Abstract
Listeria monocytogenes is a food-borne, opportunistic, bacterial pathogen causing a wide spectrum of diseases, including meningitis, septicemia, abortion, and gastroenteritis, in humans and animals. Among the 13 L. monocytogenes serovars described, human listeriosis is mostly associated with strains of serovars 4b, 1/2b, and 1/2a. Within the species L. monocytogenes, three phylogenetic lineages are described. Serovar 1/2a belongs to phylogenetic lineage I, while serovars 4b and 1/2b group in phylogenetic lineage II. To explore the role of gene expression in the adaptation of L. monocytogenes strains of these two major lineages to different environments, as well as in virulence, we performed whole-genome expression profiling of six L. monocytogenes isolates of serovars 4b, 1/2b, and 1/2a of distinct origins, using a newly constructed Listeria multigenome DNA array. Comparison of the global gene expression profiles revealed differences among strains. The expression profiles of two strains having distinct 50% lethal doses, as assessed in the mouse model, were further analyzed. Gene ontology term enrichment analysis of the differentially expressed genes identified differences in protein-, nucleic acid-, carbon metabolism-, and virulence-related gene expression. Comparison of the expression profiles of the core genomes of all strains revealed differences between the two lineages with respect to cell wall synthesis, the stress-related sigma B regulon and virulence-related genes. These findings suggest different patterns of interaction with host cells and the environment, key factors for host colonization and survival in the environment.
Listeria monocytogenes is a gram-positive, facultative, intracellular bacterium that causes severe food-borne infections, such as gastroenteritis, septicemia, abortion, and meningitis, in humans and animals (60). L. monocytogenes is able to cross the intestinal barrier, the blood-brain barrier, and the fetoplacental barrier and to invade and replicate inside epithelial and professional phagocytic cells. L. monocytogenes is widely present in nature, and it has also been isolated from numerous animals, including cattle, sheep, and goats (21). Furthermore, L. monocytogenes has the important capacity to adapt to and survive in extreme environments, such as high salt concentration (10% NaCl), a broad pH range (from 4.5 to 9.0), and a wide temperature range. Its ability to grow at temperatures between −1°C and 45°C increases the risk of contamination in dairy products, meats, seafood, and other processed food products via selective enrichment during refrigeration. Listeria can also survive long periods of drying and freezing with subsequent thawing (38, 54). L. monocytogenes is an environmental bacterium living, for example, on decomposing plants. However, the presence of virulence factors, which have most probably been acquired by a common ancestor through horizontal gene transfer (for reviews see references 7 and 56), allows L. monocytogenes to infect humans and other mammalian hosts. Most susceptible to listeriosis are immunocompromised individuals, elderly people, pregnant women, fetuses, and neonates. Listeriosis is characterized by a low infection rate but a high mortality rate, and thus, L. monocytogenes is a concern for public health and for the food industry.
The infectious process is dependent on the production of several virulence factors. Proteins necessary for adhesion and the invasion of eukaryotic cells (the internalins InlA and InlB, the autolysin Ami, the cell wall hydrolase p60 [Iap], and the pore-forming listeriolysin O [LLO]); proteins involved in intracellular life, such as those responsible for escaping from phagosomes (LLO and the phospholipases PlcA and PlcB), actin-based motility, and cell-to-cell spread (the surface protein ActA); and a protein involved in the bacterium's intracytoplasmic replication (the hexose phosphate transporter Hpt) are among the best studied to date (19). Virulence gene expression is tightly regulated. The major L. monocytogenes virulence genes are regulated by the transcriptional activator PrfA, whose regulon is under complex environmental control. It has been demonstrated that both temperature sensing and chemical components of the extracellular environment play important roles in regulating the expression of the PrfA regulon (4, 31, 40). Additionally, the transcription of a subset of L. monocytogenes virulence genes seems to be regulated by a network that can include activation by both PrfA and sigma B (33, 41, 58) and interplay between PrfA and sigma B has been suggested (40).
The species L. monocytogenes comprises 13 serovars; among those, serovars 1/2a, 1/2b, and 4b account for over 98% of all human listeriosis cases (29, 57). Even though serovar 1/2a is the most frequently isolated from food and environmental sources, most major food-borne outbreaks and a majority of sporadic cases of listeriosis are caused by serovar 4b strains (1, 28, 30, 43, 66). Moreover, the heterogeneous virulence of L. monocytogenes clinical strains and strains of different serovars has been demonstrated in a mouse model of infection and in the Caco-2 cell line (26, 49, 50). Comparative genomic analysis of the complete genome sequences of L. monocytogenes EGDe, of Listeria innocua CLIP11262 (25), and of three additional L. monocytogenes strains (42), as well as the comparison of over 100 L. monocytogenes strains of different serovars and origins, by DNA/DNA hybridization using DNA arrays (7, 8, 15, 68) revealed that differences in gene content exist between strains of different serovars and origins. Some of these differences may be implicated in the various disease potentials of L. monocytogenes strains. However, differences among strains may also be due to different gene expression/regulation of the core genes of L. monocytogenes as already shown for other bacterial species, such as Escherichia coli (16, 35) and Pseudomonas aeruginosa (55).
In order to better understand the differences occurring between strains, as well as between the two major phylogenetic lineages of the species L. monocytogenes, in niche adaptation and probably also in disease potential, we compared the transcriptional signatures of six L. monocytogenes strains under in vitro growth conditions in defined, rich medium and late exponential phase—conditions designed to maximize the number of genes expressed. The virulence of these six strains was assessed in the mouse model of infection and in the chicken embryo model. Variations in the expression of virulence-, cell wall-, carbohydrate metabolism-, and sigma B-related genes correlated with lineage designations, suggesting differences in the expression of genes for pathways related to interactions with the host.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and RNA extraction.
The Listeria monocytogenes strains used in this study are described in Table 1. The strains were grown overnight in BHI (brain heart infusion) medium (Difco), inoculated at an optical density at 600 nm (OD600) of 0.1 into 10 ml of the defined medium MCDB 202 (CryoBioSystem) supplemented with 1% glucose, and grown at 37°C until late exponential phase (OD600, 0.9).
TABLE 1.
Characteristics of Listeria monocytognes strains used for expression profiling
| Straina | Origin | Serovar | Country | Yr | Designation | Chick embryo mortality (%) | Reference(s) |
|---|---|---|---|---|---|---|---|
| CLIP80459 | Listeriosis epidemic in humans | 4b | France | 1999 | Clinical | 100 | 11 |
| CLIP90602 | Listeriosis epidemic in humans | 1/2b | France | 2002 | Listeriosis | 100 | Unpublished |
| CLIP93667 | Healthy human | 4b | France | 1992 | Human | 100 | 46, 47 |
| CLIP93666 | Healthy human | 1/2a | France | 1991 | Carriage | 20 | |
| CLIP93665 | Environment | 4b | France | 2000 | Environmental | 100 | Unpublished |
| CLIP93649 | Environment | 1/2a | France | 2000 | Industrial plant | 100 | Unpublished |
CLIP denotes strain designations of the Listeria collection at the Institut Pasteur.
For each strain, two independent cultures were used. Cells were harvested by centrifugation at 4°C and flash frozen in a dry ice-alcohol mixture. RNA extraction was done as previously described (40). Briefly, pellets were resuspended in 400 μl of buffer (10% glucose, 12.5 mM Tris [pH 7.6], 5 mM EDTA) and 60 μl of 0.5 MEDTA. Then, cells were mechanically disrupted in a FastPrep apparatus in an acid phenol (pH 4.5) glass bead (Sigma) mixture. For extraction, Trizol (Invitrogen) was used, and RNA was precipitated with 2-propanol. After centrifugation for 30 min at 4°C, the pellets were washed with 70% ethanol and dried at room temperature prior to being dissolved in 50 μl of DNase- and RNase-free deionized water treated with 0.001% diethylpyrocarbonate (MP Biomedicals). The RNA concentration was estimated by using a spectrophotometer, and the RNA quality was checked on an agarose gel containing ethidium bromide.
Macroarray construction and hybridization.
