Abstract
Metal resistance determinants have traditionally been found in cultivated bacteria. To search for genes involved in nickel resistance, we analyzed the bacterial community of the rhizosphere of Erica andevalensis, an endemic heather which grows at the banks of the Tinto River, a naturally metal-enriched and extremely acidic environment in southwestern Spain. 16S rRNA gene sequence analysis of rhizosphere DNA revealed the presence of members of five phylogenetic groups of Bacteria and the two main groups of Archaea mostly associated with sites impacted by acid mine drainage (AMD). The diversity observed and the presence of heavy metals in the rhizosphere led us to construct and screen five different metagenomic libraries hosted in Escherichia coli for searching novel nickel resistance determinants. A total of 13 positive clones were detected and analyzed. Insights about their possible mechanisms of resistance were obtained from cellular nickel content and sequence similarities. Two clones encoded putative ABC transporter components, and a novel mechanism of metal efflux is suggested. In addition, a nickel hyperaccumulation mechanism is proposed for a clone encoding a serine O-acetyltransferase. Five clones encoded proteins similar to well-characterized proteins but not previously reported to be related to nickel resistance, and the remaining six clones encoded hypothetical or conserved hypothetical proteins of uncertain functions. This is the first report documenting nickel resistance genes recovered from the metagenome of an AMD environment.
Microbial enzymes conferring metal resistance have significant biotechnological and environmental importance because of their potential use in bioremediation. Our knowledge about metal resistance mechanisms is based on cultured microorganisms. However, only 0.1 to 1% of the bacteria are culturable on standard laboratory media (2, 19, 26, 52, 56), and therefore, novel metal resistance determinants are still undiscovered in nature. Some culture-independent methods to rescue genes from environmental samples are based on the amplification or detection of sequences similar to those of previously known genes (5, 40, 53). However, they are not useful for finding different mechanisms of metal resistance. To circumvent these limitations, methods based on the functional analysis of libraries containing DNA from all the microbial community of a particular environment, named the metagenome (24), could provide a source of different and novel metal resistance genes. This powerful approach has been applied successfully for identifying genes conferring Li+-resistant phenotypes to Escherichia coli (33) and other microbial activities of interest (reviewed in references 10 and 23).
The geographical area called Rio Tinto hosts an extreme ecosystem located in southwestern Spain. The Tinto River has its origins at Peña de Hierro in the core of the Iberian Pyritic Belt, giving its name to an important mine region. The river is involved in the oxidation of mineral-containing sulfides, a process known as acid mine drainage (AMD), and as a result, its waters are highly acidic (pH of between 1.5 and 3.1) and enriched in toxic heavy metals (32). In the Tinto River, the extreme conditions are generated by the metabolic activity of chemolithotrophic acidophiles growing in the complex sulfides of the pyrite. Therefore, strains of Leptospirillum ferrooxidans, Acidithiobacillus ferrooxidans, and Acidiphilium spp., all related to the iron cycle, accounted for most of the prokaryotic microorganisms detected (20). It has been shown that acidophiles are highly resistant to different metals (13). Thus, the microbial communities from these sites are optimal for the discovery of new determinants of metal resistance.
In this study, the rhizosphere of the endemic heather Erica andevalensis, which grows at the banks of the Tinto River, was analyzed. Microbial communities from this environment were not previously explored and may exhibit interesting functional and diversity features. The prokaryotic communities in this ecological niche are expected to be more complex than those from the water column due mainly to the presence of nutrients coming from the root exudates. It has been shown that rhizosphere bacterial communities from metal-impacted sites display different metal resistances. For instance, bacteria associated with nickel hyperaccumulator plants are known to be nickel resistant (27, 44). Rhizosphere bacteria can also produce siderophores and chelators, which may reduce the toxic effects on the plants caused by excess metal concentrations (8, 38).
The presence of nickel in the Tinto River was previously reported (32). Nickel is the 24th most abundant element in the earth's crust, and it has been detected an all parts of the biosphere and also in meteorites (7). In this way, microorganisms have evolved in the presence of this metal, which is necessary in trace amounts for a variety of metabolic processes but toxic in high concentrations, causing oxidative stress in the cell (51). Several nickel resistance bacteria have been isolated mainly from heavy-metal-contaminated samples, but not many different mechanisms of resistance have been described compared with other metals. The best known mechanisms of nickel resistance are mediated by efflux pumps such as CnrCBA (cobalt-nickel resistance) from Cupriavidus metallidurans CH34 (formerly Ralstonia metallidurans CH34) (22, 31); NccCBA (nickel-cobalt-cadmium) and NreB (nickel resistance) from Achromobacter xylosoxidans 31A (21, 46); a homologue of NreB, NrsD, from Synechocystis sp. (18); YohM (resistant cobalt-nickel) from E. coli (42); and CznABC (cadmium-zinc-nickel) from Helicobacter pylori (50). Nevertheless, none of them have been reported to be isolated from uncultured microorganisms, and novel determinants may continue to be unexplored in nature.
The goal of this study was to identify genes involved in nickel resistance from a bacterial community growing in an acidic environment enriched in heavy metals. To this end, we have employed a metagenomic approach for identifying novel genes involved in nickel resistance. First, the microbially diverse populations in the rhizosphere from E. andevalensis have been characterized by cloning and sequencing of the 16S rRNA gene, and as a result, the diversity observed was typical of metal-impacted sites like AMD environments. The microbial DNA from the rhizosphere was then used to construct small-insert metagenomic libraries by using a direct lysis method for the extraction of DNA to avoid losing novel functional diversity. These libraries were screened for nickel resistance, and 13 different clones carrying resistance determinants have been identified, some of them similar to previously identified genes but others with no matches in known databases.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
E. coli DH5α and DH10B strains were routinely grown in Luria-Bertani (LB) medium at 37°C. The growth medium for transformed E. coli strains was supplemented with 50 μg ml−1 ampicillin (Ap) to maintain the pBluescript SKII(+) plasmid (pSKII+). LB plates used for metal assays contained NiSO4 at 2 mM and CoSO4 at 1.5 mM, which were the MICs established for both metals.
Isolation of DNA from rhizosphere samples.
Rhizosphere samples used in this study were recovered aseptically from plants of E. andevalensis, an endemic heather growing on the banks at the origin of the Tinto River (Huelva, Spain). The sample used in this study was collected in May 2005 at a distance of 30 cm from the river. The pH of the soil was measured in the supernatant of a 1:10 (wt/vol) dilution of rhizosphere soil in deionized water. The roots were shaken vigorously to separate the soil that was not tightly adhered to the roots. The samples recovered in situ were immediately frozen and stored at −80°C. Metagenomic DNA from the rhizosphere and soil that adhered to the roots was extracted directly using the BIO101 FastDNA Spin kit for soil (Qbiogene) according to the manufacturer's recommendations, with no further treatment. Approximately 33.3 μg of DNA per g of rhizosphere sample was obtained.
Cloning of the 16S rRNA gene and phylogenetic analysis.
