Abstract
The ability of two closely related species to maintain species boundaries in spite of retained interfertility between them is a documented driving force of speciation. Experimental evidence to support possible interspecific postzygotic isolation mechanisms for organisms belonging to the kingdom Fungi is still missing. Here we report on the outcome of a series of controlled comparative inoculation experiments of parental wild genotypes and F1 hybrid genotypes between closely related and interfertile taxa within the Heterobasidion annosum fungal species complex. Results indicated that these fungal hybrids are not genetically unfit but can fare as well as parental genotypes when inoculated on substrates favorable to both parents. However, when placed in substrates favoring one of the parents, hybrids are less competitive than the parental genotypes specialized on that substrate. Furthermore, in some but not all fungus × plant combinations, a clear asymmetry in fitness was observed between hybrids carrying identical nuclear genomes but different cytoplasms. This work provides some of the first experimental evidence of ecologically driven postzygotic reinforcement of isolation between closely related fungal species characterized by marked host specificity. Host specialization is one of the most striking traits of a large number of symbiotic and parasitic fungi; thus, we suggest the ecological mechanism proven here to reinforce isolation among Heterobasidion spp. may be generally valid for host-specialized fungi. The validity of this generalization is supported by the low number of known fungal hybrids and by their distinctive feature of being found in substrates different from those colonized by parental species.
Although the number of reports on hybrids of closely related species of fungi or oomycetes appears to be on the rise (5, 40), our understanding of the mechanisms regulating fungal hybridization is still relatively limited (26). The small number of known natural hybrids has hampered our ability to either experimentally test or at least describe which phenotypic and genotypic traits may be associated with fungal hybrids. Recently described hybrids (3, 7, 8, 17, 32, 43) and the discovery of horizontal gene transfers between closely related species (6, 9, 15, 25) are suggestive of the fact that hybridization, although not frequently seen in the fungal kingdom, is probably a relatively common event, sometimes masked either by our inability to discern extremely subtle differences among hybrid and parental species (37) or by a potentially transient nature of hybrid individuals (7). One common feature of all recently described hybrid fungi and oomycetes is their recovery from a substrate different from those on which the parents are normally found. Thus, it appears that in known cases of fungal and oomycete hybrids, successful hybrid development may be under ecological constraint and may occur only when hybrids avoid competition with parents by shifting their habitat. Although prezygotic (e.g., total intersterility between species) and intrinsic postzygotic (e.g., aneuploidy and sterility) isolation mechanisms are bound to limit hybridization events in general, more research needs to address the issue of postzygotic ecological limitations that hybrids may face. In particular, there are no data differentiating between hybrids with intrinsic genetic limitations and hybrids characterized by extrinsic ecological limitations (26).
A general consensus on the role played by hybridization in the evolution of plant and animal complexes has not been reached yet. While it is evident that unviable or completely infertile hybrids cannot contribute to the founding of new evolutionary lineages, it is becoming increasingly clear that plant and animal hybrids vary in their fitness with regard to their parents (1, 10, 29, 38). Hybrid fitness in fact has been reported to be lower than, equivalent to, or higher than that of parents, depending on the hybrid species being considered. In some instances hybrid superiority has been associated with certain habitats generally unoccupied by parental species (30); in other cases, hybrids more closely resembling one of the parents were capable of competing with that parent in its own habitat (14). From an evolutionary perspective, it is interesting to note that even apparently unfit low-fertility hybrids have been reported to have lead to the creation of hybrid species (39). It is clear that distinguishing between intrinsic unviability and ecologically driven constraints on fungal hybrids has significant implications in understanding the potential role played by the hybridization process in this kingdom and to predict the likelihood fungal hybrids may be frequent but undetected. Evidence supporting genetic viability of fungal hybrids would implicitly indicate hybridization plays a role yet unsuspected for this kingdom.
This study investigates the hybrid fitness of crosses between two pairs of closely related species within the complex Heterobasidion annosum (Fr.) Bref. sensu lato: namely, the European Heterobasidion abietinum Niemelä & Korhonen, Heterobasidion parviporum Niemelä & Korhonen, and H. annosum P and S intersterility groups (ISGs) in North America. Species within the complex are differentiated based on slight morphological differences, partial mating incompatibility, differential host specificity, and abundant genetic data. It should be noted that although the two North American taxa are still referred to as “ISGs” and are awaiting formal species description, there is no doubt they represent different species. Significant intersterility between them, differential host association, and all genetic evidence published so far indicate that the two are relatively distant taxa within the species complex (2, 16, 19, 23, 24, 31, 35, 44).
Each of the partially intersterile taxa within the Heterobasidion annosum species complex is characterized by a distinct host specialization (27). All species are parasites and mostly are known to cause root diseases of conifers. These fungi infect their hosts by means of airborne haploid meiospores, normally through freshly cut stumps or wounds, and are capable of infecting adjacent trees by colonizing grafted roots that act as bridges of contagion among individual trees in the same stand (27). However, all Heterobasidion species may survive saprotrophically on dead wood: host specificity appears to be not relevant in the case of saprotrophic growth (28, 36). Because genotypes of these fungi are able to fruit and thus complete their sexual cycle only when a substantial amount of woody substrate has been colonized and/or when the host is dead, virulence, a trait associated with more-extensive pathogenic growth (46), can be taken as a good indicator for fitness. Furthermore, the faster in planta growth rate of virulent isolates leads to more-frequent secondary infection of adjacent hosts, thus largely expanding the food base and the life span of the individual genotype.
