Abstract
Aerobic activated sludge granules are dense, spherical biofilms which can strongly improve purification efficiency and sludge settling in wastewater treatment processes. In this study, the structure and development of different granule types were analyzed. Biofilm samples originated from lab-scale sequencing batch reactors which were operated with malthouse, brewery, and artificial wastewater. Scanning electron microscopy, light microscopy, and confocal laser scanning microscopy together with fluorescence in situ hybridization (FISH) allowed insights into the structure of these biofilms. Microscopic observation revealed that granules consist of bacteria, extracellular polymeric substances (EPS), protozoa and, in some cases, fungi. The biofilm development, starting from an activated sludge floc up to a mature granule, follows three phases. During phase 1, stalked ciliated protozoa of the subclass Peritrichia, e.g., Epistylis spp., settle on activated sludge flocs and build tree-like colonies. The stalks are subsequently colonized by bacteria. During phase 2, the ciliates become completely overgrown by bacteria and die. Thereby, the cellular remnants of ciliates act like a backbone for granule formation. During phase 3, smooth, compact granules are formed which serve as a new substratum for unstalked ciliate swarmers settling on granule surfaces. These mature granules comprise a dense core zone containing bacterial cells and EPS and a loosely structured fringe zone consisting of either ciliates and bacteria or fungi and bacteria. Since granules can grow to a size of up to several millimeters in diameter, we developed and applied a modified FISH protocol for the study of cryosectioned biofilms. This protocol allows the simultaneous detection of bacteria, ciliates, and fungi in and on granules.
During the last 20 years, intensive research in the field of biological wastewater treatment and other applications has demonstrated that biofilms are often more efficient for water purification than suspended activated sludge. Today, the application of anaerobic and aerobic granular sludge in wastewater treatment is regarded as one of the most useful and promising biotechnologies. Granular sludge was described first for strictly anaerobic systems (26). In the late 1990s, the formation and application of aerobic granules was reported (30). Such granules are spherical compact aggregates of microorganisms, mainly bacteria, and extracellular polymeric substances (EPS). Granules can be described as “biofilm in suspension” and are considered to be a special case of biofilm formation, composed of self-immobilized cells (9, 16, 20). The application of granules for wastewater treatment shows many advantages. An outstanding feature is the excellent settleability (high settling velocity), which is a prerequisite to handle high liquid flows. Moreover, granular sludge provides high and stable rates of metabolism, resilience to shocks and toxins due to protection by a matrix of EPS (40), long biomass residence times, biomass immobilization inside the aggregates and, therefore, the possibility for bioaugmentation. Bioaugmentation can be regarded as an effective tool in the removal process of xenobiotica from wastewater (6, 41). Metabolic activities in the operating system can be kept at a high level because of the syntrophic associations which occur due to optimum distances between microbial partners at appropriate substrate levels (7). Various investigations of general characteristics of sludge granules, such as size, structure, settling performance, stability against shear forces, EPS content, reactor performance, and metabolism rates, were performed previously (13, 17, 24, 25, 28, 32). Spontaneous aerobic granulation of suspended aggregates is a phenomenon that has been most frequently observed in systems applying the sequencing batch reactor (SBR) concept. Theories about the crucial factors of granule development have been intensively discussed (39). However, in all studies of aerobic granules, the possible role of protozoa and fungi in the biofilm-forming process and the accompanying interaction with bacteria have, so far, been neglected. Several studies on fungi and protozoa in activated sludge systems in general demonstrated that these eukaryotic organisms fulfill a wide variety of important tasks in the biomass conversion and water clarification processes (10, 19, 21, 33, 35). It is known that protozoa or fungi are involved in the formation, structure and function of biofilms for several biofilm systems besides wastewater treatment (22). In preliminary observations of granular biofilms, we found stalked ciliates and, in some cases, fungi to be present in high numbers. The tree-like structure of ciliate colonies and the network of fungal filaments provided a distinctive enlargement of the area available for bacterial colonization. For that reason, this study focuses on the role that ciliates and fungi play in the structural formation of microbial granules derived from activated sludge. The combination of scanning electron microscopy (SEM) and confocal laser scanning microscopy (CLSM) with an adapted technique for fluorescence in situ hybridization (FISH) offers a powerful tool to visualize the detailed architecture and microbial composition of these granules.
MATERIALS AND METHODS
Reactor set-up and operation.
