Abstract
The effects of petroleum contamination on the bacterial community of a pristine microbial mat from Salins-de-Giraud (Camargue, France) have been investigated. Mats were maintained as microcosms and contaminated with no. 2 fuel oil from the wreck of the Erika. The evolution of the complex bacterial community was monitored by combining analyses based on 16S rRNA genes and their transcripts. 16S rRNA gene-based terminal restriction fragment length polymorphism (T-RFLP) analyses clearly showed the effects of the heavy fuel oil after 60 days of incubation. At the end of the experiment, the initial community structure was recovered, illustrating the resilience of this microbial ecosystem. In addition, the responses of the metabolically active bacterial community were evaluated by T-RFLP and clone library analyses based on 16S rRNA. Immediately after the heavy fuel oil was added to the microcosms, the structure of the active bacterial community was modified, indicating a rapid microbial mat response. Members of the Gammaproteobacteria were initially dominant in the contaminated microcosms. Pseudomonas and Acinetobacter were the main genera representative of this class. After 90 days of incubation, the Gammaproteobacteria were superseded by “Bacilli” and Alphaproteobacteria. This study shows the major changes that occur in the microbial mat community at different time periods following contamination. At the conclusion of the experiment, the RNA approach also demonstrated the resilience of the microbial mat community in resisting environmental stress resulting from oil pollution.
The behavior of pollutants in the environment is influenced primarily by the nature and amount of the contaminant present and the interplay among chemical, geochemical, and biological factors (31). Among biological factors, the diversity of microbial species and their metabolic capabilities constitute an important source of biocatalysis (17). The structure and dynamics of the indigenous microbial communities are major characteristics influencing biodegradation (49). The degradation of complex pollutant mixtures such as petroleum requires a combination of different bacterial taxa that, when functioning as a community, can degrade a broader spectrum of hydrocarbons than any single bacterial species alone (45, 51). Several studies of contaminated sites have shown that the impact of contamination on bacterial communities is dependent on the previous pollution history (22, 32, 51). Few investigations have been undertaken, however, to assess the impact of petroleum contamination on pristine microbial ecosystems (13, 20, 29). As a consequence, the impacts of petroleum contamination on pristine natural habitats are still poorly understood.
Microbial mats are complex microbial communities that exhibit extensive genetic and metabolic diversification, which confers a high capacity to adapt to hostile environments (15, 16, 44). The diverse microbial activities found in these mats suggest that they may have the capacity to degrade petroleum compounds and other pollutants (3). They are thus suitable models to study the impact of heavy-fuel-oil contamination on microbial communities.
Analyses based on 16S rRNA genes have been used extensively to investigate microbial diversity in contaminated environments (6, 7, 13, 20, 32, 51). These analyses provide valuable information on important changes in community structure but do not differentiate between the functional and nonfunctional components of the total bacterial populations. Since metabolically active cells usually contain higher numbers of ribosomes than quiescent cells (40), 16S rRNA levels are increasingly being used to estimate the general activity of specific organisms (12, 14, 19, 34, 38, 58). At the community level, ribosome synthesis presents limitations as a reliable growth activity indicator (36, 41, 59). However, this approach appears to be useful for targeting active microbial populations (36, 38, 39). Thus, the first response to heavy-fuel-oil pollution may be detected by changes in the diversity of 16S rRNA. An analysis of these changes would identify and characterize the populations that are firstly affected (positively or negatively) by the presence of the pollutant.
In this study, the impact of fuel oil contamination on microbial community structure and general activity has been investigated. A pristine microbial mat from the hypersaline environment of the Camargue salterns (France) was maintained as microcosms and contaminated with heavy fuel oil from the Erika. The capacity of microbial mats to degrade petroleum hydrocarbons has been reported previously (1, 7, 20, 48). In addition to determining the biodegradation potential of the Camargue microbial mats, the specific aims of this investigation were to determine (i) the impact of the heavy fuel oil on the bacterial community structure and (ii) the response of the metabolically active bacteria during long-term (1-year) incubation. The evolution of the structures of the bacterial communities following contamination was determined using a combination of terminal restriction fragment length polymorphism (T-RFLP) and clone library analyses of DNA and RNA extracts.
MATERIALS AND METHODS
Microbial mat sampling and microcosm experiment setup.
Camargue microbial mats develop in hypersaline environments (70 to 110 practical salinity units) under Mediterranean climatic conditions (9, 15). Samples of these microbial mats (1 m2 by 1.5 cm deep) were collected and equilibrated for 1 month in shallow tanks at room temperature (25°C) with an overlying 1-cm depth of 70-practical-salinity-unit synthetic seawater (KCl, 1.5 g liter−1; CaCl2·2H2O, 2.94 g liter−1; NH4Cl, 2.65 mg liter−1; NaCl, 53 g liter−1; MgSO4·7H2O, 13.28 g liter−1; MgCl2·6H2O, 10.56 g liter−1; Na2CO3, 0.53 g liter−1; and SL12 oligoelements solution [43], 1 ml liter−1). The mats were illuminated using an alternating light-darkness regimen of 8 h of darkness/16 h of artificial light (provided by eight 60-W tungsten lights, one fluorescent light emitting wavelengths of 450 and 700 to 780 nm, and one fluorescent light emitting a wavelength of 600 nm, each positioned 1 m away from the surfaces of the mats). The microbial mats were then sampled as microcosms in glass cylinders (5.5 cm in diameter) and incubated under the same laboratory conditions. After a further 15 days of equilibration, water was removed from half of the microcosms and approximately equal amounts of Erika heavy fuel oil (50 to 70 mg cm−2) were added to the surface of each microbial mat before the addition of synthetic seawater. These microcosms were used as contaminated samples, while the remainder served as controls. Incubations were carried out for 1 year. Sampling points were day 0 (just after the start of the experiment, i.e., after 30 min of incubation corresponding to the time required for the addition of heavy fuel oil and sampling) and days 5, 15, 30, 60, 90, 150, 240, and 364 of incubation. At each sampling point, one control microcosm and one contaminated microcosm were selected at random. From each microbial mat microcosm, 15 to 20 subsamples (6 mm in diameter and 1.5 cm deep) from different positions on the surface down through the full depth of the mat were collected by using Pasteur pipettes and immediately frozen at −80°C. Two subsamples corresponding to each condition and incubation time were used as duplicates to evaluate heterogeneity within microcosms. Heterogeneity among microcosms was investigated after 240 days of incubation.