Metagenomic DNA from the rhizosphere sample was used for the construction of bacterial and archaeal PCR-amplified 16S rRNA gene libraries. In order to avoid the bias of the PCR, two bacterial 16S rRNA gene-based libraries were constructed using two independent PCRs from the same source of environmental DNA. For the PCR amplification of the partial bacterial 16S rRNA gene, a 1,450-bp amplification product, the universal primers GM3 (5′-AGAGTTTGATCMTGGC-3′) and GM4 (5′-TACCTTGTTACGACTT-3′) (34) were used. For the PCR amplification of the partial archaeal 16S rRNA gene, a 590-bp amplification product, the universal primers ARC344F (5′-ACGGGGYGCAGCAGGCGCGA-3′) (39) and ARC915R (5′-GTGCTCCCCCGCCAATTCCT-3′) (49) were used. Each reaction mixture contained 10 ng of metagenomic DNA, 250 μM of each of the four deoxynucleoside triphosphates, 1.5 mM of MgCl2, 200 nM of each primer, 2.5 U of Taq DNA polymerase, and the appropriate buffer supplied by the manufacturer (Invitrogen), up to a total volume of 50 μl. The PCR amplification program used was as follows: 1 cycle of 5 min at 95°C; 35 cycles of 45 s at 95°C, 45 s at 44°C, and 2 min at 72°C; and, finally, 1 cycle of 10 min at 72°C. The amplification using the archaeal primers was identical, but the annealing temperature was increased to 52°C. PCR products were cloned using the TOPO TA cloning kit (Invitrogen). A total of 101 bacterial and 27 archaeal random clones were selected for sequencing. DNA was sequenced on both strands by using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer) and an ABI PRISM 377 sequencer (Perkin-Elmer) according to the manufacturer's instructions. Preliminary phylogenetic affiliation of the 16S rRNA gene clones was performed using the Classifier tool of the Ribosomal Data Project (RDP-II) database (9), and 85 bacterial and 27 archaeal clones were unambiguously classified by comparison to the RDP-II database (confidence level of 75%) and compared to available databases by use of BLAST (1). Sequences were screened for chimeras using the programs Bellerophon (25) and Chimera_Check (9). None of the selected sequences were found to be chimeric. Sequences were aligned with the Fast Aligner program in ARB (http://www.arb-home.de/) and were checked manually to resolve regions of ambiguity. Identification of the phylogenetic affiliation of the sequences was performed with the neighbor-joining clustering method and the Jukes-Cantor correction implemented in ARB. From the ARB database, a total of 38 of the closest eubacterial sequences and 8 archaeal sequences were included as representatives. Bootstrap analysis was used to evaluate the confidence of the tree topologies with 1,000 resampling trials.
To determine the richness of bacterial and archaeal 16S sequences in the sample, a rarefaction curve and Chao1 richness estimate collector's curve, both with a 95% confidence interval (CI), were calculated by using the DOTUR program (45), and operational taxonomic units (OTUs) were defined at different cutoffs. DOTUR used a distance matrix based on Jukes-Cantor correction constructed from the alignment of 85 bacterial sequences or 27 archaeal sequences in ARB.
Library construction.
The metagenomic libraries were constructed with the same DNA used for the 16S rRNA gene libraries. Metagenomic DNA was partially digested using Sau3AI, and fragments from 1 kb to 8 kb were collected directly from a 0.8% low-melting-point agarose gel with the QIAquick extraction gel (QIAGEN) for ligation into the dephosphorylated and BamHI-digested pSKII+ vector. DNA (100 to 125 ng) excised from the gel was mixed with the vector at a molar ratio of 1:1. Ligation mixtures were incubated overnight at 16°C using T4 DNA ligase (Roche) and transformed by electroporation into E. coli DH10B cells (Invitrogen) using Micropulser (Bio-Rad) according to the manufacturer's instructions. A total of five independent libraries were constructed. To determine the average insert size for each library, the plasmids of 184 random clones were isolated and digested using either XbaI or EcoRI and XbaI restriction enzymes (Roche). To amplify the libraries, they were grown on LB agar plates containing Ap (approximately 1.3 × 104 cells per plate) and incubated for 24 h at 37°C. Cells from each plate were mixed with 3.5 ml LB plus 10% (wt/vol) glycerol, pooled in a flask with cells from the same library, mixed again, and stored at −80°C.
Analysis of nickel resistance clones.
The inserts of the plasmids from nickel-resistant colonies were sequenced on both strands as described above. Sequences were analyzed with the Editseq, Megalign, and Seqman programs from the DNAStar package. Putative open reading frames (ORFs) were identified using two programs: ORF Finder, available at the NCBI website (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), and Artemis (http://www.sanger.ac.uk/Software/Artemis/). For translation to protein sequences, the bacterial code was selected, allowing ATG, GTG, and TTG as alternative start codons. All the predicted ORFs that were longer than 90 bp were translated and used as queries in BlastP. The sequences with significant matches were further analyzed with rpsBlast, and their putative function was annotated based on their similarities to sequences in the COG (Clusters of Orthologous Groups) and Pfam (Protein Families) databases. Those sequences with an E value of more than 0.001 in the BlastP searches and longer than 300 bp were considered to be hypothetical. Transmembrane helices were predicted with TMpred (http://www.ch.embnet.org/software/TMPRED_form.html).
ORFs conferring nickel resistance were identified by subcloning and/or in vitro transposon mutagenesis with the GPS-LS Genome Priming system (New England Biolabs) according to the manufacturer's instructions. Subcloning was accomplished by PCR amplification using the following reaction mixture: 25 ng of plasmid DNA, 500 μM of each of the four deoxynucleoside triphosphates, 2.5 U of Pfu Turbo DNA polymerase (Stratagene), and 100 nM of each forward and reverse primers (see Table S1 in the supplemental material) up to a total volume of 50 μl. The PCR amplification program used was as follows: 1 cycle of 5 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 52°C, and 5 min at 72°C; and, finally, 1 cycle of 10 min at 72°C. PCR amplification products were gel purified with the QIAquick extraction gel (QIAGEN), digested with the appropriate restriction enzymes (Roche), and ligated into pSKII+. A 200-bp region upstream of the start codon was also amplified to include their native expression sequences (promoters and ribosome binding sites). Some of the ORFs were truncated, or the 5′ region was close to the polylinker sequence of the pSKII+ vector, and they were subcloned in the same orientation as that of the original clone. The subclones were transformed into E. coli cells, and the resulting strains were screened again on LB agar plus Ni (2 mM). The nickel resistance results from the subclones are included in Table S1 in the supplemented material. In vitro transposon mutants were transformed in strain DH10B and selected with Ap (50 μg ml−1) plus kanamycin (20 μg ml−1). Two hundred transformants from each transposition were patched on LB agar containing Ap plus kanamycin, with and without Ni (2 mM), and grown overnight at 37°C. All mutagenized nickel-sensitive colonies and at least 10 resistant colonies were rescued from the LB agar without Ni. Nickel-sensitive colonies were considered to be mutagenized in the genes required for nickel resistance, and their plasmids were isolated and sequenced with specific primers for the transposon ends to determine the precise insertion. To localize insertions out of the nickel resistance genes, plasmids from resistant colonies were also sequenced.
Determination of cellular metal concentration.