For this study, we designed a Listeria multigenome DNA macroarray. It contains PCR products representing 2,816 genes predicted in the completely sequenced L. monocytogenes EGDe genome (40) and 153 additional PCR products specific for genes predicted in L. monocytogenes strain CLIP80459 that are not present in strain EGDe (15; unpublished data). This Listeria multigenome DNA array contains probes representing L. monocytogenes serovar 1/2a (EGDe) and L. monocytogenes serovar 4b (CLIP80459) genes, thus allowing comparisons of strains belonging to the two serovars.
The construction of the macroarray and the hybridization of RNA samples were done as previously described (40), with slight modifications. Briefly, for cDNA synthesis and labeling, 1 μg of total RNA was mixed with 6 μl of 5× AMV reverse transcriptase buffer (Roche), 3 μl of a mixture of dATP, dGTP, and dTTP (10 mM), and 5 μg of random hexamers [Primer Random p(dN)6, Roche], and diethylpyrocarbonate-treated water was added to a final volume of 21 μl. After heating at 90°C for 2 min and temperature adjustment to 42°C, 3 μl of [α-33P]dCTP (2,000 to 3,000 Ci mmol−1; Amersham) and 2 μl (50 U/μl) of AMV reverse transcriptase (Roche) were added. After a 2-h incubation at 42°C, the labeled cDNA was purified using a QIAquick nucleotide removal column (QIAquick nucleotide removal kit; QIAGEN). The cDNA obtained was denatured by heating at 99°C for 5 min just before hybridization. Macroarray hybridization was carried out as previously described (40). The macroarrays were then exposed to a phosphorimager screen and scanned with a Typhoon phosphorimager (Pharmacia-Amersham). Two hybridizations were performed for each independent RNA extraction. In total, four hybridizations were performed for each strain. The array design is available at the GEO database under the accession number GPL2811 (http://www.ncbi.nlm.nih.gov/geo/).
Data acquisition, preprocessing, and statistical analysis.
After the signal intensity of each spot was scanned, it was quantified using ArrayVision software (Imaging Research). Raw and normalized data are available at the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The intensity values were normalized arbitrarily by setting the mean of each array to 1. To study the similarities and variability of the expression signals among the strains, cluster analysis and principal component analysis (PCA), respectively, were used (51). The analysis was performed using the Euclidean distance and the complete linkage algorithm. In order to identify differentially expressed genes, the probability (Pr) that the normalized expression level G of a given gene g is greater in one strain than in the other, Pr(GA > GB), was calculated, where A and B represent all pair- wise combinations of the strains studied: Pr(GCLIP80459 > GCLIP90602), …, Pr(GCLIP93665 > GCLIP93649). The expression levels were assumed to be normal variables. The same approach was used to search for genes with expression levels associated with the results of the virulence profile calculating the probabilities Pr(GC > GD > GE) and Pr(GC < GD < GE), where C and E are the strains with extreme 50% lethal dose (LD50) results and D represents the set of all other strains: Pr(GCLIP80459 > Gothers > GCLIP93649) and Pr(GCLIP80459 < Gothers < GCLIP93649). A gene is called significantly differentially expressed when the Pr of being interrogated is greater than 80% (combinatorial pair-wise analysis or profile-matching analysis). In spite of the analyses differing in their appropriate context, for the sake of clarity we called the output of both analyses differential expression of genes. Along with the statistical significances, the magnitudes (M) of differences were calculated as usual with the log fold changes with the appropriate groupings of C and D: M = log2(GA/GB). With the list of differentially expressed genes, we performed a gene ontology (GO; http://www.geneontology.org) enrichment analysis using the BayGO method (62). The GO annotations were obtained at the EBI GOA-Proteomes web page (http://www.ebi.ac.uk/GOA/proteomes.html) (downloaded in February 2007). BayGO results showing GO categories with enrichment significance values of P smaller than 0.05 were further considered. The complete data set is available (see the supplemental material).
In vivo infection models.
Assessment of the virulence of L. monocytogenes in chick embryos was done as described previously (46). Briefly, L. monocytogenes strains were grown in BHI medium at 37°C to mid-log phase (OD600, 1.0). After being washed in phosphate-buffered saline (PBS), cells were suspended in PBS to an initial cell density of 3 × 107 CFU ml−1 to 3 × 108 CFU ml−1 and serially diluted. Fourteen-day-old embryos were inoculated with 100 ml of the 10−5 dilution via the chorioallantoic membrane. At least five embryos were used per strain tested. The embryos' vitality was monitored daily for 6 days using transillumination. The mean time to death was determined for each strain tested.
The LD50 of the six selected L. monocytogenes strains was determined after growth in BHI medium at 37°C to mid-log phase (OD600, 0.8). After being washed in PBS, pellets were resuspended in sterile 0.9% NaCl. LD50 experiments were carried out by injecting 300-μl serial dilutions of inoculum intravenously into the tail veins of 8-week-old female BALB/c mice (Charles River). The LD50 values were determined by the probit method (3) after the infection of groups of four mice.
RESULTS
Listeria monocytogenes strains of different origins and epidemiological characteristics show different in vivo virulences.
For this study, six L. monocytogenes strains isolated from different sites, showing different epidemiological characteristics, and belonging to the two major lineages present within this species were selected. The two clinical strains were L. monocytogenes CLIP80459 (serovar 4b), which was responsible for 32 cases of illness during an epidemic of listeriosis in France (11), and L. monocytogenes CLIP90602 (serovar 1/2b), which was responsible for 7 clustered cases of listeriosis in France in 2002 (personal communication from C. Jacquet). Strains L. monocytogenes CLIP93667 (serovar 4b) and CLIP9366 (serovar 1/2a) were isolated from healthy humans, and strains L. monocytogenes CLIP93665 (serovar 4b) and CLIP93649 (serovar 1/2a) were isolated from a cheese plant (Table 1). The growth curves of these six strains (MCDB202 medium) were compared and did not show any significant differences (data not shown). Furthermore, the gene contents of the six selected strains were tested on the multiple-genome DNA array, as well as on a focused “Listeria biodiversity” array carrying genes specific to three L. monocytogenes and one L. innocua strain (27, 63). The results showed that the six selected strains contained the core genome of 2,695 genes (see Table S1 in the supplemental material), including all known virulence genes (data not shown).
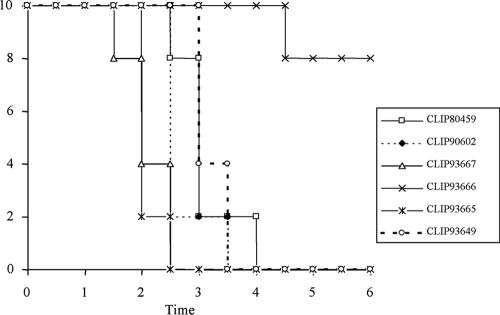
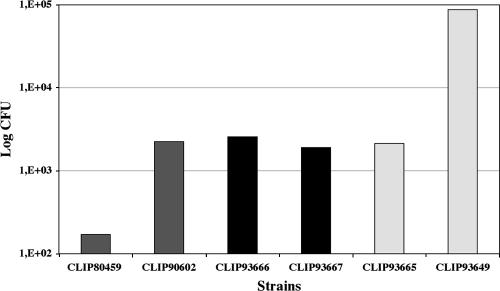
The virulence of the six L. monocytogenes isolates used in this study was determined in two models, the chicken embryo and the mouse model of infection. Infection of chick embryos resulted in 100% mortality within 3 days for all strains except for strain CLIP93666 (H1), which induced only 20% mortality 6 days after infection (Fig. 1). This strain harbors a point mutation at position 1474 of inlA generating a nonsense codon in the coding sequence which results in the translation of a truncated protein of 47 kDa. The presence of a truncated internalin may explain its reduced virulence in the chicken embryo model (46). However, the reduced virulence might also be due to other, not-yet-identified differences present in this strain, as restoration of internalin A functionality did not result in full virulence in chicken embryo assays (45). The LD50 of these six strains was also determined after intravenous injection into BALB/c mice. Two strains presented virulence phenotypes that were distinct from those of the rest of the group. The epidemic strain CLIP80459 showed a high virulence, with an LD50 of 1.7 × 102 CFU, and the environmental strain CLIP93649 showed a reduced virulence, with an LD50 of 8.7 × 104 CFU (Fig. 2). This result suggests that the chicken embryo test may allow the definition of strains expressing a truncated InlA. Furthermore, the mouse model identified one of the two epidemic strains as having a higher virulence than the environmental or carrier strains. However, the second clinical strain, CLIP90602 of serovar 1/2b, showed an LD50 of 2.2 × 103 CFU, which, like those of the four remaining strains, was intermediate compared to those of CLIP80459 and CLIP93649, suggesting that the mouse model of infection does not always reflect epidemiological characteristics (Fig. 2).