Genomic DNA and RNA extractions.
Bacterial community DNA was extracted from microbial mat subsamples of approximately 2 g with the Ultraclean soil DNA isolation kit (Mo Bio Laboratories) by using the recommended protocol with minor modifications as previously described (47).
RNA from the microbial mat samples was extracted as described by Hurt et al. (26) with minor modifications. Briefly, mat subsamples (approximately 2 g) from frozen stocks were mixed with 2 g of acid-washed sea sand (Fisher Scientific) in an RNase-free pestle and mortar, followed by the addition of 1 ml of denaturing solution (4 M guanidine isothiocyanate, 10 mM Tris-HCl [pH 7.0], 1 mM EDTA, 0.5% 2-mercaptoethanol). The mixture was frozen in liquid nitrogen and ground until thawed, and the process was repeated twice. The ground samples were transferred to 50-ml polypropylene centrifuge tubes with 9 ml of extraction buffer (100 mM sodium phosphate [pH 7.0], 100 mM Tris-HCl [pH 7.0], 100 mM EDTA [pH 8.0], 1.5 M NaCl, 1% hexadecyltrimethylammonium bromide, and 2% sodium dodecyl sulfate) and incubated for 10 min at 65°C with gentle manual mixing every 5 min before centrifugation at 1,800 × g for 5 min at 20°C. The supernatants were stored at room temperature, and pellets were suspended in 5 ml of extraction buffer. Suspended pellets were incubated for 5 min at 65°C and centrifuged at 1,800 × g for 15 min at 20°C. The supernatants were poured into prechilled tubes containing 20-ml aliquots of washing solution (10:1:2 phenol [pH 4.7]-3 M sodium acetate [pH 4.7]-24:1 chloroform-isoamyl alcohol). After incubation for 10 min on ice and centrifugation for 20 min at 10,000 × g (4°C), the aqueous phases were extracted with 20 ml of 24:1 chloroform-isoamyl alcohol and centrifuged at 2,000 × g for 20 min at 20°C. The aqueous phases were precipitated with 0.6 volumes of isopropyl alcohol for 30 min at room temperature. Crude nucleic acid pellets were washed with 1.5 ml of cold ethanol, 70% (wt/vol), and centrifuged at 16,000 × g for 20 min at 4°C. After a second washing, the nucleic acid pellets were suspended in 160 μl of diethyl pyrocarbonate-treated H2O before the DNase 1 (Sigma) reaction. The quality and quantity of extracted mixed-community RNA were evaluated on a 1.2% (wt/vol) formaldehyde agarose gel. All nucleic acid extractions were performed in duplicate from two microbial mat subsamples for each condition and incubation time.
Reverse transcription, PCR amplification, and purification.
Aliquots of RNA were reverse transcribed with Moloney murine leukemia virus reverse transcriptase (New England Biolabs) and random hexamers (Eurobio) as directed by the suppliers. Reverse transcription products were either used immediately in PCRs or conserved at −20°C. To avoid the possibility of contaminating DNA in the RNA extracts used for reverse transcription-PCR, control PCR mixtures with RNAs that were not previously subjected to reverse transcription were prepared.
The optimization of PCR conditions for each sample was performed by adjusting the amount of cDNA or genomic DNA extract used to obtain a strong band on an agarose gel with no visible nonspecific product. PCR was performed as previously described (6). The primers used were the bacterial primer 8-27F (5′-AGAGTTTGATCCTGGCTCAG-3′) (30) and the universal primer 1472-1489R (5′-TACCTTGTTACGACTTCA-3′) (57) amplifying specifically the 16S ribosomal gene. Primers were tetrahydrochloro-6-carboxyfluorescein and hexachlorofluorescein labeled for T-RFLP analysis. PCR products were then purified by using the GFX DNA and gel band purification kit as directed by the supplier (Amersham Biosciences).
T-RFLP analyses.
T-RFLP analyses were conducted as previously described (16) with minor modifications. Data sets were constructed by using peaks corresponding to a level of fluorescence higher than 100 fluorescence U (5). The size of each terminal restriction fragment was determined according to the molecular weight standard TAMRA 500 (Applied Biosystems) with an acceptable error of ±1 bp. For unclearly resolved peaks such as shouldered peaks, peak heights differing by less than 1 bp were summed. Raw data were then normalized by calculating the relative peak height as a percentage of the total signal intensity of the corresponding T-RFLP profile (always higher than 15,000 fluorescent U). This approach minimizes artifacts associated with different DNA concentrations loaded for capillary electrophoresis. As suggested by Hewson et al. (24), minor changes in amplification efficiency were minimized by considering only peaks that had a relative height of >0.5%.
Data matrices were constructed by summarizing samples as statistical observations and terminal restriction fragments as variables by using normalized peak heights to represent the abundance of each terminal restriction fragment. After square root data transformation, the degrees of similarity between replicates were estimated based on Bray-Curtis similarity coefficients. Then averages of peaks from duplicate samples were calculated. Results were completed with correspondence analyses (CA) performed using the Hill algorithm with Multi-Variate Statistical Package 3.1 software (Kovach Computing Services). Shannon evenness (E) indices were calculated for each T-RFLP profile as previously described (25). Student's t tests were performed using PAleontological STatistics version 1.19 software (http://folk.uio.no/ohammer/past/).
16S rRNA cloning and sequencing.
