Abstract
We have determined the functions of the enzymes encoded by the lnpB, lnpC, and lnpD genes, located downstream of the lacto-N-biose phosphorylase gene (lnpA), in Bifidobacterium longum JCM1217. The lnpB gene encodes a novel kinase, N-acetylhexosamine 1-kinase, which produces N-acetylhexosamine 1-phosphate; the lnpC gene encodes UDP-glucose hexose 1-phosphate uridylyltransferase, which is also active on N-acetylhexosamine 1-phosphate; and the lnpD gene encodes a UDP-glucose 4-epimerase, which is active on both UDP-galactose and UDP-N-acetylgalactosamine. These results suggest that the gene operon lnpABCD encodes a previously undescribed lacto-N-biose I/galacto-N-biose metabolic pathway that is involved in the intestinal colonization of bifidobacteria and that utilizes lacto-N-biose I from human milk oligosaccharides or galacto-N-biose from mucin sugars.
Intestinal colonization by bifidobacteria is considered to be beneficial to human health. Bifidobacteria secrete a variety of hydrolytic enzymes that act on oligosaccharides, enabling them to grow on various prebiotic oligosaccharides that function to increase probiotic bacteria in vivo (6, 7, 22). Lacto-N-biose phosphorylase (LNBP) (EC 2.4.1.211) has been found in cell extracts of Bifidobacterium bifidum (8). The enzyme specifically phosphorolyzes galacto-N-biose (Galβ1→3GalNAc) (GNB) and lacto-N-biose I (Galβ1→3GlcNAc) (LNB) to yield α-d-galactose 1-phosphate (Gal1P) and the corresponding N-acetylhexosamine, suggesting that this enzyme might play a key role in metabolizing mucin sugars (8). After purifying LNBP from B. bifidum, we found that its partial amino acid sequences (13) were highly homologous with that of the BL1641 protein of Bifidobacterium longum NCC2705, whose entire genomic sequence was available (21). Then, we cloned the LNBP gene, lnpA, from the type strain B. longum JCM1217, whose sequence was almost identical to that of the BL1641 gene (13). After subsequently cloning and sequencing the LNBP gene from B. bifidum (19), we concluded that the deduced amino acid sequences of these LNBPs shared no significant identity with those of any other proteins with known functions.
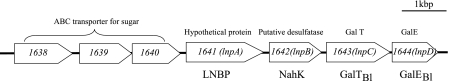
The gene cluster of B. longum NCC2705 consists of the open reading frames BL1638 to BL1644 (Fig. 1). The BL1638 to BL1640 genes were annotated to encode component proteins of an ATP-binding-cassette-type sugar transporter. The BL1641 protein was determined to be LNBP (13), whereas the BL1642, BL1643, and BL1644 proteins were annotated to be mucin desulfatase, galactose-1-phosphate uridylyltransferase (GalT2) (EC 2.7.7.10), and UDP-glucose 4-epimerase (GalE) (EC 5.1.3.2), respectively. The BL1643 and BL1644 proteins are likely related to the Leloir pathway, which sends galactose to glycolysis (Fig. 2). Promoter-like sequences were observed upstream of the BL1638 gene and between the BL1640 and BL1641 genes, and a palindromic sequence was detected downstream of the BL1644 gene. Since the full set of enzymes involved in the Leloir pathway was not found as a gene cluster in B. longum NCC2705, the cluster containing the LNBP gene (lnpA) was presumed to code for the major galactose metabolic pathway in this strain (13).
FIG. 1.
Schematic structure of the gene cluster between the BL1638 and BL1644 genes.