E. coli DH5α carrying the empty vector and clones pSM1 to pSM13 were grown in LB liquid medium containing Ap at 37°C in a shaking incubator, and growth was monitored as the optical density at 600 nm. Ni was added at 4 mM to the cultures in early stationary phase and grown for one additional hour. Cultures were washed four times extensively with ultrapure MiliQ H2O and centrifugation. Washed pellets were lyophilized, pulverized, and dissolved in H2O-HCl-HNO3-H2O2 (3:1:4:0.5) by a closed microwave digestion system for subsequent inductively coupled plasma spectroscopy-mass spectrometry (ICP-MS) analysis. Results were expressed as mg of Ni g−1 (dry weight) of cells.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained for the plasmid inserts (pSM1 to pSM13) and the 16S rRNA gene sequences have been deposited in the GenBank database under the accession numbers shown in Table 3 and Fig. 1, respectively.
TABLE 3.
Description of nickel-resistant plasmids (pSM1 to pSM13) and their observed sequence similarities
| Plasmid (insert size [bp]) | G+C content (%) | GenBank accession no. | ORFa | Length (aa)b | Closest similar protein (no. of encoded amino acids), GenBank accession no. of similar proteinc | Organism | E value | % Amino acid homology to the closest similar protein (% identity) | Putative function | No. of transmembrane helicesd |
|---|---|---|---|---|---|---|---|---|---|---|
| pSM1 (3,039) | 62.5 | DQ858211 | 1 | 71 | Hypothetical protein gsr2216 (68), NP_925162 | Gloeobacter violaceus | 1E-10 | 34/67 (50) | Small bacterial protein of unknown function, pfam04384 | 0 |
| 2 | 205 | None | Hypothetical | 0 | ||||||
| 3 | 121 | None | Hypothetical | 1 | ||||||
| 4 | 169 | None | Hypothetical | 0 | ||||||
| pSM2 (1,975) | 60.3 | DQ898541 | 1* | 403 | Protein of unknown function (503), YP_001229356 | Geobacter uraniumreducens | 1E-77 | 165/378 (43) | RmuC family, pfam02646; uncharacterized protein conserved in bacteria, COG1322 | 2 |
| 2 | 174 | None | Hypothetical | 2 | ||||||
| pSM3 (2,116) | 63.3 | DQ898542 | 1* | 110 | None | Hypothetical | 0 | |||
| 2 | 174 | None | Hypothetical | 0 | ||||||
| 3 | 109 | None | Hypothetical | 0 | ||||||
| 4* | 119 | None | Hypothetical | 0 | ||||||
| pSM4 (2,280) | 54.6 | DQ898543 | 1 | 161 | None | Hypothetical | 0 | |||
| 2 | 117 | None | Hypothetical | 0 | ||||||
| pSM5 (1,520) | 57.8 | DQ898544 | 1* | 188 | ABC transporter, ATPase subunit (282), YP_591966 | Acidobacterium sp. strain Ellin345 | 3E-54 | 110/190 (57) | ABC-type transport system, ATPase, COG1127 | 0 |
| 2* | 299 | Acyl-CoA dehydrogenase-like (392), YP_589689 | Acidobacterium sp. strain Ellin345 | 1E-134 | 222/299 (74) | Acyl-CoA dehydrogenases, COG1960 | 2 | |||
| pSM6 (1,501) | 70.3 | DQ898545 | 1* | 428 | Extensin-like protein (1,007), ZP_00768886 | Chloroflexus aurantiacus | 3E-10 | 71/145 (48) | Conserved hypothetical protein | 1 |
| pSM7 (1,140) | 47.6 | DQ898553 | 1* | 354 | Related to acyl-CoA sterol acyltransferase (580), XP_961300 | Neurospora crassa OR74A | 4E-134 | 248/377 (65) | Acyl-CoA cholesterol acyltransferase, COG5056 (exon) | 5 |
| pSM8 (1,523) | 65.4 | DQ898547 | 1* | 174 | Hypothetical protein pAL1.079 (239), YP_001210526 | Arthrobacter nitroguajacolicus | 9E-53 | 110/168 (65) | Putative inner membrane protein, pfam088491 | 1 |
| 2 | 68 | VrlI-like protein (66), ZP_01160357 | Photobacterium sp. strain SKA34 | 2E-14 | 38/60 (63) | Conserved hypothetical protein | 0 | |||
| 3 | 106 | None | Hypothetical | 1 | ||||||
| pSM9 (3,043) | 65.7 | DQ898548 | 1* | 451 | Penicillin binding protein 1A (794), YP_094959 | Legionella pneumophila | 3E-103 | 196/424 (46) | Penicillin-binding protein, COG5009 | 4 |
| 2* | 354 | Type IV pilus assembly protein PilM (372), YP_342307 | Nitrosococcus oceani | 2E-92 | 179/351 (50) | Tfp pilus assembly protein, COG4972 | 0 | |||
| pSM10 (2,001) | 57.5 | DQ898549 | 1* | 158 | Amino acid transporters (746), YP_591231 | Acidobacterium sp. strain Ellin345 | 1E-44 | 90/132 (68) | Hypothetical amino acid transporter | 3 |
| 2* | 385 | Apolipoprotein N-acyltransferase (526), YP_591416 | Acidobacterium sp. strain Ellin345 | 3E-48 | 142/391 (36) | Apolipoprotein N-acyltransferase, COG0815 | 7 | |||
| pSM11 (1,660) | 64.3 | DQ898550 | 1 | 244 | SAT (258), YP_591172 | Acidobacterium sp. strain Ellin345 | 2E-78 | 148/241 (61) | SAT, COG1045 | 1 |
| 2* | 74 | Putative O-acetylserine (thiol) lyase precursor (352), AAC27794 | Chlamydomonas reinhardtii | 3E-20 | 56/83 (67) | Cysteine synthase, COG0031 | 1 | |||
| pSM12 (1,894) | 58.9 | DQ898551 | 1 | 261 | ABC transporter, inner membrane subunit (262), YP_591897 | Acidobacterium sp. strain Ellin345 | 2E-49 | 124/254 (48) | ABC-type uncharacterized transport system, permease, COG0390 | 6 |
| 2 | 229 | ABC transporter, ATPase subunit (237), YP_591896 | Acidobacterium sp. strain Ellin345 | 1E-60 | 126/221 (57) | ABC-type metal ion transport system, ATPase, COG1135 | 1 | |||
| pSM13 (2,014) | 66.3 | DQ898552 | 1* | 671 | MobA/MobL protein (974), YP_611123 | Sphingopyxis alaskensis | 7E-43 | 125/339 (36) | Conjugal transporter protein TraA | 0 |
ORFs involved in nickel resistance are shown in boldface type, and asterisks indicate incomplete ORFs.
aa, amino acids.
Those sequences with an E value of more than 0.001 in BLAST searches were considered to be unknown proteins.
Number of transmembrane helices predicted by using the program TMPRED.
FIG. 1.