Pooled PCR products from two RNA extraction replicates were cloned into Escherichia coli by using the TOPO cloning kit (Invitrogen). Inserts of clones were amplified using specific universal primers M13F and M13R (Eurogentec) and sequenced using the BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems). Sequences were compared to sequences deposited in the GenBank DNA database by using the BLAST algorithm (2). Alignments were achieved by using ClustalX version 1.8 and corrected with Proseq version 2.91 before the design of phylogenetic trees with Mega version 3.1. Rarefaction analysis was performed and diversity indices (dominance, Shannon diversity [H], E, and Simpson [D] indices) and coverage values were calculated as previously described by Gentile et al. (19).
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to the EMBL database and assigned accession no. AM421136 to AM421215, AM421217 to AM421227, AM421231, AM421235, AM421238, and AM421245.
RESULTS
Community evolution based on T-RFLP analysis of 16S rRNA genes.
The T-RFLP fingerprints of bacterial communities incubated for different periods of time after contamination with Erika heavy fuel oil were compared with those of uncontaminated microcosms. Bacterial T-RFLP fingerprints consist of an assortment of 16S rRNA gene terminal restriction fragments, each of them defined as an operational taxonomic unit (OTU). After normalization, the average richness diversity was 44.8 ± 8.2 OTUs. E indices ranged between 0.67 and 0.96. Interestingly, the contaminated microcosm sampled at 90 days showed the lowest index, while the highest index was obtained for the control microcosm at 90 days. Bray-Curtis indices of similarity between duplicates (average of 0.67 ± 0.09) and the relationships of duplicates in the CA (data not shown) supported our experimental procedure. The validity of our procedure was further confirmed by the similarity between two pairs of subsamples from different microcosms analyzed after 240 days of incubation (average Bray-Curtis similarity index of 0.65 ± 0.05).
CA based on T-RFLP fingerprints (Fig. 1) showed that bacterial communities from contaminated microbial mat samples incubated for up to 30 days were grouped with all those from the control samples in a main “control cluster” (average Bray-Curtis similarity index of 0.40 ± 0.08). Samples from contaminated microbial mats after 60 and 90 days of exposure (60P and 90P) were distant from this main group, distributed along axis 2 (11% of the variance) at either extremity. The Bray-Curtis indices of similarity between the fingerprints of the control cluster and the contaminated microcosms were 0.31 and 0.25 for samples 60P and 90P, respectively. After 150 and 240 days of incubation, bacterial communities from the polluted microbial mat samples (150P and 240P) emerged at the extremity of the control cluster. Bray-Curtis indices of similarity between the fingerprints of the control cluster and the contaminated samples were 0.48 and 0.54, respectively. The bacterial community from the polluted microbial mat sample incubated for 364 days (364P) clustered with those from the control samples. T-RFLP profiles of contaminated and control samples at the beginning (day 0) and at the end (day 364) of the experiment presented an average Bray-Curtis similarity index of 0.42 ± 0.08.
FIG. 1.
CA based on T-RFLP data (16S rRNA genes) obtained from control (white triangles) and polluted (black circles) microcosms. The data analyzed correspond to averages for duplicate samples of HaeIII-digested 16S rRNA genes yielding 5′-end T-RFLP patterns. Each number corresponds to the day of sampling relative to the start of the experiment. Encircled symbols correspond to bacterial communities from control samples and from contaminated samples incubated up to 30 days or more than 364 days (the control cluster). Percentage values represent Bray-Curtis similarity among these samples.
Evolution of active bacterial communities based on 16S rRNA analysis.
Metabolically active bacterial communities were analyzed by T-RFLP based on 16S rRNA after selected periods of incubation: at the beginning of the experiment, when bacterial community structure appeared to be unaffected by heavy-fuel-oil contamination (immediately after fuel contamination and 5 days after); when conspicuous divergence between the control and contaminated microbial mats was observed (day 90); and at the end of the experiment (day 364), when the initial bacterial community structure was recovered. The first sampling point (day 0) corresponds to samples (0P and 0C) collected immediately after the experiment was set up (i.e., approximately 30 min after heavy-fuel-oil contamination).
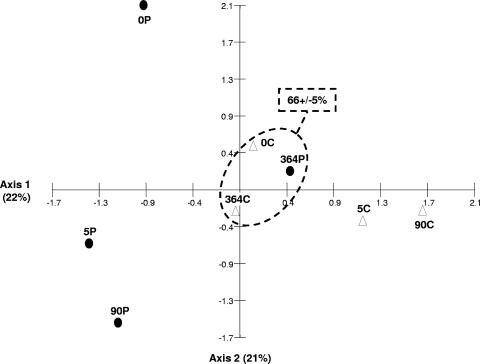
The average Bray-Curtis index of similarity between the fingerprints of the different RNA extraction duplicates was 0.68 ± 0.09. As for DNA analysis, this result supports our experimental procedure. CA based on the T-RFLP profiles showed modifications of the structure of the metabolically active bacterial community after heavy-fuel-oil contamination (Fig. 2). This divergence was observable immediately (within about 30 min, as demonstrated by sample 0P versus sample 0C) and increased after 5 and 90 days of incubation (Bray-Curtis similarity indices, 0.54 and 0.52, respectively). The bacterial community of the contaminated microbial mat at the end of the incubation period (sample 364P) was grouped with those of 0C and 364C. The average Bray-Curtis index of similarity among samples 0C, 364C, and 364P was 0.66 ± 0.05.
FIG. 2.
CA based on T-RFLP data (reverse-transcribed 16S rRNA) obtained from control (white triangles) and polluted (black circles) microcosms. The data analyzed correspond to averages for duplicate samples of HaeIII-digested reverse-transcribed 16S rRNA yielding 5′-end T-RFLP patterns. Each number corresponds to the day of sampling relative to the start of the experiment. Encircled symbols correspond to metabolically active bacterial communities from a control sample at the beginning of the experiment and from control and contaminated samples at the end of the experiment. Percentage values represent Bray-Curtis similarity among these samples.