Biomass was enriched in three lab-scale SBRs. Seed sludge was obtained from the municipal wastewater treatment plant in Garching, Germany. The SBRs were operated with different wastewater. SBR1 was fed with particulate-rich malthouse wastewater prepared by mixing barley dust with tap water as described elsewhere (39). SBR2 was operated with raw wastewater from a brewery (Bavarian State Brewery Weihenstephan, Freising, Germany) and SBR3 with synthetic wastewater according to previous recommendations (32). Different wastewater types and operational set-ups of the SBRs were chosen to investigate and to compare possibly different granule structure developments, settling properties, and growth of protozoa. SBR1 and SBR2 had a working volume of 12 liters and were operated with three 8-h cycles per day. Each cycle consisted of 6 h 45 min of aeration, 5 min of settling, 5 min of effluent withdrawal, 1 h of feeding, and 5 min of resting time. Six liters of supernatant was removed, and 6-liter volumes of malthouse and brewery wastewater were fed into SBR1 and SBR2 during each cycle. The pH of the raw wastewater was adjusted to pH 8. The reactors were aerated through air bubble diffusers at a volumetric flow rate of 10 liters/min. The average organic loading rates (CODtotal) were 2.2 kg m−3 day−1 for malthouse and 3.6 kg m−3 day−1 for brewery wastewater. Granular sludge samples (100-ml volumes) were collected from SBR1 and SBR2 and subjected to fixation procedures and microscopic investigations. Samples were taken two to three times per week 45 min prior to the end of the aeration phase. Sampling was carried out from reactor start-up to 2 weeks after granulation. Subsequently, the experiment was terminated. The working volume of SBR3 was 8 liters, with an average CODtotal of 2.4 kg m−3 day−1. SBR3 was operated with four 6-h cycles per day. Each cycle consisted of 5 h 20 min of aeration, 2 min of settling, 7 min of effluent withdrawal, 10 min of feeding, and 25 min of resting time. Four liters of supernatant was removed and 4 liters of synthetic wastewater was fed into SBR3 during each cycle. An airflow rate of 4 liters min−1 was provided during the aerobic stage of SBR3. Since SBR3 was operated as a steady-state reactor over 18 months, a 100-ml volume of sludge was discharged each day to maintain solid retention times and similar amounts of granular sludge. Granular sludge samples of SBR3 were collected two times per week 45 min prior to the end of the aeration phase. The mean pH value in SBR3 was between 7.5 and 8.0.
Sample fixation, preparation, and microbiological analysis.
The complete forming process from an activated sludge floc to a mature granule was investigated and documented by light microscopy and SEM, together with digital imaging. Sludge samples were collected as described above. One part of the samples was immediately analyzed with light microscopy, whereas another aliquot was preserved by fixation for later microscopic examinations. Light microscopy was performed using a stereo microscope (Stemi SV11; Carl Zeiss AG, Oberkochen, Germany) and an inverted microscope equipped with differential interference contrast (Axiovert S100; Carl Zeiss AG) to observe the granular biofilm development and to identify protozoa. Microscopic analyses to identify filamentous bacteria, including Gram and Neisser staining, were performed as described by Eikelboom and van Buijsen (15). Samples for SEM were fixed in 2.5% glutaraldehyde in a 25 mM concentration of pH 7.0 cacodylate buffer. Further SEM sample preparations and microscopic analyses were performed as described previously (23). Samples for subsequent FISH were fixed in 2% paraformaldehyde solution or Bouin's solution (15 volumes of saturated picric acid, 5 volumes of buffered 37% formaldehyde, 1 volume of glacial acetic acid; final concentration, 50%) in accordance with previous recommendations (18). For the differentiation of bacteria involved in the granule formation process, a combination of molecular methods and microscopic techniques was applied. FISH and epifluorescence microscopy were performed to detect bacteria colonizing the ciliate stalks and to explore the microbial distribution patterns within granules. However, microscopic visualization of the core zone of a compact granule after hybridization is not possible due to the thickness (up to 10 mm) of the specimen. Therefore, a FISH protocol (18) was modified. In order to guarantee probe accessibility to the inner parts of thick granules, and for better microscopic analysis, granular biofilms had to be sectioned prior to hybridization, in a manner similar to that used for the detection of bacteria within anaerobic granules (7). The challenge in our study was to create an optimized protocol for the detection and identification of not only bacteria but also protozoa and fungi. Furthermore, it should be possible to perform FISH simultaneously with all of these microorganisms. Fixed samples were stored overnight at 4°C and gently shaken for 2 h to let the fixative infiltrate the biofilm and to preserve the inner parts of the granules. The fixative was then washed out twice in 1× phosphate-buffered saline solution while shaking on ice for 10 min. The washing step was followed by dehydration in 50%, 80%, and 100% ethanol (10 min for each step). Single granules were carefully lifted with a pipette tip, and surplus liquid was very gently removed with a paper tissue. Granules were transferred into a 2-ml cap filled with cryomedium (NEG-50 frozen section medium; Richard Allan Scientific, Kalamazoo, MI), whereby each granule was completely immersed, avoiding the inclusion of air bubbles. This embedment was performed at room temperature for a minimum of 1 h. Afterward, samples were frozen in liquid nitrogen for several minutes and stored at −80°C. Sectioning of granules into 20- to 30-μm-thick slices was performed with a microtome-cryostat (HM 500 OM; Microm, Walldorf, Germany) at −20°C. Sections were placed on precooled (−20°C) gelatin-coated microscope slides and immediately heat fixed in a hybridization oven at 46°C for at least 30 min, until the dried samples were tightly a fixed on the glass slide. To improve the adhesion of the specimen to the microscope slide during FISH, for some granules it was necessary to cover the biofilm section additionally with 0.5 to 1.0% liquid agarose (molecular grade), and subsequently with another microscope slide, in order to obtain a thin plane surface until the agarose solidified. Subsequently, to permeabilize gram-positive bacteria, samples were treated with 4 × 104 U of lysozyme at room temperature for 20 min prior to FISH. The hybridization time was extended to 2 to 3 h in order to optimize the fluorescence signals of the oligonucleotide probes bound to ribosomes within targeted cells. For the hybridization experiments, the fluorescein-labeled oligonucleotide probes EUB338, EUB338-II, and EUB338-III, which are specific for the detection of most bacteria (3, 11), the Cy3-labeled Eukarya-specific oligonucleotide probe EUK516 (3), and the Cy3-labeled My1574, specific for fungi (5), were applied. FISH preparations were visualized with a CLSM (LSM 510; Carl Zeiss AG) as described previously (18). Hybridizations were performed with dozens of granule sections from SBR1 and SBR3.