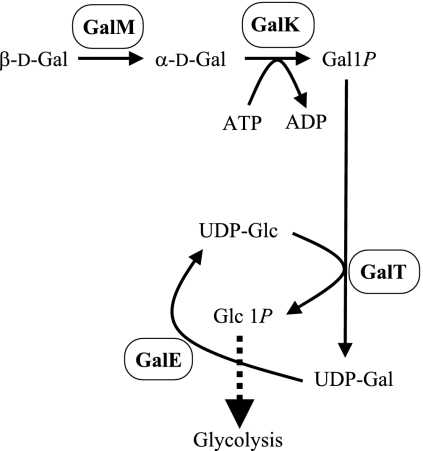
FIG. 2.
Scheme of the Leloir pathway.
To understand the role of LNBP in the growth of bifidobacteria in the human intestine, it is important to find the possible origins of GNB and LNB in humans. We think that GNB and LNB are abundantly available in mucin and human milk, respectively, as described below.
Fujita et al. found the extracellular endo-α-N-acetylgalactosaminidase from B. longum that specifically hydrolyzed α-galacto-N-bioside, which exists abundantly in mucin as core I (10). The presence of both endo-α-N-acetylgalactosaminidase and LNBP in the same organism suggests that the metabolism of GNB by this system is important for the intestinal colonization of bifidobacteria.
The intestinal growth of bifidobacteria is especially important for the health of newborn infants (2). Colonization by bifidobacteria of the intestines of breast-fed infants increases rapidly within 1 week after birth, preventing infection by pathogenic bacteria. This type of rapid growth does not occur in the intestines of bottle-fed infants (3), even when formula is supplemented with prebiotic oligosaccharides, such as lactulose (20). The difference is due to the presence in human milk of oligosaccharides other than lactose (human milk oligosaccharides [HMO]), whereas lactose is the sole saccharide in cow's milk (18). The component of HMO responsible for the growth of bifidobacteria, however, has not yet been identified, since HMO include more than 100 kinds of molecules (14, 18). Lacto-N-tetraose (Galβ1→3GlcNAcβ1→3Galβ1→4Glc) and lacto-N-fucopentaose I (Fucα1→2Galβ1→3GlcNAcβ1→3Galβ1→4Glc), which contain LNB structure, are predominant components in HMO, as well as 2′-fucosyllactose (14, 23). Though the presence of lacto-N-biosidase has not been proved, α-fucosidase has already been found in B. bifidum (12). We therefore suggested that LNB structures present in such oligosaccharides act as the bifidus factor, which enhances the growth of bifidobacteria as a carbon source (13).
In this paper, the enzymes encoded by the BL1642- to BL1644-like genes of B. longum JCM1217, the type strain, were overexpressed in Escherichia coli and characterized, as was the operon including these genes.
MATERIALS AND METHODS
Materials.
The vector pET30 was obtained from Novagen (Madison, WI). B. longum JCM1217 was obtained from the Japan Collection of Microorganisms, The Institute of Physical and Chemical Research (Wako, Japan). E. coli strains select96 (Promega, Madison, WI) and BL21(DE3) (Novagen) were used as hosts for cloning and expression.
Molecular cloning of the lnpB, lnpC, and lnpD genes.
Genomic DNA from B. longum JCM1217 was prepared using InstaGene (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. PCR primers were designed from the genomic sequence of B. longum NCC2705 (21) and the nucleotide sequence of lnpA from B. longum JCM1217 (Table 1). PCR was performed with these primers and KOD-plus polymerase (Toyobo, Osaka, Japan), using an amplification program consisting of 25 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 15 s, and extension at 68°C for 2 min. The PCR products were cloned into an appropriate vector using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA), and the nucleotide sequences of the inserts were determined using a BigDye terminator v3.1 cycle-sequencing kit and a 310 Genetic analyzer (Applied Biosystems, Foster, CA). In constructing expression plasmids, PCR was performed using primers to which were added NdeI or XhoI restriction endonuclease sites for ligation to the expression vector (Table 1). After treatment with NdeI and XhoI, the amplified fragments were ligated into the pET30 vector at the NdeI and XhoI sites using a Ligation High DNA ligation kit (Toyobo). All plasmids were designed so that a His6 tag sequence was added to the carboxyl terminus of the protein to facilitate purification. Plasmid DNA was used to transform E. coli BL21(DE3) after sequence confirmation.