With RNA-based analyses, 47.8 ± 5.3 OTUs were detected on T-RFLP profiles after normalization. E indices ranged between 0.83 and 0.91 (average, 0.87 ± 0.02); no differences between contaminated and control microcosms sampled on the same days were observed. Although DNA- and RNA-based analyses, which used different extraction protocols, showed different T-RFLP profiles (data not shown), some OTUs were detected in both analyses, allowing the comparison of the methods. The numbers of copies of 16S rRNA genes and 16S rRNA transcripts would constitute the first-level cause of differences observed in a single-time-point analysis. However, comparing the differences observed by the two approaches in a temporal evolution may give valuable information on how contamination affects a bacterial population. For example, an OTU of 67 bp was detected at a low level in a DNA-based analysis under both contaminated and control conditions (Student's t test; P for DNA-based results [PDNA], 0.1419) whereas, in the RNA-based analysis, the relative abundance of this OTU was significantly greater (Student's t test; PRNA, 0.0296) in contaminated microcosms (4.5 to 13%) than in the control microcosms (undetectable to 5.9%). In contrast, other OTUs presented similar contamination responses in both types of analysis. For example, an OTU of 64 bp was detected at lower levels in contaminated samples than in control samples in both DNA-based and RNA-based analyses. However, these differences between control and contaminated conditions were not statistically significant (Student's t test; PDNA, 0.2011, and PRNA, 0.1493). Predictive identification with the Ribosomal Data Project (http://rdp8.cme.msu.edu/html/analyses.html) matched the OTU of 67 bp to several bacterial genera (Corynebacterium, Mycobacterium, and Rhodococcus), of which related members are known to be able to degrade hydrocarbons (33, 55). The OTU of 67 bp could not be identified.
Characterization of the active bacterial communities based on 16S rRNA analysis.
To provide further information on the microorganisms involved in the response to heavy-fuel-oil contamination, 16S rRNA clone libraries were constructed from samples showing contrasting bacterial community structures as revealed by T-RFLP analyses. Sample 0C was chosen as the reference for an unpolluted metabolically active bacterial community structure, sample 0P was chosen as the reference for a community exhibiting an early impact of heavy-fuel-oil contamination, and sample 90P was chosen as the reference for a community showing late impacts. Rarefaction analysis applied to clone libraries showed similar patterns (data not shown) without saturation. Table 1 shows the dominance, H, E, and D indices and the coverage values calculated for each library. Coverage values ranged between 0.61 for the 90P library and 0.83 for the 0C library. The H index ranged from 2.592 for the 0P library to 3.069 for the 90P library, and the E index was greater for the 90P (0.7686) and 0C (0.7996) libraries and than for the 0P library (0.4770), showing a lower level of diversity among the metabolically active bacteria in the contaminated microcosm at the start of the experiment than among those in the 0C and 90P samples.
TABLE 1.
Characteristics of clone librariesa
| Sample | Total no. of clones | No. of taxa | No. of singletonsb | Dominance index | Coverage | H index | E index | D index |
|---|---|---|---|---|---|---|---|---|
| 0C | 65 | 26 | 11 | 0.0579 | 83 | 3.034 | 0.7996 | 0.9420 |
| 0P | 77 | 28 | 20 | 0.1469 | 74 | 2.592 | 0.4770 | 0.8531 |
| 90P | 51 | 28 | 20 | 0.0595 | 61 | 3.069 | 0.7686 | 0.9404 |
Diversity indices calculated for the three clone libraries from control and heavy-fuel-oil-polluted microcosms just after the launch of the experiment (samples 0C and 0P, respectively) and a polluted microcosm 90 days after contamination (sample 90P).
Singletons were those taxa represented by a single clone.
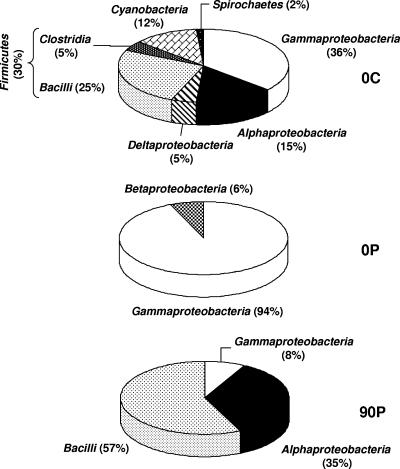
The analysis of sequences obtained from the control microcosm at the beginning of the experiment (0C) highlighted the taxonomic diversity of the microbial mats (Fig. 3). Sequences related to the classes Alphaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria were dominant (56%) in this habitat. Sequences related to members of the phyla “Cyanobacteria” (12%), “Firmicutes” (30%), and “Spirochaetes” (2%), classically found in microbial mats, were also detected (15). Only sequences related to the Betaproteobacteria (6%) and the Gammaproteobacteria (94%) were present immediately following oil contamination. Three months after contamination (sample 90P), the contaminated microcosm was dominated by sequences related to “Bacilli” (57%) and Alphaproteobacteria (35%), while sequences related to members of the Gammaproteobacteria were present at a low level (8%) and Betaproteobacteria-related sequences were undetected.
FIG. 3.
Relative abundance (%) of phylogenetic groups in 16S rRNA clone libraries from control and contaminated microcosms at the beginning of the experiment (0C and 0P, respectively) and from a contaminated microcosm after 90 days of incubation (90P).