RESULTS AND DISCUSSION
Modified FISH protocol to determine the microbial composition of heterogeneous granular biofilms.
FISH with small-subunit rRNA-directed, fluorescently labeled oligonucleotide probes is one of the most adequate and popular methods to identify bacterial species and to investigate biofilm composition and development. A modified FISH protocol as described in Materials and Methods was used to detect bacteria colonizing the ciliate stalks and to explore the microbial distribution patterns within granules. Figures 1 and 2 show oligonucleotide probe-related fluorescence signals after hybridization with the Eukarya-specific probe EUK516 for ciliates and fungi and with the Bacteria-specific probes EUB338, EUB338-II, and EUB338-III. With samples from malthouse and synthetic wastewater, all steps of the protocol worked without problems. However, proper sectioning of the bulky brewery granules from SBR2 was not possible, since they contained too many polysaccharides, which act like natural antifreeze agents. They appeared sludgy and wet and could not be sectioned with the cryotome at −20°C. However, the association of bacteria with ciliate stalks could be investigated when ciliate colonies were removed with tweezers from the surface of fixed granules and embedded in polyacrylamide prior to FISH (12).
FIG. 1.
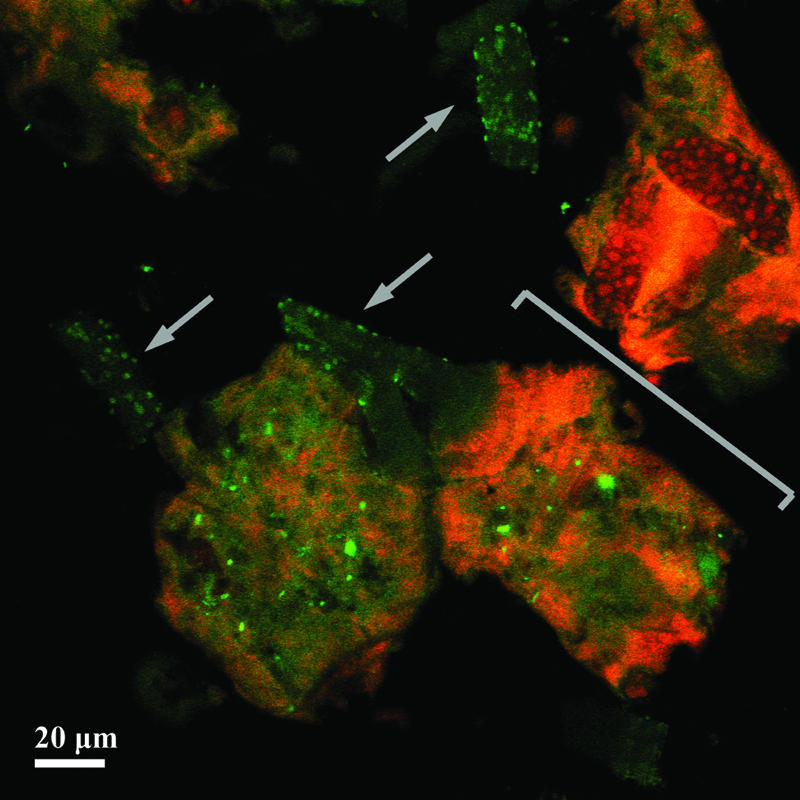
Probe-related signals after FISH applied on a single-granule microsection of 30-μm thickness. The granule was developed in malthouse-derived wastewater. Images are recorded by CLSM after FISH with the Bacteria-specific mix of the EUB338 probes (fluorescein-labeled; green) and the Eukarya-specific probe EUK516 (Cy3-labeled; red). One stack out of 36 stacks of 0.42-μm thickness (each) is shown. Arrows show the branched stalks of a colony of the peritrichous ciliate Epistylis sp. covered by bacteria. The bracket marks the truncated cell bodies (zooids) of the colony.
FIG. 2.
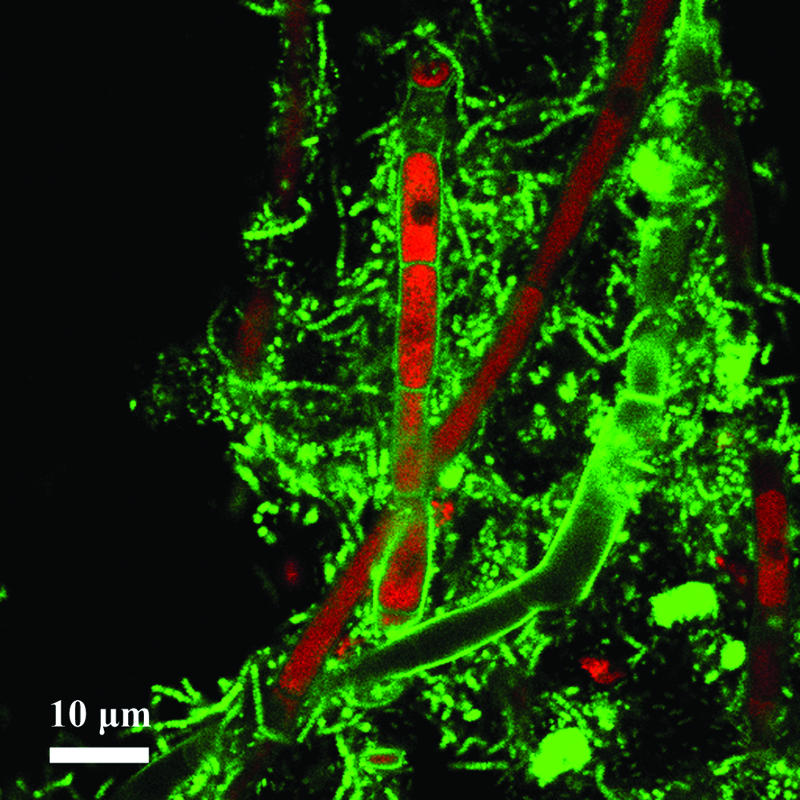
Probe-related signals after FISH applied on a single-granule microsection of 30-μm thickness. The image shows fungal filaments colonized by bacteria in a granule developed in synthetic wastewater. Probe-related signals are recorded by CLSM after FISH with the Bacteria-specific mix of the EUB338 probes (fluorescein-labeled; green) and the Eukarya-specific probe EUK516.