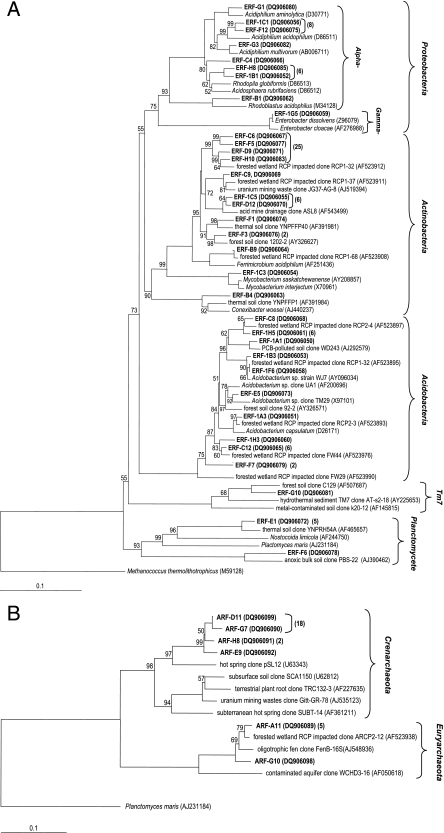
Neighbor-joining trees of the 16S rRNA gene sequences obtained from communities of Bacteria (A) and Archaea (B) from the Rio Tinto rhizosphere (shown in boldface type) and all other closest representative sequences obtained from the ARB database, which are given with their GenBank accession numbers. OTUs were defined based on 97% sequence similarity. The total number of identical to nearly identical sequences (>97% similarity) to each OTU is indicated in brackets. Bootstrap values above 50% are indicated at the nodes. The scale bar below the tree corresponds to 0.1 substitutions per nucleotide for a unit branch length. Bacterial and archaeal trees were rooted with Methanococcus thermolithotrophicus and Planctomyces maris, respectively. PCB, polychlorinated biphenyl. RCP, reject coal pile.
RESULTS
Phylogenetic analysis of the bacterial community from the rhizosphere.
The rhizosphere of the endemic heather E. andevalensis was sampled at the banks of the Tinto River. The pH of the rhizosphere sample was moderately acidic (pH 3.5). ICP-MS semiquantitative analysis of the rhizosphere samples showed the presence of the main heavy metals found in the water column, including nickel (Table 1). To extract the DNA from such a challenge ecological niche, a direct lysis method was chosen in order to access as many microbially diverse populations as possible. Diverse methods and kits for DNA purification from environmental samples were tested with unsuccessful results, possibly due to the humic substances coextracted with the nucleic acids. Only the DNA isolated with the BIO101 FastDNA Spin kit was useful for further enzymatic reactions (i.e., PCR amplification, digestion, and ligation).
TABLE 1.
Semiquantitative determination of the concentrations of the main metal elements in rhizosphere samples by ICP-MS
| Metal | Mean concn (ng g−1 of rhizosphere) ± SDa |
|---|---|
| Cr | 88.9 ± 6.2 |
| Fe | 932,276.8 ± 18,815.3 |
| Co | 32.1 ± 0.1 |
| Ni | 1,167.1 ± 31.8 |
| Cu | 1,217.6 ± 81.0 |
| Zn | 679.3 ± 16.1 |
| As | 2,587.1 ± 96.8 |
| Ag | 12.9 ± 0.5 |
| Cd | 3.2 ± 0.1 |
| Pb | 914.5 ± 56.3 |
Values represent averages and standard deviations for two independent measurements of a single sample. These values do not correspond to the absolute metal concentrations in the samples.
This DNA was used to explore the bacterial and archaeal diversity of the rhizosphere samples. The 16S rRNA gene was amplified using universal primers for these domains, and the products were subsequently cloned and sequenced. In the domain Bacteria, a total of 101 clones were sequenced. Eighty-five clones were unambiguously classified by comparison to the RDP-II database (confidence level of 75%) and analyzed further. A dendrogram showing the presence of five phylogenetic groups (Fig. 1A) was constructed. The majority of the clones recovered were affiliated with the Actinobacteria (38 clones/12 OTUs) and the Acidobacteria (21 clones/10 OTUs). The Actinobacteria group was represented by clones related to Mycobacterium spp., Conexibacter woesei, and “Ferrimicrobium acidiphilum.” The Acidobacteria group was represented by clones related to Acidobacterium spp., including Acidobacterium capsulatum. The Proteobacteria group was represented by Alphaproteobacteria (18 clones/8 OTUs) related to Acidiphilium spp., Acidosphaera rubrifaciens, Rhodopila globiformis, and Rhodoblastus acidophilus and by Gammaproteobacteria (1 clone/1 OTU) related to Enterobacter spp. There were also members of the Planctomycetes group (6 clones/2 OTUs) and the candidate division TM7 (1 clone/1 OTU), with no cultivated representatives. Most of the clones (71 clones/25 OTUs) were closely related to sequences obtained from different AMD sites, like a forested wetland site impacted by acid solutions derived from coal (6), uranium mining waste, and AMD at Iron Mountain (14). Some of these sequences were similar to the cultivable heterotrophic acidophiles Acidobacterium capsulatum, Acidiphilium spp., and Acidosphaera rubrifaciens; the anoxygenic phototroph Rhodopila globiformis; and the heterotrophic iron oxidizer “Ferrimicrobium acidiphilum” (3, 6, 14) (Fig. 1A). Other clones (12 clones/7 OTUs) observed in the phylogeny were affiliated with sequences recovered from different sources like a forest soil (15), a thermal soil (37), an anoxic bulk soil of a flooded rice microcosm (11), and a polychlorinated biphenyl-polluted soil (36). Some of the bacterial strains were similar to microorganisms that are difficult to lyse, such as Mycobacterium spp., which indicates the validity of this DNA isolation method to recover enough microbiologically diverse populations.
In the domain Archaea, a total of 27 clones were classified by comparison to the RDP-II database (confidence level of 75%) and revealed the presence of 6 clones related to the Euryarchaeota and 21 clones related to the Crenarchaeota phylogenetic groups in the rhizosphere sample (Fig. 1B). The majority of the clones recovered were affiliated with Crenarchaeota (21 clones/4 OTUs), and these clones were affiliated with clones recovered from a hot spring (4). There were also members of the Euryarchaeota (6 clones/2 OTUs), which were affiliated with clones recovered from a forested wetland impacted by rejected coal (6) and from an aquifer contaminated with hydrocarbons and chlorinated solvents (12).
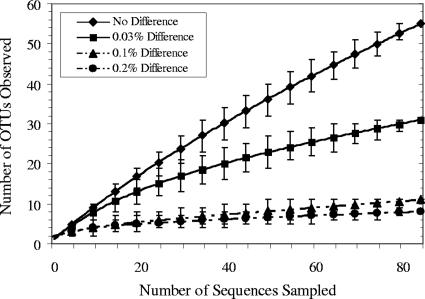
For quantifying the composition of bacterial and archaeal lineages in the rhizosphere community, we performed two analyses: a rarefaction curve and a Chao1 richness estimation collector's curve. Rarefaction curve analysis predicted that the number of bacterial OTUs observed in the Rio Tinto rhizosphere was 31 when the OTU definition was set at a 3% distance level (species level) and 8 OTUs when the OTU definition was set at 20% (phylum level) (Fig. 2). On the other hand, Chao1 richness estimation collector's curve analysis revealed that a total of 62 OTUs were expected at a 3% distance level (CI, 40 to 130) and that 10 OTUs were expected at 20% (CI, 8 to 23) (data not shown). The expected richness at the phylum level was almost reached since the rarefaction curve plateaued from approximately the 80th sequence. In the domain Archaea, five predicted OTUs were observed in the rarefaction curve analysis at a 3% distance level, and only 2 OTUs were predicted at a 5% distance level (data not shown). Distance levels of 10% and 20% were not retrieved from this analysis, possibly due to the low diversity found and/or the low number of sequences sampled. In a similar way, Chao1 analysis was discarded in the archaeal domain since the CIs at different distance levels overlapped with one another, probably due to the low diversity found. In summary, these analyses revealed that the microbial diversity in the rhizosphere may be considered low and that the number of species estimated with the sequences sampled could represent an important fraction of the total community.