FIG. 1.
Assessment of the virulence of the six studied L. monocytogenes strains in the chicken embryo model. The survival curve for the chick embryos is shown. L. monocytogenes cells were inoculated into 14-day-old chick embryos (0.3 × 102 to 1 × 102 CFU per egg) via the chorioallantoic membrane. The survival of the embryos was monitored daily for 6 days.
FIG. 2.
Virulence of six L. monocytogenes strains in BALB/c mice. LD50s of two epidemic strains (left bars), two carriage strains (middle bars), and two environmental strains (right bars) were determined after intravenous injection of increasing inocula of each strain in BALB/c mice.
Global gene expression profiles of L. monocytogenes strains show differences and correlate with virulence.
The L. monocytogenes multigenome array used in this study contains all the genes predicted in L. monocytogenes strain EGDe (serovar 1/2a) and all the genes specific for L. monocytogenes CLIP80459 (serovar 4b) with respect to EGDe. L. monocytogenes strain EGDe is a representative of lineage I and strain L. monocytogenes CLIP80459 of lineage II of the three divergent evolutionary lineages found within the species L. monocytogenes (6, 15, 48, 64, 66). These two strains thus belong to the most common serovars found in foods and in human disease. The DNA array newly constructed here allows the comparison of isolates belonging to the two major lineages, as it contains genes of serovar 1/2a and of serovar 4b strains. To analyze differences and similarities in gene expression among the six L. monocytogenes strains chosen, the expression profile of cells cultured to an OD600 of 0.9 in MCDB broth was determined for each strain and then compared using different methods.
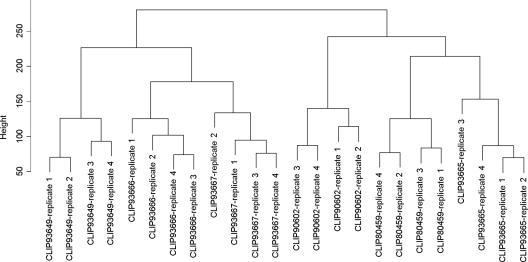
Unsupervised clustering methods offer efficient ways of finding overall patterns and tendencies in gene expression data. Thus, hierarchical clustering may disclose gene expression-based patterns that classify/split the tested strains, by their virulence potential, serovar, or lineage characteristics, for instance. However, analysis of the clusters obtained showed that, by searching for similarities in global gene expression profiles, no such obvious patterns were identified (Fig. 3). This method defined two clusters. Cluster 1 contained the two epidemic strains of serovar 4b and 1/2b and an environmental strain of serovar 4b (CLIP80459, CLIP93665, and CLIP90602). Cluster 2 contained the two carriage strains of serovar 1/2a and 4b and the environmental serovar 1/2a strain (CLIP93666, CLIP936667, and CLIP93649) (Table 1). PCA showed that only about 12% of the variance among the transcriptomes of the strains is significantly linked to the LD50 results, suggesting that there may be a specific set of genes associated with different levels of virulence as assessed in the mouse model. Our primary analysis focused on finding genes preferentially expressed in the strain defined as most virulent in the mouse model of infection, showing high Pr(GCLIP80459 > Gothers > GCLIP93649), and those preferentially expressed in the least virulent strain in the same model of infection, showing high Pr(GCLIP80459 < Gothers < GCLIP93649).
FIG. 3.
Hierarchical clustering. The analysis was performed using the Euclidean distance and the complete linkage algorithm. Numbers 1 to 4 refer to the experimental replicates.
In order to investigate whether the clustering was dependent on the strain-specific genes, we excluded those genes from the analysis and reran the PCA. The relative disposition of the strains did not change when strain-specific genes were excluded, suggesting that the source of variation in the data relates to the differential expression of common L. monocytogenes genes (data not shown). Thus, we subsequently compared the strains with respect to the expression of their core genomes to better understand the possible impact on lineage-related differences in disease potential and niche adaptation. This analysis focused on a total of 2,695 genes common among the strains (see Table S1 in the supplemental material).
Differences in proteins, nucleic acids, carbon metabolism, and virulence-related gene expression characterize the epidemic strain CLIP80459.
CLIP80459 is a clinical isolate responsible for a listeriosis outbreak and presents high virulence as assessed in the mouse model. Its expression profile was compared to those of the other five strains selected for this study. We explored whether there exist genes presenting an expression profile distinguishing this strain as did the virulence profile, with expression levels in CLIP80459 greater than in those strains presenting moderate virulence potentials and greater even than the levels in CLIP93649. A total of 78 core genome genes corresponded to these characteristics (Pr greater than 80%; see Table S2 in the supplemental material). GO term enrichment analysis was used for the functional analysis of these differentially expressed genes (9, 67). The major GO categories of the genes expressed differentially between CLIP80459 and the remaining five strains are genes coding for proteins associated with transport and protein and nucleic acid metabolism, as well as carbon utilization (Table 2). Major virulence genes also showed significant, n-fold variations. Surprisingly, prfA, encoding the master regulator of Listeria virulence gene expression (34); the two phospholipase-encoding genes plcA and plcB (24, 36, 39, 59); and the major invasion protein-encoding genes inlA and inlB (17), as well as the more recently characterized bsh gene, coding for a bile salt hydrolase and regulated by PrfA (18), showed lower expression levels in the epidemic strain CLIP80459 (Table 3).
TABLE 2.
Functional annotation of genes highly expressed in strain CLIP80459a
| Locus tag(s) | GO term identification no. | Description | P |
|---|---|---|---|
| lmo0284 lmo0448 lmo2362 | GO:0006865 | Amino acid transport | 0 |
| lmo2193 | GO:0015833 | Peptide transport | 0.04 |
| lmo2545 lmo2546 | GO:0009088 | Threonine biosynthetic process | 0 |
| lmo1522 lmo1619 | GO:0019478 | d-Amino acid catabolic process | 0 |
| lmo1221 lmo1520 lmo1660 | GO:0006418 | tRNA aminoacylation for protein translation | 0 |
| lmo1096 | GO:0004808 | tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase activity | 0.03 |
| lmo1359 | GO:0003715 | Transcription termination factor activity | 0.03 |
| lmo1235 | GO:0004072 | Aspartate kinase activity | 0.05 |
| lmo1517 | GO:0006808 | Regulation of nitrogen utilization | 0.04 |
| lmo1516 | GO:0008519 | Ammonium transporter activity | 0.04 |
| lmo0001 | GO:0006275 | Regulation of DNA replication | 0.01 |
| lmo1885 | GO:0043101 | Purine salvage | 0.02 |
| lmo1891 lmo1942 | GO:0006310 | DNA recombination | 0.05 |
| lmo1096 lmo1238 | GO:0008033 | tRNA processing | 0.05 |
| lmo1482 | GO:0030420 | Establishment of competence for transformation | 0.03 |
| lmo1600 | GO:0016832 | Aldehyde-lyase activity | 0.04 |
| lmo1600 | GO:0004106 | Chorismate mutase activity | 0.02 |
| lmo2524 | GO:0016836 | Hydro-lyase activity | 0.05 |
| lmo1952 lmo2363 | GO:0016831 | Carboxy-lyase activity | 0 |
| lmo1072 | GO:0004736 | Pyruvate carboxylase activity | 0.02 |
| lmo1917 | GO:0008861 | Formate C-acetyltransferase activity | 0.03 |
| lmo2363 | GO:0019752 | Carboxylic acid metabolic process | 0.03 |
| lmo0482 lmo1407 lmo1661 | GO:0051539 | 4 Iron, 4 sulfur cluster binding | 0 |
lmo indicates locus tags of L. monocytogenes strain EGDe accessible at http://genolist.pasteur.fr/ListiList/. GO terms are accessible at http://www.geneontology.org. P indicates values of significance for GO term enrichment.