Granule forming process. (i) Interactions of protozoa and bacteria.
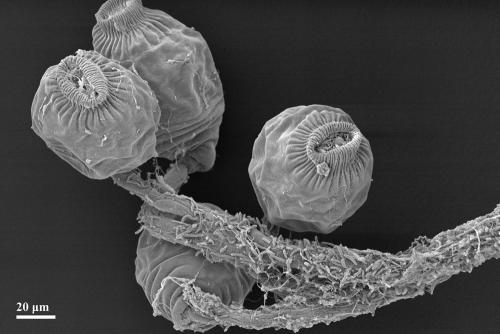
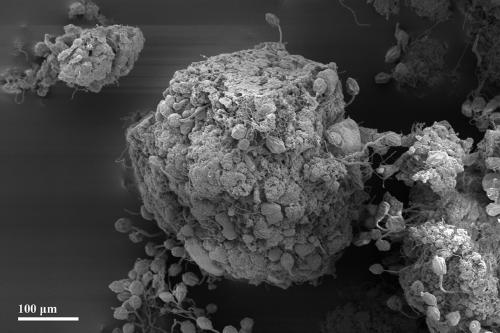
Granules formed within the first 2 weeks of operation. As revealed by microscopic analysis, the granule formation process can be divided into three consecutive phases (Fig. 3). The seed sludge comprised activated sludge flocs composed of bacteria, EPS, and sometimes particles (e.g., wheat glumes as in the case of SBR1, operated with malthouse wastewater). In phase 1, swarming ciliated protozoa of the subclass Peritrichia settled on sludge flocs and built new stalks (Fig. 3A). Subsequently, they started to proliferate and to form large colonies while their stalks were concurrently colonized by bacteria (Fig. 3B and 4). This colonization was additionally enhanced by the cilia beat of the ciliates, which provide a continuous nutrient flux toward the biofilm (19, 22). After a few days, several hundred ciliate cells covered the surface of each floc. Mostly tree-like colony-forming ciliates of the genera Opercularia and Epistylis occurred. During phase 2, flocs condensed and a huge growth of ciliate cells could be observed (Fig. 3C). During the formation of these bulky flocs, a core zone consisting of ciliate stalk remnants and EPS-producing bacteria occurred. The ciliate stalks served hereby as a “backbone” for granule development, since bacteria used them as a substratum to grow. The condensed aggregates were considered to be granule precursors. Subsequently, with the beginning of phase 3, the zooids (cell bodies) of the stalked ciliates were likewise colonized by bacterial cells and embedded in the expanding biofilm. After a while, they were completely overgrown (Fig. 5). Most ciliates died during this process. Some ciliate cells formed swarmers (unstalked free swimming cells) and left the biofilm to escape decay (Fig. 3D). Thus, smooth and compact bacterial granules were formed (Fig. 3E and Fig. 6). However, these mature granules were colonized step by step by the surviving swarming ciliate cells (Fig. 3E), which again formed new stalks and colonies used as a substratum for bacterial growth. Granule size may reach a steady-state size due to abrasion, washout, and floating of granules (7). This could be confirmed for granules of the SBR1 and SBR2 approach. The observations of all SBR approaches documented that peritrichous ciliates were crucially implemented in the granule structure-forming process, since they served as the basis for bacterial biofilm growth. Thus, the question of which specific interactions occur between ciliates and bacteria in granular biofilms arises. It is known that protozoa can excrete growth-stimulating compounds which enhance bacterial activity (34). Furthermore, it was recently reported that eukaryotes and bacteria can systematically interact with each other via small molecules (14). It remains to be investigated if such communication skills arise in the wastewater biofilm community as well and if they can enhance or stimulate granule development.
FIG. 3.
Granule development supported by ciliates. Phase 1, formation of flocs. Ciliates settle on other organisms or particles (A). Bulky growth of ciliates (e.g., Epistylis sp.). Arrow indicates beginning colonization of ciliate stalks by bacteria (B). Phase 2, granule growth and core zone development. Zooids of the ciliates become completely overgrown by bacteria and die. Arrow marks limpid zooids of the ciliate colony. A dense core zone consisting of bacteria and the remains of ciliate stalks is formed in the center of the aggregates. Thereby, the cellular remnants of the ciliates act like a “backbone” (C). Phase 3, “mature” granule. Compact granules have formed. They can serve as a new substratum for swarming ciliate cells (D) settling on the granule surfaces (E).
FIG. 4.
