. M. Malburg, Jr., G. A. Verrinder, and C. W. Forsberg. 2003. Characteristics of a cluster of xylanase genes in Fibrobacter succinogenes S85. Can. J. Microbiol. 49:171-180. [DOI] [PubMed] [Google Scholar]
- 18.Jun, H.-S., M. Qi, J. Gong, E. E. Egbosimba, and C. W. Forsberg. 2007. Outer membrane proteins of Fibrobacter succinogenes with potential roles in adhesion to cellulose and in cellulose digestion. J. Bacteriol. 189:6806-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kam, D. K., H. S. Jun, J. K. Ha, G. D. Inglis, and C. W. Forsberg. 2005. Characteristics of adjacent family 6 acetylxylan esterases from Fibrobacter succinogenes and the interaction with the Xyn10E xylanase in hydrolysis of acetylated xylan. Can. J. Microbiol. 51:821-832. [DOI] [PubMed] [Google Scholar]
- 20.Kataeva, I. A., V. N. Uversky, J. M. Brewer, F. Schubot, J. P. Rose, B. C. Wang, and L. G. Ljungdahl. 2004. Interactions between immunoglobulin-like and catalytic modules in Clostridium thermocellum cellulosomal cellobiohydrolase CbhA. Protein Eng. Des. Sel. 17:759-769. [DOI] [PubMed] [Google Scholar]
- 21.Kulkarni, N., J. Chauthaiwale, and M. Rao. 1995. Characterization of the recombinant xylanases in Escherichia coli from an alkaliphilic thermophilic Bacillus sp. NCIM-59. Enzyme Microb. Technol. 17:972-976. [Google Scholar]
- 22.Lever, M. 1972. A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 47:273-279. [DOI] [PubMed] [Google Scholar]
- 23.Lin, C., and D. A. Stahl. 1995. Comparative analyses reveal a highly conserved endoglucanase in the cellulolytic genus Fibrobacter. J. Bacteriol. 177:2543-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, J. H., L. B. Selinger, Y. J. Hu, M. M. Moloney, K. J. Cheng, and K. A. Beauchemin. 1997. An endoglucanase from the anaerobic fungus Orpinomyces joyonii: characterization of the gene and its product. Can. J. Microbiol. 43:477-485. [DOI] [PubMed] [Google Scholar]
- 25.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malburg, L. M., Jr., A. H. Iyo, and C. W. Forsberg. 1996. A novel family 9 endoglucanase gene (celD), whose product cleaves substrates mainly to glucose, and its adjacent upstream homolog (celE) from Fibrobacter succinogenes S85. Appl. Environ. Microbiol. 62:898-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malburg, S. R., L. M. Malburg, Jr., T. Liu, A. H. Iyo, and C. W. Forsberg. 1997. Catalytic properties of the cellulose-binding endoglucanase F from Fibrobacter succinogenes S85. Appl. Environ. Microbiol. 63:2449-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGavin, M., and C. W. Forsberg. 1988. Isolation and characterization of endoglucanases 1 and 2 from Bacteroides succinogenes S85. J. Bacteriol. 170:2914-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGavin, M. J., C. W. Forsberg, B. Crosby, A. W. Bell, D. Dignard, and D. Y. Thomas. 1989. Structure of the cel-3 gene from Fibrobacter succinogenes S85 and characteristics of the encoded gene product, endoglucanase 3. J. Bacteriol. 171:5587-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michalet-Doreau, B., I. Fernandez, C. Peyron, L. Millet, and G. Fonty. 2001. Fibrolytic activities and cellulolytic bacterial community structure in the solid and liquid phases of rumen contents. Reprod. Nutr. Dev. 41:187-194. [DOI] [PubMed] [Google Scholar]
- 31.Miron, J., and D. Ben Ghedalia. 1993. Digestion of cell-wall monosaccharides of ryegrass and alfalfa hays by the ruminal bacteria Fibrobacter succinogenes and Butyrivibrio fibrisolvens. Can. J. Microbiol. 39:780-786. [DOI] [PubMed] [Google Scholar]
- 32.Morrison, M., K. E. Nelson, I. K. O. Cann, C. W. Forsberg, R. I. Mackie, J. B. Russell, B. A. White, K. Amava, B. Cheng, M. Qi, H. Jun, S. Mulligan, K. Tran, H. A. Carty, H. Khouri, W. Nelson, S. Daugherty, and C. M. Fraser. 2003. The Fibrobacter succinogenes strain S85 genome sequencing project. Abstr. 3rd ASM-TIGR Conf. Microb. Genomes, p. 33.
- 33.Qi, M., K. E. Nelson, S. C. Daugherty, W. C. Nelson, I. R. Hance, M. Morrison, and C. W. Forsberg. 2005. Novel molecular features of the fibrolytic intestinal bacterium Fibrobacter intestinalis not shared with Fibrobacter succinogenes as determined by suppressive subtractive hybridization. J. Bacteriol. 187:3739-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakoda, M., and K. Hiromi. 1976. Determination of the best-fit values of kinetic parameters of the Michaelis-Menten equation by the method of least squares with the Taylor expansion. J. Biochem. (Tokyo) 80:547-555. [DOI] [PubMed] [Google Scholar]
- 35.Scott, H. W., and B. A. Dehority. 1965. Vitamin requirements of several cellulolytic rumen bacteria. J. Bacteriol. 89:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 37.Tajima, K., R. I. Aminov, T. Nagamine, H. Matsui, M. Nakamura, and Y. Benno. 2001. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 67:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Väljamäe, P., V. Sild, A. Nutt, G. Pettersson, and G. Johansson. 1999. Acid hydrolysis of bacterial cellulose reveals different modes of synergistic action between cellobiohydrolase I and endoglucanase I. Eur. J. Biochem. 266:327-334. [DOI] [PubMed] [Google Scholar]
- 39.Woodward, J., M. Lima, and N. E. Lee. 1988. The role of cellulase concentration in determining the degree of synergism in the hydrolysis of microcrystalline cellulose. Biochem. J. 255:895-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, S., G. Lao, and D. B. Wilson. 1995. Characterization of a Thermomonospora fusca exocellulase. Biochemistry 34:3386-3395. [DOI] [PubMed] [Google Scholar]
- 41.Ziemer, C. J., R. Sharp, M. D. Stern, M. A. Cotta, T. R. Whitehead, and D. A. Stahl. 2000. Comparison of microbial populations in model and natural rumens using 16S ribosomal RNA-targeted probes. Environ. Microbiol. 2:632-643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.