TABLE 1.
Primer sequences for cloning and expression
| Gene | Primer | Sequencea |
|---|---|---|
| lnpB | Cloning forward | GCGACCAACGATTATGGCAAGGGC |
| Cloning reverse | GGCGTATACCTCGGTCAGCTGATCGCTCAT | |
| Expression forward | AACGGACCCCATATGACCGAAAGCAATGAAGTTTTATTC | |
| Expression reverse | GCTGACCTCGAGCCTGGCAGCCTCCATGATGTCGGCTAC | |
| lnpC | Cloning forward | CATCGTAGCCGACATCATGGAGGCTGCC |
| Cloning reverse | CCCGTAACCAGGACAGTGGTCATGTTTCC | |
| Expression forward | GATATATACATATGAACGATCAGCTGACCGAGGTA | |
| Expression reverse | TGACCTCTCGAGCCTAGCGGCAAAACCAAGGCTTTCGA | |
| lnpD | Cloning forward | GGCAACACGGCCGTCTTCAAGCAGAAAGC |
| Cloning reverse | GGCAACACGGCCGTCTTCAAGCAGAAAGC | |
| Expression forward | GATATACATATGACTACTGTTCTGGTTACGGGC | |
| Expression reverse | CTGCTCCTCGAGCTCCGCGTCGCGGAAACCGTTGGGGTTC |
Restriction endonuclease sites are underlined.
Purification of recombinant enzymes.
Each transformant was cultivated at 30°C with shaking in 100 ml LB medium containing 50 μg/ml kanamycin until the absorbance at 600 nm reached 0.5. Protein expression was induced by the addition of isopropyl-1-thio-β-d-galactoside at a final concentration of 0.5 mM, followed by incubation for an additional 20 h at 30°C with shaking. The cells were harvested by centrifugation at 15,000 × g for 10 min, resuspended in 20 mM MOPS (morpholinepropanesulfonic acid) buffer (pH 7.5), and sonicated using a sonifier (Branson Ultrasonic Corporation, Danbury, CT). The cell debris was removed by centrifugation at 17,000 × g for 30 min, and each enzyme was purified on an Ni-nitrilotriacetic acid agarose gel (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The homogeneity of the purified enzymes was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein concentrations were determined using theoretical extinction coefficients calculated from the amino acid sequences of the proteins (11). For the proteins encoded by the lnpB, lnpC, and lnpD genes of B. longum JCM1217, these calculated extinction coefficients at 280 nm were 27,390, 77,810, and 48,820 M−1 cm−1, respectively.
HPIC.
The products of the enzymatic reactions were quantified by high-performance ion chromatography (HPIC) with a pulsed amperometric detector (DX500; Dionex Corporation, Sunnyvale, CA) using a Dionex CarboPac PA1 column (4 mm [inside diameter] by 250 mm) and a linear gradient of 0 to 1 M sodium acetate in 100 mM sodium hydroxide for 20 min at a flow rate of 1 ml/min.
Measurement of enzyme activity.
All enzymatic reactions were measured at 30°C. The activity of N-acetylhexosamine 1-kinase (NahK) was determined by quantifying the GlcNAc1P produced in 30 min in a reaction mixture containing 0.1 M Tris buffer (pH 8.5), 1 mM GlcNAc, 1 mM ATP, 1 mM MgCl2, and various concentrations of enzyme, and the reaction was stopped by boiling the mixture for 5 min. One unit of NahK activity was defined as the amount of enzyme that produced 1 μmol of GlcNAc1P per minute under these conditions.