TABLE 3.
Virulence-related genes differentially expressed in CLIP80459 and CLIP93649a
| Locus tag | Gene; function | M | P |
|---|---|---|---|
| lmo0200 | prfA; listeriolysin-positive regulatory protein | −25.1 | 0.89 |
| lmo0201 | plcA; phosphatidylinositol-specific phospholipase C | −29.7 | 0.79 |
| lmo0205 | plcB; phospholipase C | −38.6 | 0.91 |
| lmo0433 | inlA; internalin A | −26.4 | 0.86 |
| lmo0434 | inlB; internalin B | −3.85 | 0.89 |
| lmo2067 | bsh; bile salt hydrolase | −4.89 | 0.81 |
| lmo2785 | kat; catalase | −17.7 | 0.91 |
lmo indicates locus tags of L. monocytogenes strain EGDe accessible at http://genolist.pasteur.fr/ListiList/. M = log2(GCLIP80459/GCLIP93649). Negative M value indicates gene repression in CLIP80459. P = Pr(GCLIP93649 > GCLIP80459).
In contrast to the epidemic strain CLIP80459, which had a very low LD50, L. monocytogenes CLIP93649 (serovar 1/2a), which was isolated in a French cheese plant in 2000, showed the highest LD50 in the mouse model. In order to investigate whether the differences in virulence observed in the mouse model are reflected in differences in gene expression patterns, the expression profile of this strain was analyzed in detail. Eighty-two genes were statistically differentially regulated with respect to their regulation in the remaining strains (see Table S3 in the Supplemental Material). The ontology term analyses pointed to several GO terms related to environmental monitoring and stress responses like heat shock and oxidative stress as the major groups of genes characterizing this strain (Table 4).
TABLE 4.
Functional annotation of genes highly expressed in strain CLIP93649a
| Locus tag(s) | GO term identification no. | Description | P |
|---|---|---|---|
| lmo0641 | GO:0005261 | Cation channel activity | 0 |
| lmo2064 | GO:0005216 | Ion channel activity | 0.01 |
| lmo2680 | GO:0006813 | Potassium ion transport | 0.04 |
| lmo2174 | GO:0000155 | Two-component sensor activity | 0.05 |
| lmo2673 lmo2679 | GO:0006950 | Response to stress | 0.03 |
| lmo1472 | GO:0031072 | Heat shock protein binding | 0.02 |
| lmo2785 | GO:0006979 | Response to oxidative stress | 0.03 |
| lmo0205 | GO:0004629 | Phospholipase C activity | 0.05 |
| lmo0905 lmo0938 lmo2230 | GO:0006470 | Protein amino acid dephosphorylation | 0 |
| lmo1014 lmo1421 | GO:0015171 | Amino acid transporter activity | 0.01 |
| lmo1579 | GO:0000286 | Alanine dehydrogenase activity | 0.01 |
| lmo2551 | GO:0003715 | Transcription termination factor | 0.02 |
| lmo0211 | GO:0008097 | 5S rRNA binding | 0.04 |
| lmo1471 | GO:0006479 | Protein amino acid methylation | 0.04 |
| lmo1538 lmo2695 | GO:0006071 | Glycerol metabolic process | 0.01 |
| lmo2205 | GO:0004619 | Phosphoglycerate mutase activity | 0.01 |
| lmo0014 lmo0970 lmo1372 lmo1439 lmo1688 lmo1830 lmo2573 | GO:0016491 | Oxidoreductase activity | 0.02 |
| lmo0014 lmo2389 lmo2390 | GO:0006118 | Electron transport | 0.04 |
| lmo1931 | GO:0009060 | Aerobic respiration | 0.05 |
| lmo1227 lmo1639 | GO:0006284 | Base excision repair | 0.02 |
| lmo1929 | GO:0009209 | Pyrimidine ribonucleoside triphosphate biosynthetic process | 0.02 |
lmo indicates locus tags of L. monocytogenes strain EGDe accessible at http://genolist.pasteur.fr/ListiList/. GO terms are accessible at http://www.geneontology.org. P indicates values of significance for GO term enrichment.
Virulence-, sigma B-, and cell wall-related gene expression differ between L. monocytogenes lineages I and II.
Differential gene expression analysis between strains belonging to lineage I (CLIP93666 and CLIP93649) and lineage II (CLIP80459, CLIP90602, CLIP93667, and CLIP93665) was carried out in an attempt to reveal core genome features that may distinguish the two groups. This analysis identified a total of 1,034 genes, including core genome and strain-specific genes, as being differentially expressed between the two groups (see Table S4 in the supplemental material). Several of those genes that showed overexpression in lineage I compared to their level of expression in lineage II were cell wall-associated core genome genes common to serotype 1/2 and 4 strains (Table 5). Differences in cell wall-associated genes are somewhat expected as there are structural differences between the cell walls of L. monocytogenes strains belonging to serogroups 1/2 and 4 (22, 23). The somatic component of the serotype designation of Listeria is related to the teichoic acid (TA; polyribitol phosphate covalently linked to peptidoglycan) present in the cell wall. Glycosidic substitutions of the ribitol phosphate units render this component variable, structurally and antigenically (32). Such differences could account for distinct patterns of interaction with host cells, with a direct impact on the virulence of the different serovars. Serotype 1/2 and 4 L. monocytogenes strains show structural differences in the cell wall due to distinct genome content (22, 23). However, here we show that the gene expression of common cell wall-regulated genes is consistently different between the two lineages, suggesting also common features of the cell walls of 1/2b and 4b strains. Interestingly, differences are also seen in prfA and PrfA-regulated genes. As depicted in Table 5, prfA, plcB, plcA, hly, actA, and inlAB are highly expressed in lineage I strains compared to their levels of expression in lineage II strains.
TABLE 5.
Selected core genome features differentially expressed between L. monocytogenes strains belonging to lineages I and IIa
| Locus tag | Gene; function | M | P |
|---|---|---|---|
| lmo0200 | prfA; listeriolysin-positive regulatory protein | 13.5 | 0.87 |
| lmo0201 | plcA; phosphatidylinositol-specific phospholipase C | 21.3 | 0.98 |
| lmo0202 | hly; listeriolysin O precursor | 8.8 | 0.9 |
| lmo0204 | actA; actin-assembly inducing protein precursor | 0.54 | 0.96 |
| lmo0205 | plcB; phospholipase C | 19.4 | 0.84 |
| lmo0433 | inlA; internalin A | 15.3 | 0.93 |
| lmo0434 | inlB; internalin B | 2.35 | 0.95 |
| lmo1074 | Similar to TA translocation permease protein TagG | 0.67 | 0.88 |
| lmo1075 | Similar to TA translocation ATP-binding protein TagH | 1.55 | 0.96 |
| lmo0842 | Putative peptidoglycan-bound protein (LPXTG motif) | 0.24 | 0.98 |
| lmo0441 | Similar to penicillin-binding protein (d-alanyl-d-alanine carboxypeptidase) | −1.12 | 0.94 |
| lmo0540 | Similar to penicillin-binding protein | −2.54 | 0.97 |
| lmo1892 | Similar to penicillin-binding protein 2A | −3.64 | 0.96 |
| lmo2229 | Similar to penicillin-binding protein | −1.98 | 0.99 |
lmo indicates locus tags of L. monocytogenes strain EGDe accessible at http://genolist.pasteur.fr/ListiList/. M = log2(GlinI/GlinII), where linI and linII are lineages I and II. P = Pr(LinI > LinII) when M is a positive number, and P = Pr(LinII > LinI) when M is a negative number.