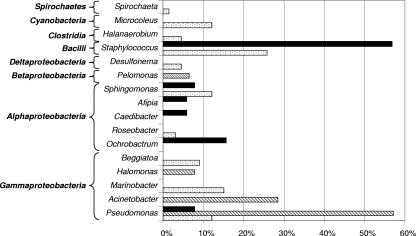
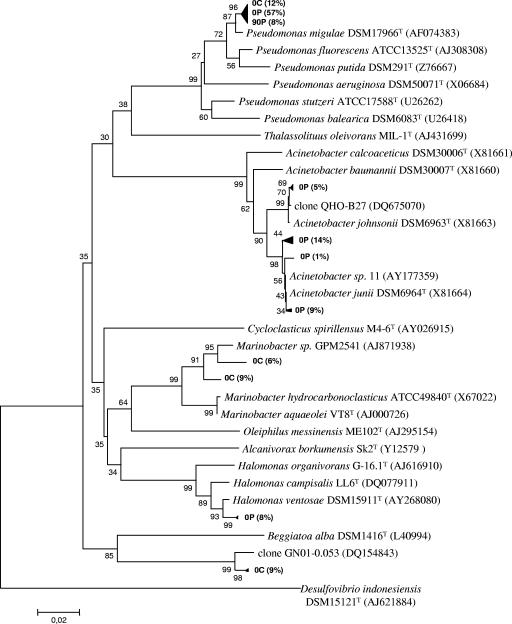
Except for the Alphaproteobacteria and Gammaproteobacteria classes, each class was represented by a single genus (Fig. 4). Gammaproteobacteria were dominated by species of Pseudomonas (77%) and Acinetobacter (29%), two genera well known to contain hydrocarbon-degrading species (18, 22). These two genera were particularly dominant in the contaminated-microcosm 0P library (57 and 29%, respectively). Furthermore, 24% of 0P sequences affiliated with the genus Acinetobacter were related (>99% homology) to Acinetobacter sp. strain 11, a strain presenting phenanthrene degradation activity, and 5% of such sequences corresponded (>99% homology) to a clone (clone QHO-B27) from a microbial community associated with petroleum reservoirs (Fig. 5). Eight percent of 16S rRNA sequences from 0P were also affiliated (>98% homology) with Halomonas ventosae, known to produce exopolysaccharides and frequently used as a bioemulsifier in hydrocarbon biodegradation (27, 37).
FIG. 4.
Phylotype distribution (%) of reverse-transcribed 16S rRNA sequences found in the clone libraries from the different microcosm samples: control (dotted bars) and polluted (striped bars) microcosms sampled just after heavy-fuel-oil contamination and a polluted microcosm sampled 90 days after contamination (solid bars).
FIG. 5.
Phylogenetic tree based on the analysis of reverse-transcribed 16S rRNA cloned sequences (673 bp aligned) from control and contaminated microcosms sampled at the beginning of the experiment (0C and 0P, respectively) and a contaminated microcosm sampled after 90 days of incubation (90P). Only clones (in bold type) and their closest relative sequences affiliated with Gammaproteobacteria are shown. The numbers in parentheses indicate the percentage of clones (difference, ≤1 bp) in the corresponding microcosm library. The tree was rooted with 16S rRNA gene sequences of the strain Desulfovibrio indonesiensis DSM15121T. The scale bar corresponds to 0.02 substitutions per nucleotide position. Percentages of 100 bootstrap resamplings that supported the branching orders in each analysis are shown above or near the relevant nodes.
DISCUSSION
The stability of the community structure in contaminated microbial mats during the first month of incubation illustrates the resistance of the mats to heavy-fuel-oil contamination. This contamination appears to have a delayed effect, occurring after 60 to up to 90 days, on the bacterial community structure. A latent period has been described previously for other pristine environments, e.g., 30 days for microbial mats (6) and 60 to 90 days for soils (11, 13). However, early changes in community structure were not evident using the T-RFLP approach based on 16S rRNA genes since DNA-based analyses also detected inactive microorganisms. A combination of 16S rRNA gene and 16S rRNA analyses is therefore essential to obtain a more reliable vision of the evolution of microbial communities, especially for the earliest modifications. Previous studies have demonstrated an increase in bacterial activity within a few minutes after stress treatments such as nutritional starvation or heat shock by using 16S rRNA-based analyses (21, 54). In our study, the modification of the structure of the metabolically active bacterial community just after heavy-fuel-oil contamination and during at least 90 days of incubation was well illustrated. This divergence in the structures of the bacterial communities may be due to increased activity of some bacterial species as well as rapid inactivation of petroleum-sensitive populations. The fuel compounds may either be used as organic substrates, have toxic effects, or induce prophages (10) or simply change the physicochemical conditions of the environment (4), inducing many stresses for other bacteria.
The Bray-Curtis indices of similarity between DNA-based T-RFLP profiles of control and contaminated samples after 150 days of incubation were higher than the average value for the control cluster. Microbial mats are considered to return to the unimpacted community structure after 150 days of incubation. The initial bacterial community structure in both contaminated and control microcosms was recovered after 1 year of incubation. With RNA-based analyses, similarities between control and contaminated metabolically active bacterial communities after 364 days of incubation were also observed. These results clearly illustrate the resilience of microbial mats after petroleum contamination. This capacity has been demonstrated previously by the similarity between pre- and postpollution bacterial community structures in several petroleum bioremediation studies involving biostimulation by the addition of fertilizer (42, 50). In our study, no biostimulation processes were used. However, CO2 fixation and N2 fixation as major ecophysiological processes occurring in microbial mats (46) allow these structures to be self-sufficient, a fortiori, in a phosphorus-enriched environment like seawater. Like solar-driven ecosystems, they have few growth requirements and no obvious benefit from biostimulation. Furthermore, the resilience of the system could be explained by biological and chemical pressures applied to the studied microbial mat. Interdependence between different bacterial groups (56) and selective pressure linked to the hypersaline environment could impose an adequate and stable community structure.
Discrepancies between analyses based on rRNA and those based on rRNA genes can be explained by the use of different extraction protocols to obtain the nucleic acids (RNA and DNA). However, other authors using the same extraction protocols for both DNA and RNA have previously observed similar differences (19, 35). In the present study, the comparison was limited to monitoring the evolution of OTUs detected in both DNA- and RNA-based analyses. This method allowed us to link quantitative and qualitative aspects of the response for a specific OTU after heavy-fuel-oil contamination. Comparisons of T-RFLP data and predictive identifications suggested that an OTU of 67 bp presented a metabolic capacity to cope with the presence of heavy-fuel-oil contamination. This OTU may potentially play a role in the microbial response after petroleum exposure despite the failure of the DNA approach to detect it as a representative of a major population in the contaminated microbial mats. Inversely, the low level of detection of the OTU of 64 bp in both analyses suggests that the corresponding population was structurally and metabolically sensitive to oil stress and/or associated with modifications of environmental parameters (e.g., oxygen limitation and light reduction, etc.).