UDP-glucose-hexose 1-phosphate uridylyltransferase (EC 2.7.7.12) (GalT) activity was determined by quantifying the Glc1P produced in 30 min in a reaction mixture containing 0.1 M MOPS buffer (pH 7.5), 0.5 mM UDP-Glc, 0.5 mM Gal1P, 1 mM MgCl2, and various concentrations of enzyme; the reaction was stopped by boiling the mixture for 1 min. One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol of Glc1P per minute under these conditions.
GalE (EC 5.1.3.2) activity was measured in a reaction mixture containing 0.1 M MOPS buffer (pH 7.5), 1 mM UDP-Gal, and various concentrations of enzyme; the reaction was stopped by boiling the mixture for 1 min. Following the addition of 50 μl of 1 N H2SO4 to hydrolyze UDP sugars, 100 μl of the reaction mixture was boiled for 30 min, and the resultant glucose, the hydrolytic product of UDP-Glc, was quantified by the glucose oxidase method using a glucose CII test (Wako Pure Chemical Industries, Osaka, Japan). One unit of GalE was defined as the amount of enzyme that produced 1 μmol of UDP-Glc per minute under these conditions. When UDP-GalNAc was the substrate, the amount of UDP-GlcNAc produced was quantified by HPIC.
Characterization of NahK.
The stability of NahK was measured by determining residual activity after incubation with 1 μM enzyme at various temperatures or in 10 mM of citrate (pH 2.2, 2.7, 3.2, 3.7, and 4.2), acetate (pH 4.3, 4.8, and 5.2), MES (morpholineethanesulfonic acid) (pH 5.7, 6.1, and 6.6), HEPES (pH 7.1, 7.5, and 8.0), Tris (pH 7.8, 8.2, and 8.7), CHES [2-(cyclohexylamino)ethanesulfonic acid] (pH 9.2, 9.6, and 10.1), and glycine (pH 10.0, 10.5, and 11.0) buffers. The optimum temperature and pH were determined by incubating 0.1 μM of enzyme at various temperatures and in 0.1 M acetate (pH 4.3, 4.8, and 5.2), MES (pH 5.7, 6.1, and 6.6), MOPS (pH 6.7, 7.2, and 7.7), HEPES (pH 7.1, 7.5, and 8.0), Tris (pH 7.8, 8.2, and 8.7), CHES (pH 9.2, 9.6, and 10.1), and glycine (pH 10.0, 10.5, and 11.0) buffers. The effects of metal ions were assayed by incubating enzyme in the presence of 1 mM of various metal ions, and substrate specificity was determined by using 1 mM of various substrates. Kinetic parameters were calculated from an s-v plot by curve fitting the experimental data to the Michaelis-Menten equation using the GraFit computer program (15).
Structural analysis of NahK products.
Following incubation of 20 mM GlcNAc or GalNAc with 22 mM ATP, 10 mM MgCl2, and 70 μg/ml NahK in 0.1 M Tris buffer (pH 8.5) at 37°C for 24 h, the reaction products were purified by Toyopearl HW40F (Tosoh, Tokyo, Japan) gel filtration column chromatography (2.6 cm [inside diameter] by 80 cm) using H2O as the solvent at a flow rate of 1 ml/min. 1H nuclear magnetic resonance spectra were determined in D2O using an Avance 800 spectrometer (Bruker Biospin, Rheinstetten, Germany) at 298 K and compared with those of authentic α-GlcNAc1P and α-GalNAc1P.
Nucleotide sequence accession number.
The nucleotide and deduced amino acid sequences for lnpA to lnpD are available in the GenBank sequence database under accession number AB303839.
RESULTS
Molecular cloning of the lnpB, lnpC, and lnpD genes.