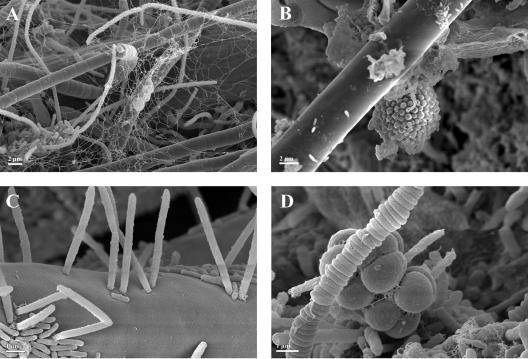
Peritrichous ciliates growing in a tree-like colony. Ciliate stalks serve as backbones in the granule-forming process since bacteria use them as a substratum on which to grow. SEM.
FIG. 5.
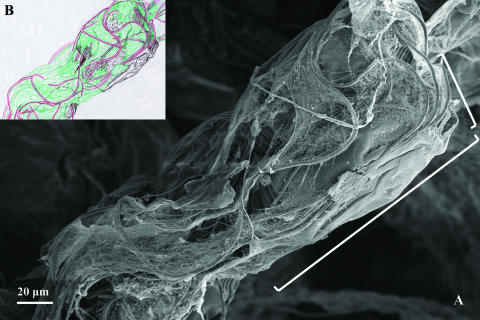
Already-dead ciliates from a SBR operated with wastewater released from a brewery. Zooids (cell bodies) and stalks of two adjacent ciliate cells are completely overgrown by bacterial filaments. SEM. Brackets mark the approximate zooid boundaries of the ciliate cell on the right side (A). The scheme (B) redrawn from panel A emphasizes the boundaries between the microorganisms as ciliate cells are colored in black, covered by bacterial filaments (red) and EPS (green).
FIG. 6.
Mature granule from an SBR operated with synthetic wastewater. The granule is spherical in shape and consists of bacteria and extracellular polymeric substances. It is colonized with many stalked ciliates of the subclass Peritrichia. SEM.
(ii) Role of fungi.
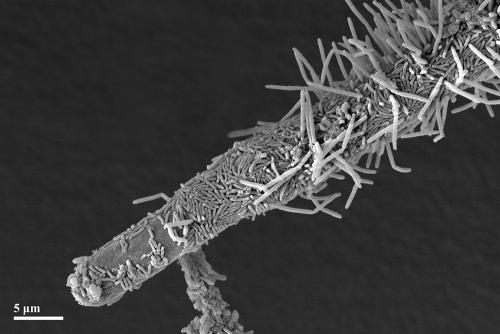
A process similar to the previous one could be observed when fungi were present in the sludge samples. Compared to the ciliate-supported process, granule formation, including fungal filaments as backbones, did not result in lysis of fungi in phase 2 and a recolonization of granule surfaces in phase 3. Instead, spores of fungi germinated once in activated sludge flocs and then grew fast and continuously. Bacteria used the fungal filaments (hyphae) as well as ciliate stalks as a substratum to grow on, but fungi were never completely overgrown. The filament tips were only slightly covered with bacteria. According to a series of SEM images, the extension of this sparsely colonized, actively growing region was 5 to 30 μm long (Fig. 7). Nevertheless, dense core zones and, later, compact granules with several protruding fungal filaments developed. Microscopic analyses of sectioned mature granules revealed that their whole inner part contained remnants of hyphae. Granule formation with the help of fungi appeared first of all in the SBR3 approach with synthetic wastewater. In the SBR2, fed with brewery wastewater, no fungi except yeast cells could be found. In the SBR1, fed with malthouse wastewater, fungal spores occurred as revealed by microscopic observations. Most of them probably do not originate from seed sludge but rather from barley dust. The major portion of these spores did not germinate or build filaments to support granule formation. Only a few fungal filaments grew for some days, but they vanished thereafter. An explanation may be that many ciliates were found in the samples, possibly competing with fungi for nutrients. In the SBR3 approach, only in some cases were ciliates and fungal filaments observed growing together in the same granule. Granules normally comprised a community of either fungi and bacteria or ciliates and bacteria. Previous studies confirmed the possible structure-enhancing role of fungi in the aerobic granule-formation process (8, 17). In these studies, filamentous fungal pellets dominated the sludge population in the first 10 to 15 days and were assumed to initiate the granular biofilm structure, serving as an immobilizing matrix for bacteria. Fungal pellets fell apart later due to cell lysis in the inner part of the pellets. In contrast to these observations, fungi were detected in our study even in mature granules. They supported the development and maintenance of mature biofilms. It is also known that some fungi improve bioconversion of activated sludge (27) and perform nitrification and denitrification at high rates (21). Future studies on the diversity and function of fungi in aerobic granules could therefore yield interesting insights into sludge degradation pathways and possible biotechnological applications.
FIG. 7.
Fungal filaments (hyphae) serving as the basis for bacterial colonization and therefore acting as a skeletal element in the granular structure. The active growing tip region is colonized only sparsely by bacterial cells. SEM.
Structure of mature granules.