An immediate decrease in the diversity of metabolically active bacteria in contaminated microcosms was clearly observed using clone library analysis. This result is supported by diversity indices (Table 1). Several studies have previously exposed a reduction in the diversity of bacterial communities associated with the selection of hydrocarbon-degrading species after petroleum exposure (8, 11, 51, 53). In some cases, an increase in bacterial diversity has also been reported (28, 60). The response to the presence of the pollutant is characterized by the replacement of classical microbial-mat and/or hypersaline-environment organisms by Gammaproteobacteria just after the contamination. Previous studies have reported that the dominance of Gammaproteobacteria is a characteristic of bacterial communities inhabiting environments contaminated with petroleum compounds (51, 52); for more details, see the article by Hernandez-Raquet et al. (23). After this immediate response, the supremacy of Gammaproteobacteria was displaced by the development of “Bacilli” and Alphaproteobacteria in fuel-exposed microcosms. These classes appear to play an important role in the late response of microbial mats to petroleum contamination. This conclusion is consistent with the findings of previous studies reporting rapid and strong selection for Gammaproteobacteria and replacement by Alphaproteobacteria in oil-contaminated microcosms within a few days (51).
There is a considerable amount of literature on microbial hydrocarbon degradation and petroleum impact on bacterial communities (6, 7, 13, 42, 53). The capacity of microbial mats to degrade petroleum compounds is well known (1, 20, 48). Given these previous studies, the present work focuses specifically on the impact of heavy-fuel-oil contamination on pristine microbial mat communities. The combination of 16S rRNA gene and 16S rRNA analyses shows a more accurate representation of the bacterial response to the pollution than a single approach. To the best of our knowledge, our study uniquely integrated this combination to monitor the evolution of biodiversity in a pristine microbial mat after heavy-fuel-oil contamination. The main observation of this study is the high degree of resilience of the microbial mat community after contamination. This impact was first characterized by late changes (occurring after more than 1 month of incubation) in the microbial community structure as demonstrated by 16S rRNA gene analyses. However, the metabolically active bacterial community showed an almost immediate response to the presence of the pollutant, a response characterized by the dominance of Gammaproteobacteria genera known for their hydrocarbon biodegradation potential. “Bacilli” and Alphaproteobacteria subsequently appeared as actors involved in the response of the microbial mat to petroleum contamination. These evolutions confirm that bacterial communities of microbial mats may adapt to fuel contamination and/or associated environmental modifications. Be that as it may, the mat recovers its original structure and metabolically active community 1 year after the contamination. Further studies establishing links between function and community structure will provide evidence on the environmental functioning of bacterial populations after heavy-fuel-oil pollution.
Acknowledgments
We acknowledge financial support by the European Commission (MATBIOPOL project, grant no. EVK3-CT-1999-00010, and FACEiT project, grant no. 018391), the Departmental Government Council of Pyrénées-Atlantiques (France), and the Ministère de l'Ecologie et du Développement Durable (LIT'EAU/Erika project, no. 01/1213857, and PNETOX, no. CV04000147). S.B. was supported partly by a doctoral grant from the Aquitaine (France) Regional Government Council.
We are grateful to the company of Salins du Midi at Salins-de-Giraud for facilitating access to the salterns for sampling and field experiments. We are especially indebted to Rodney Herbert for careful reading of the English and for helpful comments on the paper.
Footnotes
Published ahead of print on 17 August 2007.
REFERENCES
- 1.Abed, R. M. M., and J. Köster. 2005. The direct role of aerobic heterotrophic bacteria associated with cyanobacteria in the degradation of oil compounds. Int. Biodeterior. Biodegradation 55:29-37. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bender, J., and P. Phillips. 2004. Microbial mats for multiple applications in aquaculture and bioremediation. Bioresour. Technol. 94:229-238. [DOI] [PubMed] [Google Scholar]
- 4.Berge, J. A., R. G. Lichtenthaler, and F. Oreld. 1987. Hydrocarbon depuration and abiotic changes in artificially oil contaminated sediment in the subtidal. Estuar. Coast. Shelf Sci. 24:567-568. [Google Scholar]
- 5.Blackwood, C. B., T. Marsh, S. H. Kim, and E. A. Paul. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bordenave, S., A. Fourçans, S. Blanchard, M. S. Goñi, P. Caumette, and R. Duran. 2004. Structure and functional analyses of bacterial communities changes in microbial mats following petroleum exposure. Ophelia 58:195-203. [Google Scholar]
- 7.Bordenave, S., A. Fourçans, M. Goñi-Urriza, R. Guyoneaud, R. Grimaud, P. Caumette, R. Duran, R. Jézéquel, F. X. Merlin, T. Fourel, and H. Budzinski. 2004. Degradation of the “Erika” oil. Aquat. Living Resour. 17:261-267. [Google Scholar]
- 8.Bundy, J. G., G. I. Paton, and C. D. Campbell. 2002. Microbial communities in different soil types do not converge after diesel contamination. J. Appl. Microbiol. 92:276-288. [DOI] [PubMed] [Google Scholar]
- 9.Caumette, P., R. Matheron, N. Raymond, and J.-C. Relexans. 1994. Microbial mats in the hypersaline ponds of Mediterranean salterns (Salins-de-Giraud, France). FEMS Microbiol. Ecol. 13:273-286. [Google Scholar]