FIG. 2.
Rarefaction curve using bacterial 16S rRNA gene sequences from the Rio Tinto rhizosphere. Error bars represent the 95% CI.
Construction of metagenomic libraries.
Five metagenomic libraries were constructed with DNA extracted from the rhizosphere of E. andevalensis cloned in the high-copy-number vector pSKII+. Approximately 726,500 recombinant clones (16,000 recombinant clones per μg of DNA) were obtained. Restriction digestion analysis of 184 random clones showed an average insert size of 2.5 kb, ranging between 1 and 8.5 kb (Table 2). Approximately 2.15 Gb of environmental DNA was cloned in these libraries, which where subsequently amplified as described in Materials and Methods.
TABLE 2.
Nickel resistance screening of the libraries constructed with metagenomic DNA from the Rio Tinto rhizosphere
| Library | No. of clones | Avg insert size (kb) (range)a | Library size (Mb) | No. of Ni-resistant clonesb | No. of Ni-resistant clonesc/no. of unique Ni-resistant clonesd (plasmids) |
|---|---|---|---|---|---|
| A | 145,000 | 2 (1-5.5) | 290 | 884 | 10/5 (pSM1, pSM2, pSM3, pSM4 and pSM5) |
| B | 168,500 | 1.7 (1.2-3) | 286 | 630 | 18/2 (pSM11 and pSM12) |
| C | 102,000 | 2.5 (1-5) | 255 | 542 | 0/0 |
| D | 41,000 | 2 (1.3-3.3) | 82 | 351 | 6/4 (pSM6, pSM8, pSM9 and pSM10) |
| E | 270,000 | 4.5 (1-8.5) | 1,237 | 74 | 8/2 (pSM7 and pSM13) |
| Total | 726,500 | 2.5 (1-8.5) | 2,150 | 2,481 | 42/13 |
Estimated from restriction digestion analysis of 184 random clones using either EcoRI or EcoRI/XbaI enzymes.
Number of nickel resistance clones recovered after screening of the amplified library.
Number of nickel resistance clones recovered after retransformation.
Number of nickel resistance clones recovered after fragment length polymorphism analysis.
Screening for genes conferring nickel resistance.
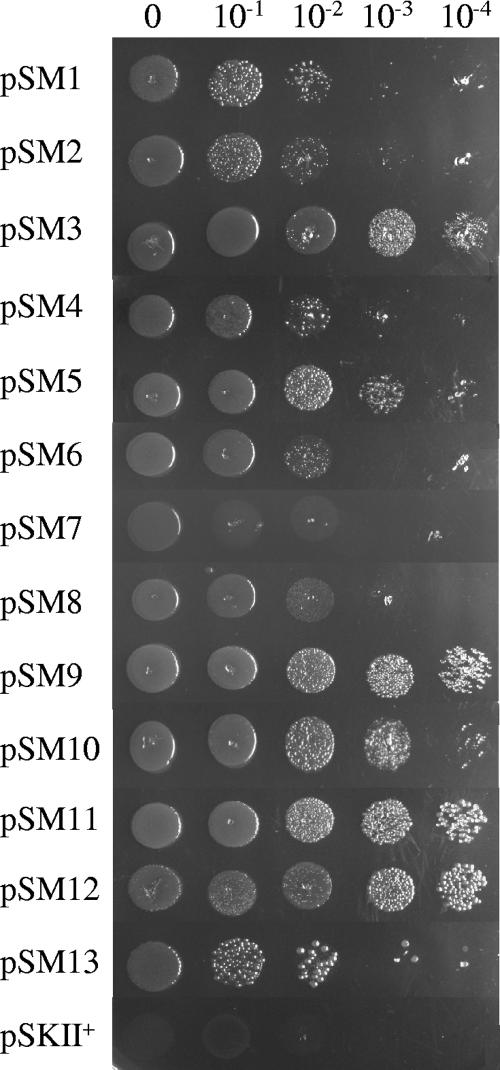
In order to obtain genes encoding nickel resistance from the metagenomic libraries, aliquots of approximately 6 × 108 cells from each amplified library were plated onto LB-Ap agar containing 2 mM Ni, a lethal concentration for strain DH10B(pSKII+). Nickel resistance colonies were detected after incubations at 37°C overnight (Table 2). To exclude chromosomal mutations, the colonies from each library were pooled, and plasmid DNA was isolated, transformed into E. coli, and selected on LB-Ap plates without Ni. One hundred colonies from each transformation were patched onto LB-Ap plates containing Ni. The plasmids isolated from the nickel-resistant colonies were digested with EcoRI and XbaI enzymes, and a total of 13 recombinant plasmids were unique in their restriction patterns (Table 2). These plasmids clearly conferred nickel resistance on the resulting recombinant E. coli strains, which exhibited different resistance profiles using the drop assay on LB-Ap plates containing Ni (Fig. 3). Clones with more elevated resistance to nickel were pSM9, pSM11, and pSM12, followed by pSM3, pSM5, and pSM10. All the clones were tested in the presence of higher concentrations of nickel, and clones pSM11 and pSM12 were able to grow at 4 mM Ni. Nickel resistance determinants frequently provide resistance to other metals such as cobalt and cadmium. All the clones were then also tested for cobalt resistance, and the pSM1, pSM9, pSM10, pSM11, and pSM12 clones were resistant to 1.5 mM Co (see Fig. S1 in the supplemental material), and clones pSM8 and pSM12 showed enhanced resistance to 0.8 mM Cd (see Fig. S2 in the supplemental material). No significant resistance to zinc and copper was observed (data not shown).
FIG. 3.
Drop assay of the 13 nickel-resistant clones obtained from the screening of the metagenomic libraries on LB plates containing 2 mM Ni using serial dilutions (0 to 10−4) from cultures grown overnight. DH5α(pSKII+) was used as a negative control. Each assay was performed at least three times using independent cultures.
Molecular analysis of plasmids conferring nickel resistance.
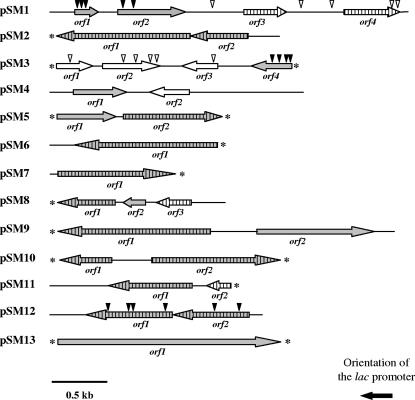
The gene organization, genetic similarities, and transmembrane domain predictions of the inserts are summarized in Fig. 4 and Table 3. The 13 plasmids harbored a total of 15 complete and 13 incomplete heterologous ORFs. A total of 11 ORFs showed very low or no similarity to sequenced genes from other organisms, which shows the novelty of some of the genes recovered from this environment. The G+C content of the sequenced inserts varied from 47.6% to 70.3%, indicating their diverse phylogenetic affiliation. The insert from pSM7, with the lowest G+C content (47.6%), could be of eukaryotic origin based on its similarity to an acyl-coenzyme A (CoA) sterol acetyltransferase gene from the ascomycete Neurospora crassa.