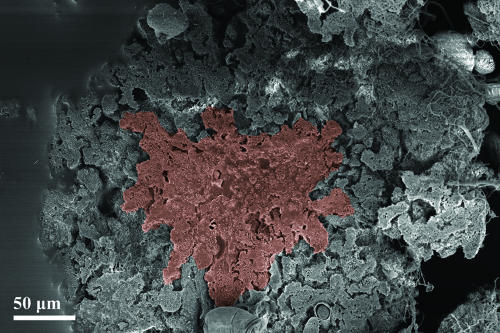
Granules may comprise several microbial layers. This has also been reported in other case studies on anaerobic and aerobic granules (2, 13, 29, 37). Different layers were sometimes composed of distinct bacterial species with various functional tasks, such as nitrification, denitrification, or ammonia oxidation (1, 4, 38). In this study on aerobic granules, different structured zones could be identified as well. SEM analysis documented that fully grown granules always comprised a core and a fringe zone (Fig. 8). The expansions of core and fringe zones differed from granule to granule, depending on the developmental phase and wastewater type. The fringe zone could be identified as a loosely structured layer consisting mainly of bacteria and stalked ciliates or fungal filaments. In the very compact and mature granule, the fringe zone contained only ciliates or fungi growing on granule surfaces. The core zone comprised a dense mixture of bacteria and EPS. However, the rather large amount of EPS allowed bacteria to occur not only in clusters but even as isolate cells within the EPS matrix. Remnants of fungal filaments and ciliate stalks were included in the outer parts of the core zone. Depending on the size of the granules, the inner part of the core zone may contain only dead cell debris, as was reported in another study (29).
FIG. 8.
Cross-section of a mature granule from malthouse wastewater. The dense core zone containing bacteria and possibly dead cell debris is recolored in red. The loose-structured fringe zone (gray) containing bacteria and ciliates encases the core zone. SEM.
Granule formation in brewery wastewater.
The above-mentioned granule-forming process could be observed with all three SBR approaches, but the settling properties of granules in the SBR2 with brewery wastewater differed from those in SBR1 and SBR3. Granules of SBR2 resulted in extremely big and fluffy granules which therefore did not settle properly and caused problems like bulking in the reactor. The SBR2 was started twice, and in both cases, granules quickly developed to the phase 1 and phase 2 stages. Granule precursors of phase 2 contained many ciliate colonies. High substrate concentrations gave rise to large granules, with a diameter of 5 to 6 mm on average. The phenomenon of granule enlargement was also reported for anaerobic granules (20). However, the forming of phase 3 biofilms did not result in fast settling and compact granules, due to very intense growth of bacterial filaments. These filaments dominated the microbial composition of granules and therefore caused bulking. The dominant growth of filamentous bacteria may be considered a stress reaction of occasional oxygen deficiency due to the sometimes very high temperatures during the summer (35°C instead of the usual 20°C). Similar observations were reported previously (31). The appearance of filamentous organisms in large amounts can depend also on the substrate composition or the organic loading rate. Energy-rich substrates are known to support the proliferation of filamentous bacteria in activated sludge (32). The CODtotal loading rate in SBR2 was higher (3.6 kg m−3 day−1) than in SBR 1 (2.2 kg m−3 day−1) and SBR3 (2.4 kg m−3 day−1).
Bacterial composition of granules.
Bacteria were found to be the main microbial components of the investigated granular biofilms. They occurred in a large morphological variety. Rods and cocci as well as spirilli and tetrad-arranged organisms could be found in the same granule (Fig. 9). They were embedded in a thick EPS matrix as single cells or dense clusters. Cocci often occurred in somewhat spherical clusters of several dozen cells (Fig. 9B). Colonization of fungal filaments and ciliate stalks was observed mainly for rod-shaped bacteria (Fig. 7 and 9C). Furthermore, large quantities of bacteria could be detected with SEM and FISH analysis in the digestion vacuoles of ciliates. The composition of the bacterial community and the distribution of distinct morphotypes differed depending on the wastewater type and operational set-up of the SBRs. In SBR2, mainly filamentous bacteria occurred (Fig. 9A). Gram and Neisser stains and FISH analyses showed that most of the filaments belonged to the genus Thiothrix or to Sphaerotilus natans. Synthetic wastewater granules were composed mainly of cocci, tetrad-arranged organisms (Fig. 9D), and a huge amount of EPS. EPS are generally important for granule stability. On the one hand, they cause hydration of the granule surfaces, and on the other hand, they enhance shear force stability of activated sludge flocs and granules (36). We assume that EPS are produced not only by bacteria but also by other organisms present in the observed biofilm community—in this case, especially by ciliates and fungi.
FIG. 9.
Various bacterial morphotypes observed with the three reactors. Different filamentous forms (A), coccoid colony on a ciliate stalk (B), different rod shaped bacteria on a fungal hypha (C), and tetrad-arranged bacteria (D). SEM.
Conclusions.
In the present study, the structural and main microbial composition of aerobic activated sludge granules originating from three differently operated SBRs was described. The combination of SEM, light microscopy, and CLSM, together with a modified FISH protocol, provides a powerful tool to explore the structure of microbial granules and may be used for other biofilm formations, too. With this newly established protocol, FISH, applied to microsections of microbial granules, can now be used to monitor the development of such granules, because bacteria, ciliates, and fungi can be detected simultaneously. This is a great advantage, since eukaryotic organisms play a crucial role in the formation of granules. Stalked ciliates of the subclass Peritrichia were always involved in the process of granule development, whereas fungi were found only in some cases. It was shown that the development from the sludge floc to the mature granule takes place in three phases. The process starts with a sludge floc which proliferates to a granule precursor and results in the formation of a compact, mature granule. Ciliates and fungi serve as the main substratum in the formation of bacterial biofilms and, therefore, act as a backbone for the granules. The compactness, size, and microbial composition of granules depended on the wastewater type and the operational setup of the SBR. Many different bacterial morphotypes and large quantities of EPS could be found in the examined biofilms. The results of our study indicate that other organisms, such as ciliates and fungi, may also be involved in the formation of EPS. This finding opens a new field in biofilm research, because current studies on EPS (29) concentrate on the formation of EPS by bacteria.