Intersterility among taxa is controlled by five loci called intersterility (IS) genes (13). For mating to occur, not only must mating alleles be different but at least two + alleles must be found at one of five IS loci. While alternate allele fixation has evolved at two of the loci, the presence of rare + alleles at one of the remaining three loci allows for illegitimate interspecific mating. In western North America, up to 20% of interspecific crosses can be fertile in laboratory tests (17, 22). Despite the leaky intersterility among species and the current presence of common habitats in which two taxa could come into contact with one another, interspecific hybrids have rarely been reported (17, 41) and only a single long-lived hybrid genotype has been found in nature, in California (17).
The purpose of this study was to understand why interspecific Heterobasidion hybrids are rarely found in nature in spite of abundant sympatry of distinct taxa by testing their fitness through inoculation studies. Parental strains and F1 hybrids were reciprocally inoculated on hosts preferred by each of the parental species and also on “new” hosts or substrates known to harbor both parental species. Our working hypothesis was that hybrids are rarely found because they are at a disadvantage compared to their parental species on the hosts on which each parent is specialized but that hybrid fitness may be comparable to that of their parents (or higher) on habitats that are not those on which the parents are specialized. This hypothesis suggests hybridization among species may be under postzygotic ecological constraint, explains the rarity of hybrids, and predicts increased hybridization if the ecological constraints are removed (e.g., by increasing the availability of novel habitats amenable to different taxa). Alternatively, the fitness of hybrids may be reduced because of intrinsic genetic problems, potentially due to the genomic divergence between parental species.
Because F1 hybrids contain each of the nuclear genomes of the parents and one of the two uniparentally inherited mitochondrial genomes (cytoplasms), several outcomes may be expected in terms of hybrid phenotype: (i) hybrid fitness may be extremely low; F1 hybrids are not competitive on most or all substrates tested, suggesting intrinsic poor viability of hybrids; (ii) F1 hybrids may match the fitness of both parents on both respective specialized substrates; hybrids have the host range of both parents, combined; (iii) F1 hybrids may be as competitive as the parent providing the cytoplasm; (iv) hybrids are less competitive than either parent on substrates already exploited by parental genotypes but can be viable on neutral or novel substrates. It should be noted that not all outcomes are mutually exclusive: for instance, both outcomes iii and iv can be true.
MATERIALS AND METHODS
Inoculation studies. (i) Experiment one.
In experiment one, the only known natural hybrid between the North American S and P ISGs of H. annosum (i.e., SP hybrid), genotype AWR 400 (Mycoteca Universitatis Taurinensis [MUT] accession no. 4001) (17), and two wild heterokaryotic genotypes (09Az200 and AWR 282), each representing one of the two parental taxa (S and P ISGs, respectively), were inoculated on ponderosa pine (Pinus ponderosa P. & C. Lawson) (P ISG host) and Sitka spruce (Picea sitchensis [Bong.] Carr.) (universal host) seedlings. The hybrid nature of AWR 400 was discovered using a taxon-specific competitive-priming PCR technique and confirmed by heterozygosity using both a variety of isozyme markers and DNA restriction fragment length polymorphisms (17). All three genotypes came from northeastern California and had been found on several trees. The fact that selected fungal genotypes were found on several adjacent trees was taken as an indication of their significant natural fitness.
Pine wood wedges of about 1.5 by 1 by 0.5 cm were autoclaved twice for 50 min in malt extract broth (20 g malt extract, 1 liter distilled water) before being placed partially submerged in petri dishes (20× dish). Each plate was inoculated by placing a plug of mycelium in the center of the plate. Plates were incubated in the dark at 20°C for 6 weeks to allow colonization of the wedges.
Seedlings were kept in the nursery bed for 2 years and then transplanted in seedling cartridges and kept in the greenhouse for 2 years. Seedlings came from the same nursery and were all transplanted in a University of California at Berkeley greenhouse several months prior to the experiment. Each seedling was inoculated by wounding the bark, the cambium, and the outer xylem with a sterile scalpel and by inserting fungus-colonized wedges. Sterile wedges were used to mock inoculate control trees. Parafilm was used to tighten and protect the inoculations, and seedlings were kept in the greenhouse at the University of California at Berkeley for 6 months.
Seedlings were organized in five blocks (replications). For each fungal genotype and for controls, 12 pines and 20 spruces per block were inoculated according to a random design. A total of 240 pines and 400 spruces were thus employed in this study.
Cumulative mortality of seedlings was recorded by inspecting the greenhouse on a weekly basis. Seedlings dead within 2 weeks from inoculation were excluded from the analysis, since such deaths were not likely to have been directly caused by the pathogen. To ensure that the recorded mortality was caused by the inoculated fungal strains, stems of dead seedlings were placed on a moistened filter paper disk in a petri dish. After an incubation of 10 to 15 days at room temperature, stems were examined with the aid of a stereoscope for the presence of colonies of H. annosum.