- 10.Cochran, P. K., C. A. Kellogg, and J. H. Paul. 1998. Prophage induction of indigenous marine lysogenic bacteria by environmental pollutants. Mar. Ecol. Prog. Ser. 164:125-133. [Google Scholar]
- 11.Duarte, G. F., A. S. Rosado, L. Seldin, W. De Araujo, and J. D. Van Elsas. 2001. Analysis of bacterial community structure in sulfurous-oil-containing soils and detection of species carrying dibenzothiophene desulfurization (dsz) genes. Appl. Environ. Microbiol. 67:1052-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duineveld, B. M., G. A. Kowalchuk, A. Keijzer, J. D. Van Elsas, and J. A. Van Veen. 2001. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl. Environ. Microbiol. 67:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, F. F., A. S. Rosado, G. V. Sebastian, R. Casella, P. L. O. A. Machado, C. Holmström, S. Kjelleberg, J. D. Van Elsas, and L. Seldin. 2004. Impact of oil contamination and biostimulation on the diversity of indigenous bacterial communities in soil microcosms. FEMS Microbiol. Ecol. 49:295-305. [DOI] [PubMed] [Google Scholar]
- 14.Felske, A., A. Wolterink, R. Van Lis, W. M. De Vos, and A. D. Akkermans. 2000. Response of a soil bacterial community to grassland succession as monitored by 16S rRNA levels of the predominant ribotypes. Appl. Environ. Microbiol. 66:3998-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fourçans, A., T. G. De Oteyza, A. Wieland, A. Sole, E. Diestra, J. Van Bleijswijk, J. O. Grimalt, M. Kühl, I. Esteve, G. Muyzer, P. Caumette, and R. Duran. 2004. Characterization of functional bacterial groups in a hypersaline microbial mat community (Salins-de-Giraud, Camargue, France). FEMS Microbiol. Ecol. 51:55-70. [DOI] [PubMed] [Google Scholar]
- 16.Fourçans, A., A. Sole, E. Diestra, A. Ranchou-Peyruse, I. Esteve, P. Caumette, and R. Duran. 2006. Vertical migration of phototrophic bacterial populations in a hypersaline microbial mat from Salins-de-Giraud (Camargue, France). FEMS Microbiol. Ecol. 57:367-377. [DOI] [PubMed] [Google Scholar]
- 17.Galvão, T. C., V. De Lorenzo, and W. W. Mohn. 2005. Exploring the microbial biodegradation and biotransformation gene pool. Trends Biotechnol. 23:497-506. [DOI] [PubMed] [Google Scholar]
- 18.Geissdorfer, W., R. G. Kok, A. Ratajczak, K. J. Hellingwerf, and W. Hillen. 1999. The genes rubA and rubB for alkane degradation in Acinetobacter sp. strain ADP1 are in an operon with estB, encoding an esterase, and oxyR. J. Bacteriol. 181:4292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentile, G., L. Giuliano, G. D'Auria, F. Smedile, M. Azzaro, M. De Domenico, and M. M. Yakimov. 2006. Study of bacterial communities in Antarctic coastal waters by a combination of 16S rRNA and 16S rDNA sequencing. Environ. Microbiol. 8:2150-2161. [DOI] [PubMed] [Google Scholar]
- 20.Grötzschel, S., J. Köster, R. M. M. Abed, and D. De Beer. 2002. Degradation of petroleum model compounds immobilized on clay by a hypersaline microbial mat. Biodegradation 13:273-283. [DOI] [PubMed] [Google Scholar]
- 21.Hansen, M. C., A. K. Nielsen, S. Molin, K. Hammer, and M. Kilstrup. 2001. Changes in rRNA levels during stress invalidate results from mRNA blotting: fluorescence in situ rRNA hybridization permits renormalization for estimation of cellular mRNA levels. J. Bacteriol. 183:4747-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Head, I. M., D. M. Jones, and W. F. Röling. 2006. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4:173-182. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez-Raquet, G., H. Budzinski, P. Caumette, P. Dabert, K. Le Ménach, G. Muyzer, and R. Duran. 2006. Molecular diversity studies of bacterial communities of oil polluted microbial mats from the Etang de Berre (France). FEMS Microbiol. Ecol. 58:550-562. [DOI] [PubMed] [Google Scholar]
- 24.Hewson, I., and J. A. Fuhrman. 2004. Richness and diversity of bacterioplankton species along an estuarine gradient in Moreton Bay, Australia. Appl. Environ. Microbiol. 70:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill, T. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 26.Hurt, R. A., X. Qiu, L. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer, A., K. Mody, and B. Jha. 2006. Emulsifying properties of a marine bacterial exopolysaccharide. Enzyme Microb. Technol. 38:220-222. [Google Scholar]
- 28.Juck, D., T. Charles, L. G. Whyte, and C. W. Greer. 2000. Polyphasic microbial community analysis of petroleum hydrocarbon-contaminated soils from two northern Canadian communities. FEMS Microbiol. Ecol. 33:241-249. [DOI] [PubMed] [Google Scholar]
- 29.Kaufmann, K., M. Christophersen, A. Buttler, H. Harms, and P. Höhener. 2004. Microbial community response to petroleum hydrocarbon contamination in the unsaturated zone at the experimental field site Værlφse, Denmark. FEMS Microbiol. Ecol. 48:387-399. [DOI] [PubMed] [Google Scholar]
- 30.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackenbradt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 31.Leahy, J. G., and R. R. Colwell. 1990. Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 54:305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacNaughton, S. J., J. R. Stephen, A. D. Venosa, G. A. Davis, Y. J. Chang, and D. C. White. 1999. Microbial population changes during bioremediation of an experimental oil spill. Appl. Environ. Microbiol. 65:3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGowan, L., R. Herbert, and G. Muyzer. 2004. A comparative study of hydrocarbon degradation by Marinobacter sp., Rhodococcus sp. and Corynebacterium sp. isolated from different mat systems. Ophelia 58:271-281. [Google Scholar]