FIG. 4.
Schematic organization of the ORFs identified in plasmids pSM1 to pSM13. The arrows indicate the locations and the directions of transcription of the ORFs in the different plasmids. Those ORFs involved in nickel resistance are indicated by gray arrows. Arrows shaded with vertical bars indicate the presence of predicted transmembrane helices. Asterisks indicate incomplete ORFs. Plasmids pSM1, pSM3, and pSM12 were mutagenized by in vitro transposition, and vertical arrowheads indicate transposon insertions that either abolish the resistance phenotype (filled in black) or do not affect the resistance phenotype (not filled).
Three plasmids, pSM6, pSM7, and pSM13, harbored only one predicted ORF. The remaining plasmids contained more than one ORF, and as determined by subcloning (pSM2, pSM3, pSM4, pSM5, pSM8, pSM9, pSM10, and pSM11) (see Table S1 in supplemental material) or transposon mutagenesis (pSM1, pSM3, and pSM12), one or two ORF products were involved in nickel resistance (Fig. 4). In pSM1, pSM2, pSM5, pSM8, and pSM12, the ORF products providing nickel resistance may constitute an operon, since they are transcribed in the same direction. In fact, in pSM12, the stop codon from orf2 overlaps with the start codon from orf2.
As shown in Table 3, 4 of the 20 ORF products identified as being involved in nickel resistance did not reveal a significant similarity with any other protein in the databases. Five ORF products were similar to conserved hypothetical proteins whose functions are uncertain. Eleven ORF products were similar to known proteins, but only orf1 from pSM11 encoded an amino acid sequence that was similar to a protein known to confer nickel resistance, a serine O-acetyltransferase (SAT). Recently, the SAT enzyme from the plant Thlaspi goesingense was reported to be involved in intracellular Ni and Co protection when overexpressed in E. coli cells (17). On the other hand, pSM5 and pSM12 ORFs encoded components of ATP-binding cassette (ABC) transporters, which may be involved in nickel translocation outside the cells, but they were distantly related to nickel uptake members of this transporter family. ABC transporters use the energy of ATP hydrolysis to transport specific substrates across the cellular membrane. Typical ABC transporters are constituted by a different number of two types of subunits: a permease that contains between 6 and 11 membrane-spanning regions and an ATPase that contains an ATP-binding domain, divided into the Walker A (GXXGXGXGKS/T, where X represents any amino acid) and Walker B (hhhhD, where h represents a hydrophobic amino acid) motifs, which are separated by approximately 90 to 120 amino acids. In addition, there is a third conserved motif called a linker peptide (LSGGQQ/R/KQR) preceding the Walker B motif. In the case of pSM5, the orf1 product was similar to the ATPase subunit of an ABC transporter and contained the ISGGMRRR linker peptide motif preceding the Walker B motif LLLYD, which is typical of these proteins, but the Walker A motif was absent. The incomplete orf2 product from pSM5, which is also required for nickel resistance, was similar to an acyl-CoA dehydrogenase-like protein containing two putative transmembrane domains but not similar to the characteristic inner membrane subunit of ABC transporters. pSM12 contains two complete ORFs, orf1 and orf2, whose products were similar to the inner membrane subunit and to the ATPase subunit of an ABC transporter, respectively. Sequence analysis of the orf1 product revealed six transmembrane motifs characteristic of the inner membrane subunit of these transporters and spanning positions 12 to 32, 66 to 86, 93 to 117, 125 to 147, 196 to 222, and 224 to 246 along the sequence. The deduced amino acid sequence of orf2 contained the three highly conserved domains common to ATPase components of classical ABC-type transport systems: Walker A, which corresponds to the short sequence GPSGAGKS, and the linker peptide LSGGEAQR, preceding the Walker B motif VLLLD. The remaining ORFs from plasmids pSM7, pSM9, pSM10, and pSM13 were similar to well-characterized proteins not previously reported to be involved in metal resistance, such as, for instance, acyl-CoA sterol acyltransferase, penicillin-binding protein, or the conjugal transporter protein TraA.
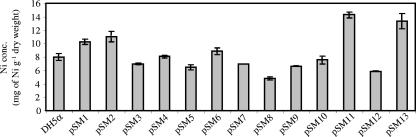
Determination of cellular nickel content.
To gain insights into the possible mechanisms of nickel resistance, the cellular concentration of nickel from clones pSM1 to pSM13 was measured by ICP-MS 1 h after the addition of nickel to a culture of growing cells (Fig. 5). Based on their nickel accumulation properties, the clones were classified into two groups: those favoring metal accumulation (i.e., intracellular sequestration and reduction in the sensitivity of cellular targets) and those preventing metal accumulation (i.e., active transport efflux pumps, extracellular sequestration, and exclusion by permeability barrier). The first group is constituted by the clones accumulating more or similar amounts of nickel compared to the control (pSM1, pSM2, pSM4, pSM6, pSM10, pSM11, and pSM13). orf1 from pSM11 encodes a protein highly similar to an SAT, which has been reported, as described above, to be involved in nickel intracellular protection (17). Thus, cells expressing SAT enzymes can accumulate the metal, and in fact, the nickel content in pSM11 cells was higher than that in the control cells. ORFs from other plasmids of this group were neither similar to known genes nor similar to other genes involved in metal resistance. The second group is constituted by those clones accumulating less nickel than the control (pSM3, pSM5, pSM7, pSM8, pSM9, and pSM12). This group includes the ORFs encoding ABC transporter components from pSM5 and pSM12. They could constitute an efflux system involved in nickel translocation outside the cell, which is in agreement with the lower nickel content of these clones.
FIG. 5.
Test for cellular content of nickel in E. coli clones pSM1 to pSM13 and DH5α(pSKII+) with empty vector after 1 h of growth with 4 mM Ni. Values are the averages of two independent ICP-MS measurements. Error bars indicate standard deviations.
DISCUSSION
Metagenomic technology is a powerful tool that has been applied to the discovery of novel natural products and enzymes of biotechnological interest. To our knowledge, this is the first work in which this methodology is employed in exploring the variety of mechanisms conferring nickel resistance to microorganisms from an AMD environment (rhizosphere). To this end, we searched for nickel resistance genes from the microbial community of the Rio Tinto rhizosphere, a natural environment highly enriched in toxic metals and metalloids.
The metagenomic libraries from the rhizosphere of E. andevalensis contain DNA from a community constituted mainly by heterotrophic acidophiles similar to those found in AMD systems and by members of the Actinobacteria and Alphaproteobacteria from the Tinto River described previously (20). However, the most representative bacteria from the Tinto River, the chemolithotrophic bacteria Leptospirillum ferrooxidans and Acidithiobacillus ferrooxidans (20), were not detected in the rhizosphere, probably due to their sensitivity to organic compounds (28). In contrast, we have identified members of the Crenarchaeota group, which has not been detected in the Tinto River (E. Gonzalez-Toril, personal communication). In conclusion, the microbially diverse communities in the rhizosphere were more complex than those in the river and mainly represented by heterotrophic acidophiles.