Basal levels of the sigB operon genes (rsbR-rsbS-rsbT-rsbU-rsbV-rsbW-sigB-rsbX) are provided by transcription initiating at a promoter positioned upstream of the rsbR gene. A second SigB-dependent promoter is located between rsbU and rsbV and is induced by all SigB-activating environmental and metabolic stimuli described thus far. Several virulence-related genes, such as inlA and inlB, are also under the influence of sigma B. Like the virulence genes mentioned above, sigB was overexpressed in lineage I, as were rsbV, rsbW, and rsbX. We thus specifically analyzed the expression of the genes reported to be under the control of sigma B as described by Kazmierczak and colleagues (33). This analysis additionally showed that 20 genes described as belonging to the sigma B regulon were up-regulated in lineage I (Table 6).
TABLE 6.
Sigma B-related genes overexpressed in L. monocytogenes strains belonging to lineage Ia
| Locus tag | Gene; function | M | P |
|---|---|---|---|
| lmo1421 | Similar to glycine betaine/carnitine/choline ABC | 1.95 | 0.94 |
| lmo1426 | opuCC; transporter (membrane protein) | 3.64 | 0.83 |
| lmo1427 | opuCB; similar to glycine betaine/carnitine/choline ABC | 0.908 | 0.67 |
| lmo1428 | opuCA; similar to glycine betaine/carnitine/choline ABC | 1.94 | 0.81 |
| lmo0893 | Anti-anti-sigma factor (antagonist of RsbW) | 2.67 | 0.84 |
| lmo0894 | RNA polymerase sigma-37 factor (sigma B) | 2.29 | 0.79 |
| lmo0895 | Sigma B activity negative regulator RsbW | 2.34 | 0.78 |
| lmo0896 | Indirect negative regulation of sigma B-dependent gene | 1.54 | 0.81 |
| lmo2230 | Expression (serine phosphatase) similar to arsenate reductase | 5.33 | 0.88 |
| lmo0783 | Similar to mannose-specific phosphotransferase system component IIB | 19.3 | 0.94 |
| lmo0784 | Similar to mannose-specific phosphotransferase system component IIA | 9.94 | 0.96 |
| lmo2398 | Low-temperature-requirement C protein | 3.21 | 0.94 |
| lmo2602 | Phosphotransferase system component IIA cation-transporting ATPase | 2 | 0.99 |
| lmo1539 | Similar to glycerol uptake facilitator | 0.748 | 0.92 |
| lmo0669 | Transporter (ATP-binding protein) similar to oxidoreductase | 2.93 | 1 |
| lmo1694 | Similar to CDP-abequose synthase | 3.09 | 0.99 |
| lmo2695 | Similar to dihydroxyacetone kinase | 2.3 | 0.86 |
| lmo2205 | Similar to phosphoglyceromutase 1 | 9.5 | 0.82 |
| lmo2085 | Putative peptidoglycan-bound protein (LPXTG motif) | 2.81 | 0.94 |
| lmo0880 | Similar to cell wall-associated protein precursor (LPXTG motif) | 2.17 | 0.83 |
lmo indicates locus tags of L. monocytogenes strain EGDe accessible at http://genolist.pasteur.fr/ListiList/. M = log2(GlinI/GlinII), where linI and linII are lineages I and II. P = Pr(LinI > LinII).
Penicillin binding- and antibiotic response-related genes were also identified as markers that differentiate the two groups. Four of the genes overexpressed in this category in lineage II (lmo0441, lmo0540, lmo1892, and lmo222) (Table 5) present the characteristics of penicillin-binding proteins. Penicillin-binding proteins are transpeptidases involved in different aspects of cell wall synthesis in bacteria, and their overexpression in lineage II corroborates distinct cell wall activities in the two groups under the studied conditions.
Motility- and chemotaxis-related genes were overexpressed in lineage II (Table 7), suggesting that the signals for the expression of these genes may be perceived in a different way by the two groups. Distinct patterns of regulation may act upon the pathogen's efficiency in cell invasion and its interaction with the host—in particular, the immune system.
TABLE 7.
Motility- and chemotaxis-related genes overexpressed in L. monocytogenes strains belonging to lineage IIa
| Locus tag | Gene; function | M | P |
|---|---|---|---|
| lmo0678 | Similar to flagellar biosynthesis protein FliR | −0.613 | 0.72 |
| lmo0679 | Similar to flagellar biosynthesis protein FlhB | −0.964 | 0.78 |
| lmo0680 | Similar to flagellum-associated protein FlhA | −0.129 | 0.63 |
| lmo0681 | Similar to flagellar biosynthesis protein FlhF | −0.417 | 0.78 |
| lmo0682 | Similar to flagellar hook-basal body protein FlgG | −0.478 | 0.8 |
| lmo0683 | Similar to chemotactic methyltransferase CheR | −0.986 | 0.83 |
| lmo0685 | Similar to motility protein (flagellar motor rotation) MotA | −0.323 | 0.88 |
| lmo0686 | Similar to motility protein (flagellar motor rotation) MotB | −1.49 | 0.88 |
| lom0689 | Similar to CheA activity-modulating chemotaxis protein CheV | −0.933 | 0.89 |
| lmo0690 | Flagellin protein | −7.61 | 0.74 |
| lmo0691 | Chemotaxis response regulator CheY | −0.868 | 0.81 |
| lmo0692 | Two-component sensor histidine kinase CheA | −2.57 | 0.78 |
| lmo0693 | Similar to flagellar motor switch protein FliY C-terminal part | −0.248 | 0.77 |
| lmo0697 | Similar to flagellar hook protein FlgE | −2.44 | 0.8 |
| lmo0698 | Weakly similar to flagellar switch protein | −0.353 | 0.82 |
| lmo0699 | Similar to flagellar switch protein FliM | −0.55 | 0.83 |
| lmo0700 | Similar to flagellar motor switch protein fliY | −0.676 | 0.8 |
| lmo0705 | Similar to flagellar hook-associated protein FlgK | −0.817 | 0.9 |
| lmo0706 | Similar to flagellar hook-associated protein 3 FlgL | −0.689 | 0.913 |
| lmo0707 | Similar to flagellar hook-associated protein 2 FliD | −1.44 | 0.88 |
| lmo0710 | Similar to flagellar basal-body rod protein FlgB | −1.19 | 0.919 |
| lmo0711 | Similar to flagellar basal-body rod protein FlgC | −0.514 | 0.88 |
| lmo0712 | Similar to flagellar hook-basal body complex protein FliE | −0.239 | 0.9 |
| lmo0713 | Similar to flagellar basal-body M-ring protein FliF | −0.636 | 0.87 |
| lmo0714 | Similar to flagellar motor switch protein FliG | −1.02 | 0.902 |
lmo indicates locus tags of L. monocytogenes strain EGDe accessible at http://genolist.pasteur.fr/ListiList/. M = log2(GlinI/GlinII), where linI and linII are lineages I and II. P = Pr(LinII > LinI).
Strain-specific genes expressed are mainly cell wall related.
As reported recently, strain-specific genes are differentially expressed during intracellular growth of L. monocytogenes (10). Thus, we investigated whether strain-specific genes were also expressed under our study conditions. Genes considered to be strain specific were those defined as present only in the L. monocytogenes EGDe or in the L. monocytogenes CLIP80459 genome sequence according to the best BLASTP hits and an amino acid sequence similarity lower than 70% over two-thirds of the length of the protein. Table 8 shows 18 genes (out of a total of 151 L. monocytogenes serovar 4b strain-specific genes present on the array) that were significantly expressed in lineage II strains under the study conditions. Fourteen of these genes code for proteins related to cell wall biosynthesis or cell wall-associated proteins.
TABLE 8.