In the same experiment, each of the three fungal genotypes was also inoculated in 240 white fir (Abies concolor [Gord. & Glend.] Lindl. ex Hildebr.) seedlings (S ISG host). However, due to the low levels of mortality, no differences between the three isolates were observed, and results are not discussed.
(ii) Experiment two.
In experiment two, the three strains above were inoculated in the xylem of primary roots in a natural stand of white fir at Blodgett Forest, Sierra Nevada, CA (38°53′42″N, 120°37′59"W). A total of 60 roots were inoculated in two experimental blocks of equal size. The inoculum was prepared as described above, with 2-cm-long, 4-mm-diameter dowels instead of wedges. Roots were excavated, and a 5-mm-diameter hole was drilled at 1 m from the root collar, the dowel was placed inside the hole, and grafting wax was applied to seal the hole. Each root was also mock inoculated with a sterile wood dowel. Inoculations occurred in the summer of 1998, and the experiment lasted 1 year. At the end of the trial, approximately 2 m of each root was excavated, taken to a workshop, and sectioned in 5-cm disks. Disks were sealed in plastic bags to ensure 100% relative humidity and incubated at room temperature for 10 to 15 days. At that time all disks were examined with the aid of a stereoscope for the presence of colonies of H. annosum. Because each disk represents a 5-cm increment in the linear growth of the pathogen, if colonies were found on both sides of the disk, that accounted for the entire 5 cm; if colonies were found on only one side of the disk, that disk would account for a 2.5-cm increment in the growth of the pathogen.
(iii) Experiment three.
In experiment three, two hybrids were obtained in the laboratory by mating isolate J79r of H. abietinum (MUT accession no. 3989) and isolate 430.1 (MUT accession no. 3988) of H. parviporum. Inocula of the two isolates were placed about 1 cm apart in the middle of a petri dish filled with 1.5% malt extract agar. After 3 weeks, a small piece of mycelium was taken from the side of each of the two isolates and transferred into a new petri dish, resulting in hybrids with identical nuclear genomes but one with a mitochondrial genome of H. abietinum and the other with a mitochondrial genome of H. parviporum. The hybrid nature of the isolates was revealed by the presence of clamp connections in the mycelia, and mitochondrial characterization was done using a taxon-specific competitive-priming PCR technique as previously described (21). Inoculum and inoculation techniques were as described for experiment one.
Three-year-old seedlings of silver fir (Abies alba Mill.), the host on which H. abietinum is specialized, and Norway spruce (Picea abies [L.] Karst.), the host on which H. parviporum is specialized, were inoculated with the two hybrids and the two parental genotypes. Seedlings were organized in four blocks, and for each fungal genotype and for the negative control, 20 silver firs and 20 Norway spruces were inoculated according to a random design. A total of 400 silver fir trees and 400 Norway spruce trees were thus employed in this experiment.
Genotypes were represented as follows: H. parviporum, Pa; H. abietinum, A; H. parviporum × H. abietinum hybrid, carrying H. parviporum mitochondrion, (PaxA)p; H. parviporum × H. abietinum hybrid, carrying H. abietinum mitochondrion, (PaxA)a.
Six months after inoculation, the experiment was ended. The low mortality of white fir seedlings observed in experiment one indicated that host species respond differently to infection by this pathogen and that disease in some plant species does not necessarily lead to mortality of inoculated seedlings. Based on these observations, we decided to measure the extent of fungal colonization in inoculated seedlings besides tallying dead plants. Measurements of growth were obtained by sectioning the stems of the seedlings in 3-mm sections and placing them in concentric order on a moist disk of filter paper in a petri dish. After incubation for 15 days at room temperature, disks were examined for absence or presence of colonies of H. annosum.
Statistical analyses.
Blocks appeared to have no effect in any of the experiments, and data were thus pooled together before analysis. Arcsine and log transformation of data was accomplished for mortality rates in experiment one and for colonization values in experiment three, respectively, to normalize the distribution of the residuals and to correct for nonhomogeneity of variances (Kolmogorov-Smirnov test, P > 0.05; Levene test, P > 0.05).
One-way analysis of variance (ANOVA) was performed either by using mortality rates for each block as replicates (experiments one and three) or by comparing values of root or seedling colonization in experiments two and three. Analysis of each tree species was performed independently, since comparison of susceptibilities among host species was not of interest for this study. In experiment three, values from parental genotypes were compared both to values from the two hybrid genotypes and to values from hybrids pooled together. The Student-Newman-Keuls (SNK) test was used for posthoc comparison of means.
A P value of 0.05 was assumed as the significant cutoff in all experiments.
RESULTS
Experiment one.