Acknowledgments
This work was supported by German Research Foundation (DFG) projects LU 421/3-2 and LU 421/3-3, the University of Innsbruck, and the “Verein zur Förderung der wissenschaftlichen Ausbildung und Tätigkeit von Südtirolern an der Landesuniversität Innsbruck.”
We thank Norbert Schwarzenbeck and Ewelina Zima for providing samples and for helpful technical support with the SBRs; Gerhard Wanner for support with scanning electron microscopy; Dorothea Begert, Andreas Hofmann, Martina Dörner, Silvia Dobler, and Susanne Cornfine for excellent technical assistance; and Hilde Lemmer for helpful discussions.
Footnotes
Published ahead of print on 17 August 2007.
REFERENCES
- 1.Abreu, A. A., J. C. Costa, P. Araya-Kroff, E. C. Ferreira, and M. M. Alves. 2007. Quantitative image analysis as a diagnostic tool for identifying structural changes during a revival process of anaerobic granular sludge. Water Res. 41:1473-1480. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J., T. Daidou, S. Tsuneda, and A. Hirata. 2002. Characterization of denitrifying phosphate-accumulating organisms cultivated under different electron acceptor conditions using polymerase chain reaction-denaturing gradient gel electrophoresis assay. Water Res. 36:403-412. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoi, Y., S. Tsuneda, and A. Hirata. 2004. Transition of bacterial spatial organization in a biofilm monitored by FISH and subsequent image analysis. Water Sci. Technol. 49:365-370. [PubMed] [Google Scholar]
- 5.Baschien, C. 2003. Development and evaluation of rRNA targeted in situ probes and phylogenetic relationships of freshwater fungi. Ph.D. thesis. Technical University of Berlin, Berlin, Germany.
- 6.Bathe, S., T. V. K. Mohan, S. Wuertz, and M. Hausner. 2004. Bioaugmentation of a sequencing batch biofilm reactor by horizontal gene transfer. Water Sci. Technol. 49:337-343. [PubMed] [Google Scholar]
- 7.Batstone, D. J., J. Keller, and L. L. Blackall. 2004. The influence of substrate kinetics on the microbial community structure in granular anaerobic biomass. Water Res. 38:1390-1404. [DOI] [PubMed] [Google Scholar]
- 8.Beun, J. J., A. Hendriks, M. C. M. van Loosdrecht, E. Morgenroth, P. A. Wilderer, and J. J. Heijnen. 1999. Aerobic granulation in a sequencing batch reactor. Water Res. 33:2283-2290. [DOI] [PubMed] [Google Scholar]
- 9.Beun, J. J., M. C. M. van Loosdrecht, and J. J. Heijnen. 2002. Aerobic granulation in a sequencing batch airlift reactor. Water Res. 36:702-712. [DOI] [PubMed] [Google Scholar]
- 10.Curds, C. R., A. Cockburn, and J. M. Vandyke. 1968. An experimental study of the role of the ciliated protozoa in the activated sludge process. Water Pollut. Control 1968:312-329. [Google Scholar]
- 11.Daims, H., A. Brühl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 12.Daims, H., S. Luecker, and M. Wagner. 2006. Daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8:200-213. [DOI] [PubMed] [Google Scholar]
- 13.de Kreuk, M. K., J. J. Heijnen, and M. C. van Loosdrecht. 2005. Simultaneous COD, nitrogen, and phosphate removal by aerobic granular sludge. Biotechnol. Bioeng. 90:761-769. [DOI] [PubMed] [Google Scholar]
- 14.Dudler, R., and L. Eberl. 2006. Interactions between bacteria and eukaryotes via small molecules. Curr. Opin. Biotechnol. 17:268-273. [DOI] [PubMed] [Google Scholar]
- 15.Eikelboom, D. H., and H. J. J. van Buijsen. 1999. Handbuch für die mikroskopische Schlamm-Untersuchung, 4th ed. Hirthammer Verlag, München, Germany.