- 34.Miskin, I. P., P. Farrimond, and I. M. Head. 1999. Identification of novel bacterial lineages as active members of microbial populations in a freshwater sediment using a rapid RNA extraction procedure and RT-PCR. Microbiology 145:1977-1987. [DOI] [PubMed] [Google Scholar]
- 35.Moeseneder, M. M., J. M. Arrieta, and G. J. Herndl. 2005. A comparison of DNA- and RNA-based clone libraries from the same marine bacterioplankton community. FEMS Microbiol. Ecol. 51:341-352. [DOI] [PubMed] [Google Scholar]
- 36.Molin, S., and M. Givskov. 1999. Application of molecular tools for in situ monitoring of bacterial growth activity. Environ. Microbiol. 1:383-391. [DOI] [PubMed] [Google Scholar]
- 37.Mulligan, C. N. 2005. Environmental applications for biosurfactants. Environ. Pollut. 133:183-198. [DOI] [PubMed] [Google Scholar]
- 38.Nogales, B., E. R. Moore, W. R. Abraham, and K. N. Timmis. 1999. Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl-polluted moorland soil. Environ. Microbiol. 1:199-212. [DOI] [PubMed] [Google Scholar]
- 39.Nogales, B., E. R. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nomura, M., R. Gourse, and G. Baughman. 1984. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 53:75-117. [DOI] [PubMed] [Google Scholar]
- 41.O'Donnell, A. G., and H. E. Görres. 1999. 16S rDNA methods in soil microbiology. Curr. Opin. Biotechnol. 10:225-229. [DOI] [PubMed] [Google Scholar]
- 42.Ogino, A., H. Koshikawa, T. Nakahara, and H. Uchiyama. 2001. Succession of microbial communities during a biostimulation process as evaluated by DGGE and clone library analyses. J. Appl. Microbiol. 91:625-635. [DOI] [PubMed] [Google Scholar]
- 43.Overmann, J., U. Fischer, and N. Pfennig. 1992. A new purple sulfur bacterium from saline littoral sediments, Thiorhodovibrio winogradskyi gen. nov. and sp. nov. Arch. Microbiol. 157:329-335. [Google Scholar]
- 44.Paerl, H. W., J. L. Pinckney, and T. F. Steppe. 2000. Cyanobacterial-bacterial mat consortia: examining the functional unit of microbial survival and growth in extreme environments. Environ. Microbiol. 2:11-26. [DOI] [PubMed] [Google Scholar]
- 45.Pelz, O., M. Tesar, R. M. Wittich, E. R. B. Moore, K. N. Timmis, and W. R. Abraham. 1999. Towards elucidation of microbial community metabolic pathways: unravelling the network of carbon sharing in a pollutant-degrading bacterial consortium by immunocapture and isotopic ratio mass spectrometry. Environ. Microbiol. 1:167-174. [DOI] [PubMed] [Google Scholar]
- 46.Pinckney, J. L., and H. W. Paerl. 1997. Anoxygenic photosynthesis and nitrogen fixation by a microbial mat community in a Bahamian hypersaline lagoon. Appl. Environ. Microbiol. 63:420-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Precigou, S., P. Goulas, and R. Duran. 2001. Rapid and specific identification of nitrile hydratase (NHase)-encoding genes in soil samples by polymerase chain reaction. FEMS Microbiol. Lett. 204:155-161. [DOI] [PubMed] [Google Scholar]
- 48.Raghukumar, C., V. Vipparty, J. J. David, and D. Chandramohan. 2001. Degradation of crude oil by marine cyanobacteria. Appl. Microbiol. Biotechnol. 57:433-436. [DOI] [PubMed] [Google Scholar]
- 49.Rhee, S. K., X. Liu, L. Wu, S. C. Chong, X. Wan, and J. Zhou. 2004. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Röling, W. F., M. G. Milner, D. M. Jones, F. Fratepietro, R. P. Swannell, F. Daniel, and I. M. Head. 2004. Bacterial community dynamics and hydrocarbon degradation during a field-scale evaluation of bioremediation on a mudflat beach contaminated with buried oil. Appl. Environ. Microbiol. 70:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Röling, W. F., M. G. Milner, D. M. Jones, K. Lee, F. Daniel, R. J. Swannell, and I. M. Head. 2002. Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl. Environ. Microbiol. 68:5537-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi, Y., M. D. Zwolinski, M. E. Schreiber, J. M. Bahr, G. W. Sewell, and W. J. Hickey. 1999. Molecular analysis of microbial community structures in pristine and contaminated aquifers: field and laboratory microcosm experiments. Appl. Environ. Microbiol. 65:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song, H. G., and R. Bartha. 1990. Effects of jet fuel spills on the microbial community of soil. Appl. Environ. Microbiol. 56:646-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tolker-Nielsen, T., M. H. Larsen, H. Kyed, and S. Molin. 1997. Effects of stress treatments on the detection of Salmonella typhimurium by in situ hybridization. Int. J. Food Microbiol. 35:251-258. [DOI] [PubMed] [Google Scholar]
- 55.Uyttebroek, M., J. J. Ortega-Calvo, P. Breugelmans, and D. Springael. 2006. Comparison of mineralization of solid-sorbed phenanthrene by polycyclic aromatic hydrocarbon (PAH)-degrading Mycobacterium spp. and Sphingomonas spp. Appl. Microbiol. Biotechnol. 72:829-836. [DOI] [PubMed] [Google Scholar]
- 56.Van Gemerden, H. 1993. Microbial mats: a joint venture. Mar. Geol. 113: 3-25. [Google Scholar]
- 57.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weller, R., J. W. Weller, and D. M. Ward. 1991. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed cDNA. Appl. Environ. Microbiol. 57:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wellington, E. M. H., A. Berry, and M. Krsek. 2003. Resolving functional diversity in relation to microbial community structure in soil: exploiting genomics and stable isotope probing. Curr. Opin. Microbiol. 6:295-301. [DOI] [PubMed] [Google Scholar]
- 60.Zucchi, M., L. Angiolini, S. Borin, L. Brusetti, N. Dietrich, C. Gigliotti, P. Barbieri, C. Sorlini, and D. Daffonchio. 2003. Response of bacterial community during bioremediation of an oil-polluted soil. J. Appl. Microbiol. 94:248-257. [DOI] [PubMed] [Google Scholar]