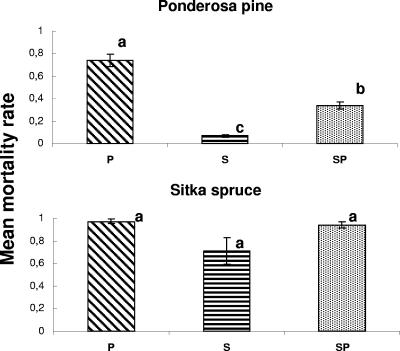
No control seedlings died; hence, negative controls were excluded from the analysis. Conversely, significant mortality was observed among seedlings inoculated with Heterobasidion genotypes. A total of 63 (35%) pines and 262 (87%) spruce seedlings were cumulatively tallied as dead by the end of the experiment. Upon incubation in a moist chamber, H. annosum conidiophores (asexual fruiting structures) could be seen on the xylem of all dead seedlings using a ×30 magnification dissecting scope, confirming death incurred because of stem colonization by the pathogen. In the ponderosa pine trial, the P ISG genotype caused the highest rate of mortality while the S genotype was responsible for the lowest rate of mortality. There were significant differences in mortality levels caused by the three genotypes (df = 2; F = 77.863; P < 0.001). The SNK test indicated that all three genotypes were different and placed the hybrid individual in an intermediate position (Fig. 1).
FIG. 1.
Experiment one: mean mortality rate of seedlings per block (± standard error) for tree species and Heterobasidion genotype. P, S, and SP refer to the North American P ISG, S ISG, and SP hybrid genotypes, respectively. Shared letters indicate nonsignificant differences between Heterobasidion genotypes within each of the two inoculation trials, as determined by separate ANOVAs.
In the Sitka spruce trial, the P and SP hybrid genotypes killed, respectively, an average of 97% and 94% of seedlings. The S genotype on average killed 71% of seedlings. Mortality rates were not significantly different (df = 2; F = 3.246; P = 0.075).
Only 19 (8%) white fir seedlings died: because of the low mortality attained, no significant differences were detected among genotypes.
Experiment two.
The three fungal genotypes S, P, and SP hybrid AWR 400 significantly colonized the xylem of white fir roots at Blodgett Research Forest. No fungal colonization was found associated with mock inoculations. The S genotype colonized on average 35 cm (standard deviation = 6) of root xylem, the P genotype colonized an average 30 cm (standard deviation = 3), and the hybrid genotype colonized on average 35 cm (standard deviation = 3) of root xylem. There was no statistical difference in the extent of colonization among the three genotypes.
Experiment three.
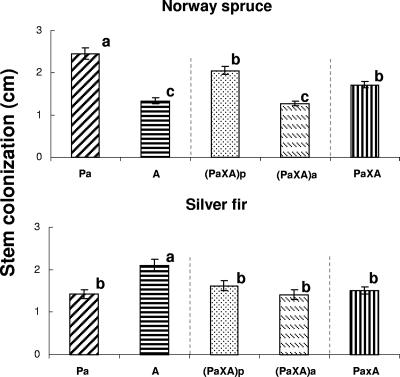
In the Norway spruce trial (H. parviporum host), colonization of stems of seedlings was greatest for the H. parviporum haploid parent, followed by the (PaxA)p genotype, the H. abietinum haploid parent, and the (PaxA)a hybrid. ANOVA detected significant differences in the extent of colonization among the isolates (df = 3; F = 32.423; P < 0.001), and the SNK test grouped the isolates in descending order as follows: greatest colonization by Pa, followed by (PaxA)p, followed by the (PaxA)a and A genotypes (Fig. 2). There were significant differences in the colonization extent among the parental genotypes and hybrids pooled together (df = 2; F = 26.300; P < 0.001). The SNK test placed the hybrid individual in an intermediate position.
FIG. 2.
Experiment three: mean extent of colonization of stems (± standard error), expressed in cm, for tree species and Heterobasidion genotype. Pa, A, (PaxA)p, and (PaxA)a refer to the following genotypes, respectively: H. parviporum, H. abietinum, H. parviporum × H. abietinum carrying H. parviporum mitochondrion, and H. parviporum × H. abietinum carrying H. abietinum mitochondrion. PaxA indicates the two hybrid genotypes combined. Shared letters indicate nonsignificant differences between Heterobasidion genotypes within each of the two inoculation trials, as determined by separate ANOVAs.
In the silver fir trial (H. abietinum-host), colonization of stems of seedlings was largest for the H. abietinum haploid parent, followed by the two hybrids and by the H. parviporum haploid parent. ANOVA detected significant differences in the extent of colonization among the isolates (df = 3; F = 6.995; P < 0.001), and only growth of the H. abietinum parent was statistically different from values recorded for the other isolates. The colonization extent differed significantly among the parental genotypes and hybrids pooled together (df = 2; F = 9.097; P < 0.001), and the SNK test grouped the isolates as follows: greatest colonization by the A genotype, followed by (PaxA), whose colonization was equivalent to that of Pa.
No mortality was detected in the silver fir trial, while only the H. parviporum parent caused a mean mortality of 5% plant per block in the Norway spruce trial.
DISCUSSION
The differential levels of pathogenicity among Heterobasidion species on a series of hosts including ponderosa pine, silver fir, and Norway spruce is well documented by both field surveys and experimental trials (12, 17, 33, 36, 45, 46). Each of the three tree species above is known to preferentially harbor host-specialized pathogen species. Our experimental assays were sensitive enough to differentiate virulence levels of host-specialized versus non-host-specialized species both for North American and European host-pathogen combinations. For all host species, the host-specialized pathogen species was significantly more virulent than the other species concomitantly tested, and this was true for the North American P ISG genotype on ponderosa pine, for H. parviporum on Norway spruce, and for H. abietium on silver fir. On the contrary, no differences among fungal species were observed on substrates known to be neutral (i.e., capable of harboring all Heterobasidion species), such as Sitka spruce in experiment one.