Considering this diversity, the construction and screening of metagenomic libraries were appropriate to retrieve genes involved in metal resistance from these communities. A total of 13 nickel resistance genes and operons from rhizosphere DNA have been identified. Despite the diversity of actinomycetes from the metagenomic DNA utilized to construct the libraries, only the pSM6 insert shows a G+C content of 70.3%, which is expected for genes derived from actinomycetes. On the other hand, pSM5-, pSM10-, and pSM12-encoded proteins are similar to proteins from Acidobacterium sp., which is in agreement with the observed G+C content, 58%, expected for this bacterium. The exception is the insert from pSM11, which encodes a protein that is highly similar to an SAT from Acidobacterium sp., with a G+C content of 64.3%. The insert from pSM7 shows the lowest G+C content (47.6%) and could be of eukaryotic origin based on the similarity of its ORF to the acyl-CoA sterol acyltransferase from Neurospora crassa.
In general, six metal resistance mechanisms are postulated: exclusion by permeability barrier, intra- and extracellular sequestration, active transport efflux pumps, enzymatic detoxification, and reduction in the sensitivity of cellular targets to metal ions (43, 47). However, most nickel resistance determinants known to date are efflux pumps isolated from cultivable bacteria, and new nickel resistance systems are still being discovered in E. coli (42), Pseudomonas putida S4 (54), Enterobacter spp. (30), and H. pylori (50).
An approach to gain insights into metal resistance mechanisms of the discovered genes was to determine the cellular nickel content of cells exposed to a high concentration of this metal. According to comparisons of the level of nickel accumulation in the nickel-resistant clones to that in the control strain, two main groups have been identified, and a representative of a previously known nickel-resistant mechanism has been assigned to each group.
One of the most important mechanisms of metal resistance is the active transport by efflux pumps, which prevents metal accumulation inside the cells. In the case of nickel resistance, some of these efflux systems have previously been described, including members of the resistance-nodulation-cell division and major facilitator superfamilies (22, 31). pSM5 and pSM12 clones could be involved in an active efflux mechanism of resistance based on the findings that (i) they accumulate less nickel than the control in the presence of this metal and (ii) their encoded products are similar to components of ABC transporters, which are involved in the export and import of many different compounds. The proteins encoded by pSM5 and pSM12 genes are distantly related to the well-studied NikABCDE and NikMNQO systems of ABC transporters involved in specific nickel uptake (35, 41). orf1 and orf2 products from pSM12 could constitute a complete functional ABC transporter since they are similar to their basic components: an inner membrane subunit and an ATPase subunit. However, the products encoded by the pSM5 insert are both truncated but still involved in nickel resistance; also, only ORF1 is similar to an ABC transporter component. An example of a functional truncated protein is the ABC transporter LmrA, a primary drug transporter in Lactococcus lactis that was engineered by removing its ATP-binding domain. The truncated protein remained active as a proton/multidrug symporter without the requirement for ATP (55). The resistance to other metals observed in pSM12 cells (cobalt and cadmium) and pSM5 cells (cadmium) led us to speculate that these genes may constitute a new group of ABC transporters related to metal efflux, although currently, this family includes only metal uptake but no metal efflux pumps (48).
It has been reported that the constitutive activation of SAT, causing elevated levels of glutathione, enables the hyperaccumulation of nickel in plants of the Thlaspi genus, including Thlaspi goesingense (16). The overexpression of the sat gene from this nickel hyperaccumulator plant in E. coli produces enhanced levels of glutathione and resistance to both nickel and cobalt but does not increase either zinc or cadmium resistance (17). orf1 of pSM11 encoded a protein that is highly similar to a bacterial SAT. Similarly to the previously reported SAT from Thlaspi goesingense, the one encoded by orf1 of pSM11 produces resistance to nickel and cobalt but not to zinc (data not shown) or cadmium (see Fig. S2 in the supplemental material). In addition, cells carrying plasmid pSM11 hyperaccumulate nickel compared to those harboring the empty vector (Fig. 5). Thus, the nickel resistance mechanism of the pSM11 clone could be similar to that produced by SAT enzymes. Nickel accumulation was also observed in the pSM13 clone. The gene harbored by pSM13 is similar to the putative conjugal transfer protein TraA and could be involved in a mechanism of resistance mediated by nickel sequestration since TraA activity is known to be dependent on the presence of the divalent cations Mg2+ and Mn2+ (29), although nickel cations in this role have not been reported.
The cross-resistance to other metal ions, such as cadmium and cobalt, observed in both categories of clones is consistent with other nickel resistance determinants reported so far, such as the above-mentioned SATs and the efflux systems encoded by cnr (cobalt-nickel resistance), ncc (nickel-cobalt-cadmium), yohM (resistance cobalt-nickel), and czn (cadmium-zinc-nickel) (21, 22, 31, 42, 46, 50).
Our knowledge about metal resistance mechanisms has been based on culturable bacteria, and in this study, we have shown how a metagenomic approach can be useful to retrieve novel genes or activities not previously related to nickel resistance. Many of these genes are completely unknown, and they will need further investigation to determine their modes of action. Future work will explore the diversity of mechanisms of resistance to other abundant metals in the microbial communities from the Rio Tinto AMD environment by prospecting these metagenomic libraries and additional libraries hosted in other bacterial species to enrich the recovery of genes whose activities may not be readily expressed in E. coli cells. In conclusion, the variety of nickel resistance genes detected using this culture-independent method can provide insight into novel metal resistance strategies developed by microorganisms in their natural environment.
Supplementary Material
Acknowledgments
We thank M. Paz Martín for the ICP-MS determination of metal concentrations and Marina Postigo for DNA sequencing. We are very grateful to Manuel Gómez for bioinformatics assistance and to Elena González-Toril for helping with the ARB program. We also thank Víctor de Lorenzo, Ricardo Amils, and Mónica M. Belinchón for critical reading of the manuscript. We are in debt to Juan Pérez-Mercader for his unconditional encouragement.
S.M. and C.G.D.F. are supported by Instituto Nacional de Técnica Aeroespacial (INTA) postdoctoral and technician fellowships, respectively. J.E.G.-P. benefits from a Ramón y Cajal contract from the Ministerio de Educación y Ciencia (MEC-Spain) at INTA. This work was supported by the Centro de Astrobiología (CSIC/INTA), associated with the NASA Astrobiology Institute, and a grant from the MEC-Spain (CGL2006-14238).
Footnotes
Published ahead of print on 3 August 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139-152. [DOI] [PubMed] [Google Scholar]
- 4.Barns, S. M., C. F. Delwiche, J. D. Palmer, and N. R. Pace. 1996. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc. Natl. Acad. Sci. USA 93:9188-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhadra, B., A. K. Nanda, and R. Chakraborty. 2006. Inducible nickel resistance in a river isolate of India phylogenetically ascertained as a novel strain of Acinetobacter junii. World J. Microbiol. Biot. 22:225-232. [Google Scholar]
- 6.Brofft, J. E., J. V. McArthur, and L. J. Shimkets. 2002. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 4:764-769. [DOI] [PubMed] [Google Scholar]
- 7.Buchwald, V. F. 1975. Handbook of iron meteorites, their history, composition and structure. University of California Press, Berkeley, CA.