- 16.El-Mamouni, R., R. Leduc, J. W. Costerton, and S. R. Guiot. 1995. Influence of the microbial content of different precursory nuclei on anaerobic granulation dynamics. Water Sci. Technol. 32:173-177. [Google Scholar]
- 17.Etterer, T., and P. A. Wilderer. 2001. Generation and properties of aerobic granular sludge. Water Sci. Technol. 43:19-26. [PubMed] [Google Scholar]
- 18.Fried, J., W. Ludwig, R. Psenner, and K. H. Schleifer. 2002. Improvement of ciliate identification and quantification: a new protocol for fluorescence-in-situ-hybridization (FISH) in combination with silver stain techniques. Syst. Appl. Microbiol. 25:555-571. [DOI] [PubMed] [Google Scholar]
- 19.Fried, J., and H. Lemmer. 2003. On the dynamics and function of ciliates in sequencing batch biofilm reactors. Water Sci. Technol. 47:189-196. [PubMed] [Google Scholar]
- 20.Grotenhuis, J. T. C., M. Smit, A. A. M. Van Lammeren, A. J. M. Stams, and A. J. B. Zehnder. 1991. Localization and quantification of extracellular polymers in methanogenic granular sludge. Appl. Microbiol. Biotechnol. 36:115-119. [Google Scholar]
- 21.Guest, R. K., and D. W. Smith. 2002. A potential new role for fungi in a wastewater MBR biological nitrogen reduction system. J. Environ. Eng. Sci. 1:433-437. [Google Scholar]
- 22.Hartmann, C., Ö. Özmutlu, H. Petermeier, J. Fried, and A. Delgado. 2007. Analysis of the flow field induced by the sessile peritrichous ciliate Opercularia asymmetrica. J. Biomech. 40:137-148. [DOI] [PubMed] [Google Scholar]
- 23.Huber, R., W. Eder, S. Heldwein, G. Wanner, H. Huber, R. Rachel, and K. O. Stetter. 1998. Thermocrinis ruber gen. nov., sp. nov., a pink-filament-forming hyperthermophilic bacterium isolated from Yellowstone National Park. Appl. Environ. Microbiol. 64:3576-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hulshoff Pol, L. W., S. I. de Castro Lopes, G. Lettinga, and P. N. L. Lens. 2004. Anaerobic sludge granulation. Water Res. 38:1376-1389. [DOI] [PubMed] [Google Scholar]
- 25.Inizan, M., A. Freval, J. Cigana, and J. Meinhold. 2005. Aerobic granulation in a sequencing batch reactor (SBR) for industrial wastewater treatment. Water Sci. Technol. 52:335-343. [PubMed] [Google Scholar]
- 26.Lettinga, G., L. W. Hulshoff Pol, I. W. Koster, W. M. Wiegant, W. J. de Zeeuw, A. Rinzema, D. C. Grin, R. E. Roersma, and S. W. Hobma. 1984. High-rate anaerobic wastewater treatment using the UASB reactor under a wide range of temperature conditions. Biotechnol. Genet. Eng. Rev. 2:253-284. [Google Scholar]
- 27.Mannan, S., A. Fakhru'l-Raui, and M. Z. Alam. 2005. Use of fungi to improve bioconversion of activated sludge. Water Res. 39:2935-2943. [DOI] [PubMed] [Google Scholar]
- 28.McSwain, B. S., R. L. Irvine, and P. A. Wilderer. 2004. The effect of intermittent feeding on aerobic granule structure. Water Sci. Technol. 49:19-25. [PubMed] [Google Scholar]
- 29.McSwain, B. S., R. L. Irvine, M. Hausner, and P. A. Wilderer. 2005. Composition and distribution of extracellular polymeric substances in aerobic flocs and granular sludge. Appl. Environ. Microbiol. 71:1051-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgenroth, E., T. Sherden, M. C. M. van Loosdrecht, J. J. Heijnen, and P. A. Wilderer. 1997. Aerobic granular sludge in a sequencing batch reactor. Water Res. 31:3191-3194. [Google Scholar]
- 31.Mosquera-Corral, A., M. K. de Kreuk, J. J. Heijnen, and M. C. van Loosdrecht. 2005. Effects of oxygen concentration on N-removal in an aerobic granular sludge reactor. Water Res. 39:2676-2686. [DOI] [PubMed] [Google Scholar]
- 32.Moy, B. Y. P., J.-H. Tay, S.-K. Toh, Y. Liu, and S. T. L. Tay. 2002. High organic loading influences the physical characteristics of aerobic sludge granules. Lett. Appl. Microbiol. 34:407-412. [DOI] [PubMed] [Google Scholar]
- 33.Nicolau, A., N. Dias, M. Mota, and N. Lima. 2001. Trends in the use of protozoa in the assessment of wastewater treatment. Res. Microbiol. 152:621-630. [DOI] [PubMed] [Google Scholar]
- 34.Ratsak, C. H., B. W. Kooi, and H. W. van Verseveld. 1994. Biomass reduction and mineralization increase due to the ciliate Tetrahymena pyriformis grazing on the bacterium Pseudomonas fluorescens. Water Sci. Technol. 29:119-128. [Google Scholar]
- 35.Roessink, R., and D. H. Eikelboom. 1997. Characterization of suspended solids in/out airlift biofilm reactors. Water Sci. Technol. 36:237-245. [Google Scholar]
- 36.Sheng, G. P., H. Q. Yu, and X. Y. Li. 2006. Stability of sludge flocs under shear conditions: roles of extracellular polymeric substances (EPS). Biotechnol. Bioeng. 93:1095-1102. [DOI] [PubMed] [Google Scholar]
- 37.Tay, J.-H., V. Ivanov, S. Pan, and S. T. L. Tay. 2002. Specific layers in aerobically grown microbial granules. Lett. Appl. Microbiol. 34:254-257. [DOI] [PubMed] [Google Scholar]
- 38.Tsuneda, S., Y. Ejiri, T. Nagano, and A. Hirata. 2004. Formation mechanism of nitrifying granules observed in an aerobic upflow fluidized bed (AUFB) reactor. Water Sci. Technol. 49:27-34. [PubMed] [Google Scholar]
- 39.Wilderer, P. A., and B. S. McSwain. 2004. The SBR and its biofilm application potentials. Water Sci. Technol. 50:1-10. [PubMed] [Google Scholar]
- 40.Wingender, J., T. R. Neu, and H.-C. Flemming. 1999. What are bacterial extracellular polymeric substances?, p. 1-20. In J. Wingender, T. R. Neu, and H.-C. Flemming (ed.), Microbial extracellular polymeric substances: characterization, structure, and function. Springer, Berlin, Germany.
- 41.Wuertz, S., S. Okabe, and M. Hausner. 2004. Microbial communities and their interactions in biofilm systems: an overview. Water Sci. Technol. 49:327-336. [PubMed] [Google Scholar]