The levels of virulence measured in each the assay for parental genotypes matched those previously reported (12, 33, 34, 45, 46), thus indicating that the individuals selected for this study can be regarded as representative of each fungal species. Furthermore, virulence levels recorded in the trials for the parental species matched the expected values in all eight cases (two North American species on two substrates and two European species on two substrates). The failure of the white fir inoculation is not unprecedented; in a previous paper by Worrall et al. (46), white fir seedlings were inoculated in two separate experiments but only one was successful in identifying differences in virulence among S ISG and P ISG isolates.
It should also be highlighted that for H. annosum sensu lato, as for many other basidiomycetes fungi, mating generally occurs by simple cell anastomosis and somatogamy of two haploid thalli (colonies) generated by germination and growth of haploid meiospores. Karyogamy does not occur until much later, and during most of their existence, individuals are characterized by a ploidy defined as hetero- or dikaryotic (n+n) rather than diploid. In dikaryotic individuals, thus, the two haploid nuclei are paired but not fused. Thanks to this feature, all F1 hybrids between two given parents are genetically identical and a single individual can be used to test hybrid fitness.
Inoculation experiments indicated that the North American hybrid genotype, despite bearing an entire nuclear genome of the pine-specialized P ISG, was not capable of matching the virulence level of the P ISG genotype in the pine inoculation experiment. Thus, on hosts characterized by the presence of a host-specialized pathogen species, it appears that hybrids are likely to be outcompeted by that species.
While inoculations on pines focused on a host on which one parental genotype is specialized, trials on Sitka spruce seedlings and in the xylem of white fir roots bypassing the selective effect of the cambium represented potential neutral ground or universal habitats on which both parental genotypes can thrive. Previous inoculations had clearly indicated that both North American taxa can equally affect Sitka spruce seedlings (W. Otrosina, personal communication), and high-performance liquid chromatography (not shown) indicated that a common generalized response to infection, and not a genotype-specific one, was being detected upon inoculation of white fir xylem. Were the hybrid generally unfit, we would expect it to have performed poorly on these unselective hosts. The ability of the hybrid to match both parents both in the extent of colonization of roots of white fir and in mortality levels caused among Sitka spruce seedlings clearly indicated that in some habitats hybrids are as competitive as their parents.
This finding is in agreement with reports regarding the ability of hybrid fungal taxa to exploit new hosts or new habitats. In the case of Heterobasidion, stumps may provide such new habitats. While pine trees are strictly colonized by the P ISG, pine stumps can be colonized by both North American taxa (36): this sympatry not only allows mating to occur, but according to our working hypothesis, it may place hybrid offspring in a relatively neutral habitat in which their chances to outcompete parental genotypes are significantly greater. It is interesting to note that the natural hybrid genotype was found in a large stump and in four adjoining trees (17).
Even if designed using unrelated genotypes, experiments one and two presented the advantage of a comparative analysis among three individuals that had survived the hurdles of real-world selection pressure. Because host specialization is a trait common to all individuals within each taxon (17, 46), the results remain generally valid. Nonetheless, three limitations are intrinsic to the above experiments: (i) a natural hybrid with the P mitochondrion was missing; (ii) the parental taxa, although interfertile, are quite distant in evolutionary terms, and the effects of such genomic disparity are hard to assess (20); and (iii) hybrid and parental genotypes were unrelated.
Experiment three circumvented the limitations of the other two experiments and compared haploid parents to two heterokaryotic hybrid offsprings, each carrying one of the two mitochondrial genomes. Four isolates were thus inoculated on both Norway spruce, the host on which H. parviporum is specialized, and silver fir, the host on which H. abietinum is specialized. Furthermore, the European H. abietinum and H. parviporum species represent two taxa in much closer evolutionary relationship to each other than their North American counterparts (19). Because the two species selected for this study are extremely close in evolutionary terms, the results of this experiment may also have relevance in highlighting potential mechanisms of reinforcement of genetic isolation, and hence of speciation, in fungi. It should also be noted that in spite of differences in ploidy between parents (n) and F1 hybrids (n+n), it has been shown that both haploid and heterokaryons (n+n) can be equally fit (18).
The results of experiment three indicated H. parviporum × H. abietinum hybrids to be significantly less fit than at least one of the parents on each host. The parental pathogen genotype specialized on each host resulted as the most effective genotype for its specific host. Hence, on hosts preferred by parental genotypes, hybrids are likely to be outcompeted by one of their parents. However, hybrids did not appear to be generally unfit, since they were at least able to match one of their parents in plant colonization rather than displaying levels of virulence lower than those of both parents.