- 8.Burd, G. I., D. G. Dixon, and B. R. Glick. 2000. Plant growth-promoting bacteria that decrease heavy metal toxicity in plants. Can. J. Microbiol. 46:237-245. [DOI] [PubMed] [Google Scholar]
- 9.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel, R. 2005. The metagenomics of soil. Nat. Rev. Microbiol. 3:470-478. [DOI] [PubMed] [Google Scholar]
- 11.Derakshani, M., T. Lukow, and W. Liesack. 2001. Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 67:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dopson, M., C. Baker-Austin, P. R. Koppineedi, and P. L. Bond. 2003. Growth in sulfidic mineral environments: metal resistance mechanisms in acidophilic micro-organisms. Microbiology 149:1959-1970. [DOI] [PubMed] [Google Scholar]
- 14.Druschel, G. K., B. J. Baker, T. M. Gihring, and J. F. Banfield. 2004. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem. Trans. 5:13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman, J. L., M. W. Persans, K. Nieman, C. Albrecht, W. Peer, I. J. Pickering, and D. E. Salt. 2004. Increased glutathione biosynthesis plays a role in nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Cell 16:2176-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freeman, J. L., M. W. Persans, K. Nieman, and D. E. Salt. 2005. Nickel and cobalt resistance engineered in Escherichia coli by overexpression of serine acetyltransferase from the nickel hyperaccumulator plant Thlaspi goesingense. Appl. Environ. Microbiol. 71:8627-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Dominguez, M., L. Lopez-Maury, F. J. Florencio, and J. C. Reyes. 2000. A gene cluster involved in metal homeostasis in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 182:1507-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Toril, E., E. Llobet-Brossa, E. O. Casamayor, R. Amann, and R. Amils. 2003. Microbial ecology of an extreme acidic environment, the Tinto River. Appl. Environ. Microbiol. 69:4853-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grass, G., B. Fan, B. P. Rosen, K. Lemke, H.-G. Schlegel, and C. Rensing. 2001. NreB from Achromobacter xylosoxidans 31A is a nickel-induced transporter conferring nickel resistance. J. Bacteriol. 183:2803-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grass, G., C. Grosse, and D. H. Nies. 2000. Regulation of the cnr cobalt and nickel resistance determinant from Ralstonia sp. strain CH34. J. Bacteriol. 182:1390-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:245-249. [DOI] [PubMed] [Google Scholar]
- 25.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 26.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Idris, R., R. Trifonova, M. Puschenreiter, W. W. Wenzel, and A. Sessitsch. 2004. Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl. Environ. Microbiol. 70:2667-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, D. B. 1998. Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol. Ecol. 27:307-317. [Google Scholar]
- 29.Kurenbach, B., D. Grothe, M. E. Farias, U. Szewzyk, and E. Grohmann. 2002. The tra region of the conjugative plasmid pIP501 is organized in an operon with the first gene encoding the relaxase. J. Bacteriol. 184:1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, Y.-K., H.-H. Chang, H.-J. Lee, H. Park, K. H. Lee, and M.-H. Joe. 2006. Isolation of a novel plasmid, pNi15, from Enterobacter sp. Ni15 containing a nickel resistance gene. FEMS Microbiol. Lett. 257:177-181. [DOI] [PubMed] [Google Scholar]
- 31.Liesegang, H., K. Lemke, R. A. Siddiqui, and H. G. Schlegel. 1993. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J. Bacteriol. 175:767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Archilla, A. I., I. Marin, and R. Amils. 2001. Microbial community composition and ecology of an acidic aquatic environment: the Tinto River, Spain. Microb. Ecol. 41:20-35. [DOI] [PubMed] [Google Scholar]
- 33.Majernik, A., G. Gottschalk, and R. Daniel. 2001. Screening of environmental DNA libraries for the presence of genes conferring Na+ (Li+)/H+ antiporter activity on Escherichia coli: characterization of the recovered genes and the corresponding gene products. J. Bacteriol. 183:6645-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 35.Navarro, C., L. F. Wu, and M. A. Mandrand-Berthelot. 1993. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol. Microbiol. 9:1181-1191. [DOI] [PubMed] [Google Scholar]
- 36.Nogales, B., E. R. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norris, T. B., J. M. Wraith, R. W. Castenholz, and T. R. McDermott. 2002. Soil microbial community structure across a thermal gradient following a geothermal heating event. Appl. Environ. Microbiol. 68:6300-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajkumar, M., R. Nagendran, K. J. Lee, W. H. Lee, and S. Z. Kim. 2006. Influence of plant growth promoting bacteria and Cr6+ on the growth of Indian mustard. Chemosphere 62:741-748. [DOI] [PubMed] [Google Scholar]
- 39.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhee, S.-K., X. Liu, L. Wu, S. C. Chong, X. Wan, and J. Zhou. 2004. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodionov, D. A., P. Hebbeln, M. S. Gelfand, and T. Eitinger. 2006. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188:317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigue, A., G. Effantin, and M.-A. Mandrand-Berthelot. 2005. Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J. Bacteriol. 187:2912-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouch, D. A., B. T. Lee, and A. P. Morby. 1995. Understanding cellular responses to toxic agents: a model for mechanism-choice in bacterial metal resistance. J. Ind. Microbiol. 14:132-141. [DOI] [PubMed] [Google Scholar]
- 44.Schlegel, H. G., J. P. Cosson, and A. J. M. Baker. 1991. Nickel-hyperaccumulating plants provide a niche for nickel-resistant bacteria. Bot. Acta 104:18-25. [Google Scholar]
- 45.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt, T., and H. G. Schlegel. 1994. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J. Bacteriol. 176:7045-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silver, S. 1992. Plasmid-determined metal resistance mechanisms: range and overview. Plasmid 27:1-3. [DOI] [PubMed] [Google Scholar]
- 48.Silver, S., and L. T. Phung. 2005. A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 32:587-605. [DOI] [PubMed] [Google Scholar]
- 49.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nu-cleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 50.Stahler, F. N., S. Odenbreit, R. Haas, J. Wilrich, A. H. M. V. Vliet, J. G. Kusters, M. Kist, and S. Bereswill. 2006. The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect. Immun. 74:3845-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stohs, S. J., and D. Bagchi. 1995. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18:321-336. [DOI] [PubMed] [Google Scholar]
- 52.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trajanovska, S., M. L. Britz, and M. Bhave. 1997. Detection of heavy metal ion resistance genes in gram-positive and gram-negative bacteria isolated from a lead-contaminated site. Biodegradation 8:113-124. [DOI] [PubMed] [Google Scholar]
- 54.Tripathi, V. N., and S. Srivastava. 2006. Extracytoplasmic storage as the nickel resistance mechanism in a natural isolate of Pseudomonas putida S4. Can. J. Microbiol. 52:287-292. [DOI] [PubMed] [Google Scholar]
- 55.Venter, H., R. A. Shilling, S. Velamakanni, L. Balakrishnan, and H. W. Van Veen. 2003. An ABC transporter with a secondary-active multidrug translocator domain. Nature 426:866-870. [DOI] [PubMed] [Google Scholar]
- 56.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.