Strain-specific genes expressed in L. monocytogenes strains belonging to lineage IIa
| Locus tag | Gene function; description or name | M | P |
|---|---|---|---|
| lm4b0014 | Weakly similar to similar to autolysin (amidase) | 0.62 | 0.81 |
| lm4b0015 | Weakly similar to similar to autolysin (amidase) | 0.39 | 0.86 |
| lm4b0290 | Similar to internalin D | 0.32 | 0.97 |
| lm4b0349 | Similar to internalin, putative peptidoglycan-bound protein (LPXTG motif) | 0.34 | 0.97 |
| lm4b0372 | Similar to internalin, peptidoglycan-bound protein (LPxTG motif) | 0.99 | 0.94 |
| lm4b0484 | Similar to transcription regulator (VirR from Streptococcus pyogenes) | 0.49 | 0.96 |
| lm4b0687 | Similar to internalin proteins, putative peptidoglycan-bound protein (LPXTG motif) | 0.48 | 0.93 |
| lm4b0943 | Similar to ABC transporter, ATP-binding protein | 0.60 | 0.99 |
| lm4b1092 | Similar to autolysin (N-acetylmuramoyl-l-alanine amidase) | 15.3 | 0.98 |
| lm4b1101 | Similar to galactosamine-containing minor TA biosynthesis protein GgaA | 0.64 | 0.88 |
| lm4b1102 | Similar to Bacillus subtilis TagF protein (probable CDP-glycerol glycerophosphotransferase) | 1.75 | 0.92 |
| lm4b1103 | Similar to TA biosynthesis protein B precursor | 0.16 | 0.82 |
| lm4b1887 | Similar to hypothetical proteins containing ChW repeats | 0.77 | 0.91 |
| lm4b2360 | Similar to putative transcription regulator | 0.91 | 0.99 |
| lm4b2526 | Autolysin, amidase | 5.82 | 0.91 |
| lm4b2631 | Similar to internalin; putative peptidoglycan-bound protein (LPXTG motif) | 1.22 | 0.96 |
| lm4b2727 | Similar to glycosyltransferase; gltA | 2.15 | 0.94 |
| lm4b2728 | Similar to glycosyltransferase; gltB | 2.29 | 0.94 |
lmo indicates locus tags of L. monocytogenes strain EGDe accessible at http://genolist.pasteur.fr/ListiList/. M = log2(GlinI/GlinII), where linI and linII are lineages I and II. P = Pr(LinII > LinI).
In lineage I, 13 specific genes (out of 139 L. monocytogenes serovar 1/2a strain-specific genes present on the array) were significantly expressed. Among those, genes coding for cell wall-associated proteins, like the clusters lmo1076 to 1088 and lmo1090 to 1091 involved in cell wall biosynthesis, again were the most prominent ones (Table 9). These results are in agreement with the fact that both groups differ in cell wall-related activities with a possible impact on host-pathogen interaction. The carbohydrate metabolism-related gene bvrA (lmo2788), encoding an antiterminator of the BglG family (4), implicated in the virulence of L. monocytogenes and absent in lineage II strains (14) was also expressed in lineage I (Table 9).
TABLE 9.
Strain-specific genes expressed in L. monocytogenes strains belonging to lineage Ia
| Locus tag | Gene function; description or name | M | P |
|---|---|---|---|
| lmo1077 | Similar to TA biosynthesis protein B | 0.27 | 0.87 |
| lmo1080 | Glycosyl transferase | 0.63 | 0.93 |
| lmo1081 | Glucose-1-phosphate thymidylyltransferase | 1.59 | 0.96 |
| lmo1082 | dTDP-4-dehydrorhamnose 3,5-epimerase | 2.09 | 0.91 |
| lmo1083 | dTDP-glucose 4,6-dehydratase | 1.89 | 0.88 |
| lmo1084 | dTDP-4-dehydrorhamnose reductase | 3.87 | 0.92 |
| lmo1085 | Similar to TA biosynthesis protein B | 0.47 | 0.98 |
| lmo1086 | Similar to CDP-ribitol pyrophosphorylase | 4.66 | 0.88 |
| lmo1087 | Similar to glucitol dehydrogenase | 1.64 | 0.89 |
| lmo1088 | Similar to TA biosynthesis protein B precursor; tagB | 0.67 | 0.93 |
| lmo1090 | Glycosyl transferase | 0.54 | 0.92 |
| lmo1091 | Glycosyl transferase domain protein; putative | 0.47 | 0.85 |
| lmo2788 | Transcription antiterminator; BglG family | 0.82 | 1 |
lmo indicates locus tags of L. monocytogenes strain EGDe accessible at http://genolist.pasteur.fr/ListiList/. M = log2(GlinI/GlinII), where linI and linII are lineages I and II. P = Pr(LinI > LinII).
DISCUSSION
All L. monocytogenes strains found in foods are considered to be pathogenic; however, the relative virulence of individual L. monocytogenes isolates can vary substantially in selected animal models (5, 53, 66). The genetic basis underlying these virulence differences is not yet understood. Comparative genomics by the hybridization of over 100 L. monocytogenes strains revealed important differences in gene content among different isolates (8, 15, 68), giving some clues as to how virulence differences might have evolved. However, complete understanding of the variations in virulence was not obtained. Global gene expression profiling has been shown to be a powerful tool for comparing differences in gene expression/regulation that might be related to virulence. Transcriptional profiling and comparison of two clinical isolates of Borrelia burgdorferi, the etiologic agent of Lyme disease, identified differences in the transcriptomes that provided clues to their pathogenesis (44). Comparison of the transcriptomes of two cystic fibrosis-causing epidemic strains of Pseudomonas aeruginosa that displayed enhanced virulence and antimicrobial resistance and of a laboratory strain revealed differences in the transcriptomes that were related to the phenotypes (55). A similar approach applied to Escherichia coli (16, 35) led to the discovery of selection-driven transcriptional profile differences and distinct differences in the expression of virulence and stress-related genes.
We used the comparison of global gene expression profiles and two different in vivo infection models to learn more about strain- and lineage-specific differences, some of which might also be related to virulence differences in L. monocytogenes strains. Six L. monocytogenes strains with different epidemiological backgrounds (epidemic, carrier, and environmental isolates) belonging to the two major disease-related lineages I and II were chosen for this first multiple-strain transcriptome comparison of L. monocytogenes. The virulence of these strains as assessed in the mouse infection model and in the chicken embryo varied according to the model used. The infection of chick embryos identified as having very low virulence one strain (CLIP93666) that was previously shown to be impaired in its ability to invade target cells via the interaction of InlA and its receptor E-cadherin, due to a truncated inlA gene (47), whereas the other five strains did not show significant virulence differences in this model. The murine model distinguished the epidemic serovar 4b strain (CLIP80459) from one environmental strain (CLIP93649) as having very high virulence and very low virulence, respectively, and these were clearly distinct from the four remaining strains for which virulence was not distinguished in this model. Interestingly, comparison of global gene expression and virulence as assessed in the mouse model showed a good correlation, indicating that differences in virulence may indeed be reflected in gene expression. Due to the many differences identified in genes encoding functions of the basic metabolism, such as carbohydrate metabolism and amino acid and cell wall biosynthesis, and those encoding functions expressed under specific circumstances, such as stress-related and virulence genes expected to be important for adaptation to the environment and the infected host, we specifically focused the analysis on the transcriptional data for these two groups of genes.
Like many other pathogens that can live saprophytically in the environment, L. monocytogenes must have tightly regulated virulence gene expression. In this study, we observed a clear repression of the prfA- and PrfA-dependent genes (plcA, plcB, hly, inlA, and inlB) in CLIP80459 in comparison to their levels of expression in the other group of strains. Interestingly, strain CLIP80459 was the most virulent one in the murine model of infection and was responsible for an epidemic of listeriosis in France from the end of 1999 to the beginning of 2000 (11). This observation is in agreement with our previous results showing that the virulence gene expression of the epidemic L. monocytogenes serovar 4b strain PAM 14 was lower than that of the L. monocytogenes EGDe strain (40). Furthermore, only low levels of haemolysin and lecithinase have been reported in clinical isolates of L. monocytogenes when grown in rich medium (53, 61), which might indicate that it is an advantage not to express these genes under in vitro or “environmental” conditions. However, another recent study comparing the invasion capacity of and the expression of the inlA and inlB genes in 27 clinical and 37 nonclinical L. monocytogenes strains in Caco-2 and HepG2 cells also identified significant differences in their invasion capacities that were correlated with gene expression. Clinical strains showed a lower invasion capacity and lower expression levels of inlA and inlB and induced lower interleukin-8 levels in HepG2 cells than nonclinical strains (65). Taken together, these findings show that in vitro gene expression and invasion into human cell lines correlate and suggest that the lower capacity of clinical strains to invade HepG2 cells, which is correlated with lower inlAB expression and the lower induction of interleukin-8, is possibly a mechanism of immune evasion used by specific L. monocytogenes strains.