In the case of Norway spruce, a clear fitness asymmetry was noticed when colonization levels of the two hybrids characterized by different cytoplasms were compared. A significant fitness advantage was displayed by the hybrid carrying the cytoplasm of H. parviporum, the parental species specialized on Norway spruce. Such asymmetry underlines the presence of a significant genotype-habitat interaction in the determination of hybrid fitness, as reported for other organisms (11, 47). It should be noted that the cytoplasm did not affect fitness of hybrids on the other host tested, thereby indicating that the correlation of cytoplasm with hybrid fitness should not be regarded as a generalized phenomenon as previously reported (33); however, more experiments are needed to fully understand the effect of cytoplasm on the fitness of Heterobasidion hybrids. Differences in fitness between parents and hybrids carrying the same cytoplasm is a relevant and novel finding for the fungi and for Heterobasidion in particular. Previous studies (33) had identified the presence of asymmetry between hybrids carrying different cytoplasms but were not able to differentiate between parents and hybrids carrying the same cytoplasm. Our results indicate greater fitness of the parent and are congruent with the low frequency of detection of hybrid genotypes in this intensively studied species complex.
The difference in colonization of Norway spruce between H. parviporum and the hybrid carrying the H. parviporum mitochondrion is statistically significant but may appear small to the reader. It should be emphasized that small differences measured in laboratory experiments can translate into a large ecological advantage/disadvantage in nature (4). To further prove this point, it should be noted that Norway spruce seedlings were killed only by the H. parviporum fungal genotype. The hybrid carrying the H. parviporum mitochondrion was only slightly less effective as a colonizer of Norway spruce stems, but nonetheless it was not capable of causing significant mortality, behaving much like the H. abietinum genotype and the hybrid carrying the H. abietinum mitochondrion.
Recent studies have described hybrids present in habitats or hosts unexploited by parental species (8, 42). Results presented in this study are among the first to experimentally elucidate the fitness of hybrids between fungal species, such as Heterobasidion spp., characterized by marked host specialization. We have experimentally shown that Heterobasidion hybrids were competitive with their parents only when their virulence was tested in substrates regarded as neutral or not selective towards the pathogen species. We propose that in fungal species complexes characterized by differential host specificity, hybrids may not be generally unfit but may be at an ecological disadvantage with regard to their parents in a habitat specific for one of the parents. According to this scenario, hybrid success may depend on the availability of new niches or habitats, in which competition with parental individuals may be less stringent.
There are several applied implications related to the findings of this study. Although we cannot predict the consequences of hybridization among different Heterobasidion species, we can expect it to cause shifts in both virulence levels and host ranges. We have shown that hybridization is genetically possible and that it is favored by novel ecological situations; anthropogenic disturbances are likely to create such novel conditions. The creation of stumps and the abundant wounding of standing trees during logging operations may favor hybridization. In a similar fashion, newly attained sympatry of allopatrically differentiated species caused by human-induced movement of the pathogen may lead to hybridization events. This knowledge dictates extreme care when managing land or trading forest products affected by this pathogen.
Intense present-day agricultural and forestry practices, combined with the rising levels of international trade in plants and plant products, is likely to have increased the availability of such new unexploited habitats. As a result, it is not surprising that the success of hybrid taxa in fungi appears to be on the rise (5). We further infer that fungal hybridization may occur more frequently than originally thought but may often be undetected because of the difficult task of correctly identifying closely related species and because of our lack of understanding of the types of habitat that may constitute viable hybrid zones.
Acknowledgments
We are grateful to the anonymous reviewers for critically reading the manuscript.
Footnotes
Published ahead of print on 27 July 2007.
REFERENCES
- 1.Arnold, M. L., and S. A. Hodges. 1995. Are natural hybrids fit or unfit relative to their parents? Tree 10:67-71. [DOI] [PubMed] [Google Scholar]
- 2.Asiegbu, F. A., S. Abu, J. Stenlid, and M. Johansson. 2004. Sequence polymorphism and molecular characterization of laccase genes of the conifer pathogen Heterobasidion annosum. Mycol. Res. 108:136-148. [DOI] [PubMed] [Google Scholar]
- 3.Bovers, M., F. Hagen, E. E. Kuramae, M. R. Diaz, L. Spanjaard, F. Dromer, H. L. Hoogveld, and T. Boekhout. 2006. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 6:599-607. [DOI] [PubMed] [Google Scholar]
- 4.Brasier, C. M. 1987. The dynamics of fungal speciation, p. 231-260. In A. D. M. Rayner, C. M. Brasier, and D. Moore (ed.), Evolutionary biology of the fungi. Cambridge University Press, Cambridge, United Kingdom.
- 5.Brasier, C. M. 2000. The rise of the hybrid fungi. Nature 405:134-135. [DOI] [PubMed] [Google Scholar]
- 6.Brasier, C. M. 2001. Rapid evolution of introduced plant pathogens via interspecific hybridization. Bioscience 51:123-133. [Google Scholar]
- 7.Brasier, C. M., S. A. Kirk, N. D. Pipe, and K. W. Buck. 1998. Rare interspecific hybrids in natural populations of the Dutch elm disease pathogens Ophiostoma ulmi and O-novo-ulmi. Mycol. Res. 102:45-57. [Google Scholar]
- 8.Brasier, C. M., D. E. L. Cooke, and J. M. Duncan. 1999. Origin of a new Phytophthora pathogen through interspecific hybridization. Proc. Natl. Acad. Sci. USA 96:5878-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brasier, C. M., M. Paoletti, K. W. Buck, A. Et-Touil, L. Bernier, and S. A. Kirk. 2006. Fitness effects of interspecific gene transfer in Ophiostoma. Abstr. 8th Int. Mycol. Congr., abstr. S42PS1-0657, p. 254.
- 10.Burgess, K. S., and B. C. Husband. 2004. Maternal and paternal contributions to the fitness of hybrids between red and white mulberry (Morus Moraceae). Am. J. Bot. 91:1802-1808. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, D. R., and N. M. Waser. 2001. Genotype-by-environment interaction and the fitness of plant hybrids in the wild. Evolution 55:669-676. [DOI] [PubMed] [Google Scholar]
- 12.Capretti, P., V. Goggioli, and L. Mugnai. 1994. Intersterility groups of Heterobasidion annosum in Italy: distribution, hosts and pathogenicity tests, p. 218-226. In M. Johansson and J. Stenlid (ed.), Root and butt rots of forest trees. Proceedings of the 8th International Conference. Swedish University of Agricultural Sciences, Uppsala, Sweden.
- 13.Chase, T. E., and R. C. Ullrich. 1990. Five genes determining intersterility in Heterobasidion annosum. Mycologia 82:73-81. [Google Scholar]
- 14.Cruzan, M. B., and M. L. Arnold. 1993. Ecological and genetic associations in an Iris hybrid zone. Evolution 47:1432-1445. [DOI] [PubMed] [Google Scholar]
- 15.Friesen, T. L., E. H. Stukenbrock, Z. Liu, S. Meinhardt, H. Ling, J. D. Faris, J. B. Rasmussen, P. S. Solomon, B. A. McDonald, and R. P. Oliver. 2006. Emergence of new disease as a result of interspecific virulence gene transfer. Nat. Genet. 22:953-956. [DOI] [PubMed] [Google Scholar]
- 16.Garbelotto, M., T. D. Bruns, F. W. Cobb, and W. J. Otrosina. 1993. Differentiation of intersterility groups and geographic provenances among isolates of Heterobasidion annosum detected by random amplified polymorphic DNA assays. Can. J. Bot. 71:565-569. [Google Scholar]
- 17.Garbelotto, M., A. Ratcliff, T. D. Bruns, F. W. Cobb, and W. Otrosina. 1996. Use of taxon-specific competitive-priming PCR to study host specificity, hybridization, and intergroup gene flow in intersterility groups of Heterobasidion annosum. Phytopathology 86:543-551. [Google Scholar]
- 18.Garbelotto, M. M., H. K. Lee, G. Slaughter, T. Popenuck, F. W. Cobb, and T. D. Bruns. 1997. Heterokaryosis is not required for virulence of Heterobasidion annosum. Mycologia 89:92-102. [Google Scholar]
- 19.Garbelotto, M., W. J. Otrosina, F. W. Cobb, and T. D. Bruns. 1998. The European S and F intersterility groups of Heterobasidion annosum may represent sympatric protospecies. Can. J. Bot. 76:397-409. [Google Scholar]
- 20.Garbelotto, M., P. Gonthier, R. Linzer, G. Nicolotti, and W. Otrosina. 2004. A shift in nuclear state as the result of natural interspecific hybridization between two North American taxa of the basidiomycete complex Heterobasidion. Fungal Genet. Biol. 41:1046-1051. [DOI] [PubMed] [Google Scholar]
- 21.Gonthier, P., M. Garbelotto, G. C. Varese, and G. Nicolotti. 2001. Relative abundance and potential dispersal range of intersterility groups of Heterobasidion annosum in pure and mixed forests. Can. J. Bot. 79:1057-1065. [Google Scholar]
- 22.Harrington, T. C., J. J. Worrall, and D. M. Rizzo. 1989. Compatibility among host-specialized isolates of Heterobasidion annosum from western North America. Phytopathology 79:290-296. [Google Scholar]
- 23.Harrington, T. C., J. Stenlid, and K. Korhonen. 1998. Evolution in the genus Heterobasidion, p. 63-74. In C. Delatour, J. J. Guillaumin, B. Lung-Escarmant, and B. Marçais (ed.), Root and butt rots of forest trees. Proceedings of the 9th International Conference, Carcans-Maubuisson, France.
- 24.Karlsson, J. O., and J. Stenlid. 1991. Pectic enzyme profiles of intersterility groups in Heterobasidion annosum. Mycol. Res. 95:531-536. [Google Scholar]
- 25.Kim, M. S., N. B. Klopfenstein, J. W. Hanna, and G. I. McDonald. 2006. Characterization of North American Armillaria species: genetic relationships determined by ribosomal DNA sequences and AFLP markers. Forest Pathol. 36:145-164. [Google Scholar]
- 26.Kohn, L. M. 2005. Mechanisms of fungal speciation. Annu. Rev. Phytopathol. 43:279-308. [DOI] [PubMed] [Google Scholar]
- 27.Korhonen, K., and J. Stenlid. 1998. Biology, p. 43-70. In S. Woodward, J. Stenlid, R. Karjalainen, and A. Hüttermann (ed.), Heterobasidion annosum, biology, ecology, impact and control. CAB International, New York, NY.