Interestingly, overexpression of motility genes was observed in lineage II L. monocytogenes strains. Flagella are of great importance for the virulence of several pathogens (12, 20, 37, 52). Even though they mediate or facilitate the adhesion and invasion of eukaryotic host cells by L. monocytogenes (2, 13), the role of flagella in vivo is still not clear. The repression of virulence-related genes and overexpression of motility- and chemotaxis-related genes possibly characterize free-living L. monocytogenes.
The gene expression of lineage I strains compared to that of lineage II strains revealed many differentially expressed genes, highlighting important metabolic differences between the two serotypes. TA biosynthesis genes common to serotype 1/2 and 4 were clearly differentially expressed, pointing not only to structural differences in the TA composition but also to differential regulation of the biosynthesis of TA. Glycosylated TA components are important antigenic determinants in L. monocytogenes, even though their specific role in infection has not been elucidated (32). Due to their surface exposure and immunogenicity, their importance in interactions between bacteria and their host cells, with other free-living organisms, and with the extracellular matrix is thus likely. In agreement with these observations, a number of genes encoding cell wall-associated proteins were also detected as being differentially expressed between the two groups. We also detected the differential expression of sigB and SigB-regulated genes. In L. monocytogenes, SigB confers stress resistance and contributes to pathogenesis since it affects prfA transcription, also differentially expressed between the two groups (33). Links between environmental stress responses and virulence suggest that SigB might have a central role in the survival and pathogenesis of L. monocytogenes. The diversity of genes affected by this global regulator could reflect the way strains respond to environmental stress. Thus, one of the major themes that emerges from the analysis of the differential gene expression between lineages I and II is that they seem to interact differently with the milieu or their host. This is in agreement with their dissimilar patterns of distribution in foods, the environment, and the infected hosts. However, gene expression profiling in in vitro conditions in rich medium does not necessarily reflect the conditions encountered in the environment or the host. Thus, our results represent a first basis of gene expression profiles, allowing it to be shown that differences among strains exist in vitro and suggesting that differences may exist also in vivo. The exciting experiment for the future will be the comparison of a greater number of strains in vivo and in conditions encountered in the food environment.
In conclusion, this study presents the first comparison of the gene expression profiles of distinct strains of L. monocytogenes, providing new insights into strain-specific differences in gene expression and new results for the better understanding of the response of L. monocytogenes to the different environments it encounters. Our results suggest that differential gene regulation of core genes of the L. monocytogenes genome may have an impact on its clinical behavior, suggesting niche adaptation. Differences in the gene expression profiles of representative strains of the two major lineages within the species L. monocytogenes that are responsible for human disease reflect diverse possibilities of interaction with their hosts.
Supplementary Material
Acknowledgments
We received financial support from the Institut Pasteur (GPH no. 9), INSERM, INRA, and the Ministère de l'Agriculture et de la Pêche (DGAL no. A03/02). P.S. received financial support from ARILAIT-Recherches.
We thank Marie-Agnès Dillies for initial help in data analysis.
Footnotes
Published ahead of print on 17 August 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barbour, A. H., A. Rampling, and C. E. Hormaeche. 2001. Variation in the infectivity of Listeria monocytogenes isolates following intragastric inoculation of mice. Infect. Immun. 69:4657-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bigot, A., H. Pagniez, E. Botton, C. Frehel, I. Dubail, C. Jacquet, A. Charbit, and C. Raynaud. 2005. Role of FliF and FliI of Listeria monocytogenes in flagellar assembly and pathogenicity. Infect. Immun. 73:5530-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bliss, C. I. 1938. The calculation of the dosage-mortality curve. Ann. Appl. Biol. 22:134-167. [Google Scholar]
- 4.Brehm, K., M. T. Ripio, J. Kreft, and J. A. Vazquez-Boland. 1999. The bvr locus of Listeria monocytogenes mediates virulence gene repression by β-glucosides. J. Bacteriol. 181:5024-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosch, R., B. Catimel, G. Milon, C. Buchrieser, E. Vindel, and J. Rocourt. 1993. Virulence heterogeneity of Listeria monocytogenes strains from various sources (food, human, animal) in immunocompetent mice and its association with typing characteristics. J. Food Prot. 56:296-301. [DOI] [PubMed] [Google Scholar]
- 6.Brosch, R., J. Chen, and J. B. Luchansky. 1994. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol. 60:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchrieser, C. Biodiversity of the species Listeria monocytogenes and the genus Listeria. Microbes Infect., in press. [DOI] [PubMed]
- 8.Call, R. D., M. K. Boroucki, and T. E. Besser. 2003. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J. Clin. Microbiol. 41:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavalieri, D., and C. De Filippo. 2005. Bioinformatic methods for integrating whole-genome expression results into cellular networks. Drug Discov. Today 10:727-734. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Valk, H., V. Vaillant, C. Jacquet, J. Rocourt, F. Le Querrec, F. Stainer, N. Quelquejeu, O. Pierre, V. Pierre, J.-C. Desenclos, and V. Goulet. 2001. Two consecutive nationwide outbreaks of listeriosis in France, October 1999-February 2000. Am. J. Epidemiol. 154:944-950. [DOI] [PubMed] [Google Scholar]
- 12.Dietrich, G., A. Bubert, I. Gentschev, Z. Sokolovic, A. Simm, A. Catic, S. H. E. Kaufmann, J. Hess, A. A. Szalay, and W. Goebel. 1998. Delivery of antigen-encoding plasmid DNA into the cytosol of macrophages by attenuated suicide Listeria monocytogenes. Nat. Biotechnol. 16:181-185. [DOI] [PubMed] [Google Scholar]
- 13.Dons, L., E. Erikson, Y. Jin, M. E. Rottenberg, K. Kristenson, C. N. Larsen, J. Bresciani, and J. O. Olsen. 2004. Role of flagellin and the two-component system CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect. Immun. 72:3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doumith, M., C. Buchrieser, P. Glaser, C. Jacquet, and P. Martin. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doumith, M., C. Cazalet, N. Simoes, L. Frangeul, C. Jaquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowd, S. E., and H. Ishizaki. 2006. Microarray based comparison of two Escherichia coli O157:H7 lineages. BMC Microbiol. 6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dramsi, S., M. Lebrun, and P. Cossart. 1996. Molecular and genetic determinants involved in invasion of mammalian cells by Listeria monocytogenes. Curr. Top. Microbiol. Immunol. 209:61-77. [DOI] [PubMed] [Google Scholar]
- 18.Dussurget, O., D. Cabanes, P. Dehoux, M. Lecuit, C. Buchrieser, P. Glaser, and P. Cossart. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45:1095-1106. [DOI] [PubMed] [Google Scholar]
- 19.Dussurget, O., J. Pizarro-Cerda, and P. Cossart. 2004. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58:587-610. [DOI] [PubMed] [Google Scholar]
- 20.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37:123-127. [DOI] [PubMed] [Google Scholar]
- 21.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiedler, F. 1988. Biochemistry of the cell surface of Listeria strains: a locating general view. Infection 16(Suppl. 2):S92-S97. [DOI] [PubMed] [Google Scholar]
- 23.Fiedler, F., and G. J. Ruhland. 1987. Structure of Listeria monocytogenes cell walls. Bull. Inst. Pasteur 85:287-300. [Google Scholar]
- 24.Geoffroy, C., J. Raveneau, J. L. Beretti, A. Lecroisey, J. A. Vazquez-Boland, J. E. Alouf, and P. Berche. 1991. Purification and characterization of an extracellular 29-kilodalton phospholipase C from Listeria monocytogenes. Infect. Immun. 59:2382-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 26.Hof, H., and J. Rocourt. 1992. Is any strain of Listeria monocytogenes detected in food a health risk? Int. J. Food Microbiol. 16:173-182. [DOI] [PubMed] [Google Scholar]
- 27.Hong, E., M. Doumith, S. Duperrier, I. Giovannacci, A. Morvan, P. Glaser, C. Buchrieser, C. Jacquet, and M. Martin. 2007. Genetic diversity of Listeria monocytogenes populations present in patients and in pork products at the store distribution level in France in 2000-2001. Int. J. Food Microbiol. 114:187-194. [DOI] [PubMed] [Google Scholar]
- 28.Jacquet, C., M. Doumith, J. I. Gordon, P. M. Martin, P. Cossart, and M. Lecuit. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094-2100. [DOI] [PubMed] [Google Scholar]
- 29.Jacquet, C., E. Gouin, D. Jeannel, P. Cossart, and J. Rocourt. 2002. Expression of ActA, Ami, InlB, and listeriolysin O in Listeria monocytogenes of human and food origin. Appl. Environ. Microbiol. 68:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 31.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 32.Kamisango, K., H. Fujii, H. Okumura, I. Saiki, Y. Araki, Y. Yamamura, and I. Azuma. 1983. Structural and immunochemical studies of teichoic acid of Listeria monocytogenes. J. Biochem. 93:1401-1409. [DOI] [PubMed] [Google Scholar]
- 33.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lampidis, R., R. Gross, Z. Sokolovic, W. Goebel, and J. Kreft. 1994. The virulence regulator protein of Listeria ivanovii is highly homologous to PrfA from Listeria monocytogenes and both belong to the Crp-Fnr family of transcription regulators. Mol. Microbiol. 13:141-151. [DOI] [PubMed] [Google Scholar]
- 35.Le Gall, T., P. Darlu, P. Escobar-Paramo, B. Picard, and E. Denamur. 2005. Selection-driven transcriptome polymorphism in Escherichia coli/Shigella species. Genome Res. 15:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leimeister-Wächter, M., E. Domann, and T. Chakraborty. 1991. Detection of a gene encoding a phosphatidylinositol-specific phospholipase C that is coordinately expressed with listeriolysin in Listeria monocytogenes. Mol. Microbiol. 5:361-366. [DOI] [PubMed] [Google Scholar]
- 37.Liu, S. L., T. Ezaki, H. Miura, K. Matsui, and E. Yabuuchi. 1988. Intact motility as a Salmonella Typhi invasion-related factor. Infect. Immun. 56:1967-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lou, Y., and A. E. Yousef. 1998. Characteristics of Listeria monocytogenes important to food processors. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis and food safety. Marcel Dekker Inc., New York, NY.
- 39.Mengaud, J., B. C. Braun, and P. Cossart. 1991. Identification of phosphatidylinositol-specific phospholipase C activity in Listeria monocytogenes: a novel type of virulence factor? Mol. Microbiol. 5:367-372. [DOI] [PubMed] [Google Scholar]
- 40.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 41.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. σB contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norrung, B., and J. K. Andersen. 2000. Variations in virulence between different electrophoretic types of Listeria monocytogenes. Lett. Appl. Microbiol. 30:228-232. [DOI] [PubMed] [Google Scholar]
- 44.Ojaimi, C., V. Mulay, D. Liveris, R. Iyer, and I. Schwartz. 2005. Comparative transcriptional profiling of Borrelia burgdorferi clinical isolates differing in capacities for hematogenous dissemination. Infect. Immun. 73:6791-6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olier, M., D. Garmyn, S. Rousseaux, J. P. Lemaitre, P. Piveteau, and J. Guzzo. 2005. Truncated internalin A and asymptomatic Listeria monocytogenes carriage: in vivo investigation by allelic exchange. Infect. Immun. 73:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olier, M., F. Pierre, J. P. Lemaitre, C. Divies, A. Rousset, and J. Guzzo. 2002. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148:1855-1862. [DOI] [PubMed] [Google Scholar]
- 47.Olier, M., F. Pierre, S. Rousseaux, J. P. Lemaitre, A. Rousset, P. Piveteau, and J. Guzzo. 2003. Expression of truncated internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect. Immun. 71:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pine, L., S. Kathariou, F. Quinn, V. George, J. D. Wenger, and R. E. Weaver. 1991. Cytopathogenic effects in enterocyte-like Caco-2 cells differentiate virulent from avirulent Listeria strains. J. Clin. Microbiol. 29:990-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pine, L., R. E. Weaver, G. M. Carlone, P. A. Pienta, J. Rocourt, W. Goebel, S. Kathariou, W. Bibb, and G. B. Malcolm. 1987. Listeria monocytogenes ATCC 35152 and NCTC 7973 contain a nonhemolytic, nonvirulent variant. J. Clin. Microbiol. 25:2247-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quackenbush, J. 2002. Microarray data normalization and transformation. Nat. Genet. 32(Suppl.):496-501. [DOI] [PubMed] [Google Scholar]
- 52.Richardson, K. 1991. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect. Immun. 59:2727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ripio, M. T., G. Dominguez-Bernal, M. Suarez, K. Brehm, P. Berche, and J. Vasquez-Boland. 1996. Transcriptional activation of virulence genes in wild type strains of Listeria monocytogenes in response to a change in extracellular medium composition. Res. Microbiol. 147:371-384. [DOI] [PubMed] [Google Scholar]
- 54.Roberts, A. J., and M. Wiedmann. 2003. Pathogen, host, and environmental factors contributing to the pathogenesis of listeriosis. Cell. Mol. Life Sci. 60:904-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salunkhe, P., C. H. Smart, J. A. Morgan, S. Panagea, M. J. Walshaw, C. A. Hart, R. Geffers, B. Tummler, and C. Winstanley. 2005. A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance. J. Bacteriol. 187:4908-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmid, M. W., E. Y. Ng, R. Lampidis, M. Emmerth, M. Walcher, J. Kreft, W. Goebel, M. Wagner, and K. H. Schleifer. 2005. Evolutionary history of the genus Listeria and its virulence genes. Syst. Appl. Microbiol. 28:1-18. [DOI] [PubMed] [Google Scholar]
- 57.Schuchat, A., B. Swaminathan, and C. V. Broome. 1991. Epidemiology of human listeriosis. Clin. Microbiol. Rev. 4:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwab, U., Y. Hu, M. Wiedmann, and K. J. Boor. 2005. Alternative sigma factor sigmaB is not essential for Listeria monocytogenes surface attachment. J. Food Prot. 68:311-317. [DOI] [PubMed] [Google Scholar]
- 59.Vazquez-Boland, J.-A., C. Kocks, S. Dramsi, H. Ohayon, C. Geoffroy, J. Mengaud, and P. Cossart. 1992. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect. Immun. 60:219-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vazquez-Boland, J.-A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vega, Y., C. Dickneite, M. T. Ripio, R. Bockmann, B. Gonzalez-Zorn, S. Novella, G. Dominguez-Bernal, W. Goebel, and J. A. Vazquez-Boland. 1998. Functional similarities between the Listeria monocytogenes virulence regulator PrfA and cyclic AMP receptor protein: the PrfA* (Gly145Ser) mutation increases binding affinity for target DNA. J. Bacteriol. 180:6655-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vencio, R. Z., T. Koide, S. L. Gomes, and C. A. Pereira. 2006. BayGO: Bayesian analysis of ontology term enrichment in microarray data. BMC Bioinformatics 7:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Volokhov, D. V., S. Duperrier, A. A. Neverov, J. George, C. Buchrieser, and A. D. Hitchins. 2007. The presence of the internalin gene in natural atypically hemolytic Listeria innocua strains suggests descent from L. monocytogenes. Appl. Environ. Microbiol. 73:1928-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Werbrouck, H., K. Grijspeerdt, N. Botteldoorn, E. Van Pamel, N. Rijpens, J. Van Damme, M. Uyttendaele, L. Herman, and E. Van Coillie. 2006. Differential inlA and inlB expression and interaction with human intestinal and liver cells by Listeria monocytogenes strains of different origins. Appl. Environ. Microbiol. 72:3862-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yue, L., and W. C. Reisdorf. 2005. Pathway and ontology analysis: emerging approaches connecting transcriptome data and clinical endpoints. Curr. Mol. Med. 5:11-21. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, C., M. Zhang, J. Ju, J. Nietfeldt, J. Wise, P. M. Terry, M. Olson, S. D. Kachman, M. Wiedmann, M. Samadpour, and A. K. Benson. 2003. Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: identification of segments unique to lineage II populations. J. Bacteriol. 185:5573-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.