- 28.Korhonen, K., P. Capretti, R. Karjalainen, and J. Stenlid. 1998. Distribution of intersterility groups in Europe, p. 93-104. In S. Woodward, J. Stenlid, R. Karjalainen, and A. Hüttermann (ed.), Heterobasidion annosum, biology, ecology, impact and control. CAB International, New York, NY.
- 29.Johnston, J. A., M. L. Arnold, and L. A. Donovan. 2003. Hybrid fitness at seed and seedling life history stages in Louisiana irises J. Ecol. 91:438-446. [Google Scholar]
- 30.Lewontin, R. C., and L. C. Birch. 1966. Hybridization as a source of variation for adaptation to new environments. Evolution 20:315-336. [DOI] [PubMed] [Google Scholar]
- 31.Maijala, P., T. C. Harrington, and M. Raudaskoski. 2003. A peroxidase gene family and gene trees in Heterobasidion and related genera. Mycologia 95:209-221. [PubMed] [Google Scholar]
- 32.Newcombe, G., B. Stirling, S. McDonald, and H. D. Bradshaw. 2000. Melampsora × columbiana, a natural hybrid of M-medusae and M-occidentalis. Mycol. Res. 104:261-274. [Google Scholar]
- 33.Olson, A., and J. Stenlid. 2001. Plant pathogens. Mitochondrial control of fungal hybrid virulence. Nature 411:438. [DOI] [PubMed] [Google Scholar]
- 34.Olson, A., M. Lind, and J. Stenlid. 2005. In vitro test of virulence in the progeny of a Heterobasidion interspecific cross. For. Pathol. 35:321-331. [Google Scholar]
- 35.Otrosina, J. O., T. E. Chase, F. W. Cobb, Jr., and K. Korhonen. 1993. Population structure of Heterobasidion annosum from North America and Europe Can. J. Bot. 71:1064-1071. [Google Scholar]
- 36.Otrosina, W. J., T. E. Chase, and F. W. Cobb. 1992. Allozyme differentiation of intersterility groups of Heterobasidion annosum isolated from conifers in the western United States. Phytopathology 82:540-545. [Google Scholar]
- 37.Pei, M. H., D. J. Royle, and T. Hunter. 1999. Hybridization in larch-alternating Melampsora epitea (M-larici-epitea). Mycol. Res. 103:1440-1446. [Google Scholar]
- 38.Pfennig, K. S., A. J. Chunco, and A. C. R. Lackey. 2007. Ecological selection and hybrid fitness: hybrids succeed on parental resources. Evol. Ecol. Res. 9:341-354. [Google Scholar]
- 39.Riesberg, L. H. 1991. Homoploid reticulate evolution in Helianthus (Asteraceae): evidence from ribosomal genes. Am. J. Bot. 78:1218-1237. [Google Scholar]
- 40.Schardl, C. L., and K. D. Craven. 2003. Interspecific hybridization in plant-associated fungi and oomycetes: a review. Mol. Ecol. 12:2861-2873. [DOI] [PubMed] [Google Scholar]
- 41.Schulze, S. 1999. Rapid detection of European Heterobasidion annosum intersterility groups and intergroup gene flow using taxon-specific competitive priming PCR (TSCP-PCR). J. Phytopathol. 147:125-127. [Google Scholar]
- 42.Slippers, B., J. Stenlid, and M. J. Wingfield. 2005. Emerging pathogens: fungal host jumps following anthropogenic introduction. Trends Ecol. Evol. 20:420-421. [DOI] [PubMed] [Google Scholar]
- 43.Tsai, H. F., J. S. Liu, C. Staben, M. J. Christensen, G. C. M. Latch, M. R. Siegel, and C. L. Schardl. 1994. Evolutionary diversification of fungal endophytes of tall fescue grass by hybridization with Epichloë species. Proc. Natl. Acad. Sci. USA 91:2542-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warner, R., P. Gonthier, W. Otrosina, G. Laflamme, G. Bussières, J. Bruhn, and M. Garbelotto. 2005. Phylogenetic analysis of Heterobasidion annosum P ISG from an inter- to an intracontinental scale, p. 64-72. In M. Manka and P. Łakomy (ed.), Root and butt rots of forest trees. Proceedings of the 11th International Conference, Poznan-Bialowieza, Poland.
- 45.Werner, A., and P. Łakomy. 2002. Intraspecific variation in Heterobasidion annosum for mortality rate on Pinus sylvestris and Picea abies seedlings growth in pure culture. Mycologia 94:856-861. [PubMed] [Google Scholar]
- 46.Worrall, J. J., J. R. Parmeter, and F. W. Cobb, Jr. 1983. Host specialization of Heterobasidion annosum. Phytopathology 73:304-307. [Google Scholar]
- 47.Wu, C. A., and D. R. Campbell. 2005. Cytoplasmic and nuclear markers reveal contrasting patterns of spatial genetic structure in a natural Ipomopsis hybrid zone. Mol. Ecol. 14:781-792. [DOI] [PubMed] [Google Scholar]