After amplifying the lnpB, lnpC, and lnpD genes from B. longum JCM1217 genomic DNA, as described in Materials and Methods, we sequenced the DNA from the 3′ terminus of the BL1640 gene homolog to the 3′ terminus of the BL1645 gene homolog from B. longum JCM1217 containing lnpA, which had been cloned previously (13). The sequences were highly homologous, except for the noncoding region between the BL1640 gene homolog and lnpA. The deduced amino acid sequences of the lnpB, lnpC, and lnpD genes were 99.2%, 96.7%, and 97.6% identical, respectively, to the deduced amino acid sequences of the BL1642, BL1643, and BL1644 genes, respectively, which had been registered on the database of Genome Information Broker (http://gib.genes.nig.ac.jp/). The recombinant enzymes encoded by the lnpB, lnpC, and, lnpD genes were purified from E. coli transformants as described previously.
Activity of LnpB (the BL1642 homolog).
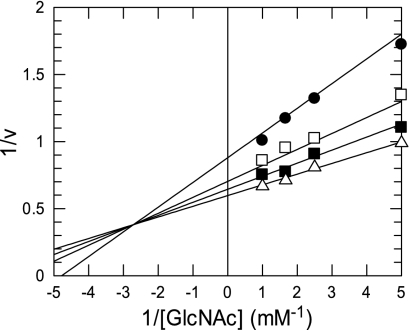
Aside from the activity of BL1642, annotated as a mucin desulfatase, no homology with other desulfatases was observed. BL1642, however, was highly homologous to a hypothetical protein whose gene was adjacent to a mucin desulfatase gene from Prevotella sp. strain RS2. Incubation of 4 μM LnpB protein with 10 mM GlcNAc 6-sulfate in 0.1 M MOPS buffer (pH 7.5) at 30°C for 30 min showed that the protein did not have hydrolytic activity, strongly suggesting that LnpB is not a desulfatase. Since a BLAST search showed that BL1642 was moderately similar to some kinases, we assessed the kinase activity of LnpB for GlcNAc in the presence of ATP. Thin-layer chromatography analysis revealed a new spot in the position of a sugar phosphate (data not shown), later confirmed to be α-GlcNAc1P, not GlcNAc6P, by both HPIC and nuclear magnetic resonance analysis, indicating that the protein encoded by the lnpB gene has a novel kinase activity. This protein had similar activity on GalNAc, also yielding α-GalNAc1P, and weak activities on several monosaccharides (Table 2). We therefore propose the name NahK for this enzyme. NahK also accepted GTP and ITP as phosphate donors (Table 3). The optimum temperature for this reaction was 40°C (measured for 30 min), and the optimum pH was 8.5. The enzyme was stable at pH 5.0 to 9.5 at 30°C and lost half of its activity during incubation for 30 min at 50°C in 0.1 M Tris buffer (pH 8.5). Similar to other kinases, NahK required magnesium ions for activity (Table 4). Its Km values against GlcNAc and GalNAc were 0.118 and 0.065 mM, respectively, and its kcat values were 1.21 and 0.752 s−1, respectively, making its kcat/Km values for GlcNAc and GalNAc almost the same (10.3 and 11.6 mM−1 s−1). NahK was determined to act by a sequential bi bi (two substrates-two products) mechanism (Fig. 3), with the reaction occurring after the binding of both ATP and N-acetylhexosamine. Its Km value against ATP was 0.172 mM at GlcNAc saturation.
TABLE 2.
Substrate acceptor specificity of NahK
| Acceptor | Sp act (μmol·min−1·mg protein−1) |
|---|---|
| N-Acetyl-d-glucosamine | 1.49 |
| N-Acetyl-d-galactosamine | 0.89 |
| N-Acetyl-d-mannosamine | 0.22 |
| d-Talose | 0.049 |
| d-Mannose | 0.024 |
| 2-Deoxyglucose | 0.007 |
| d-Glucosamine | 0.004 |
| d-Mannosamine | 0.002 |
| d-Glucose | 0.001 |
| d-Galactosamine | 0.0003 |
| d-Galactose | 0.0002 |
| d-Allose | NDa |
| d-Lyxose | ND |
| l-Rhamnose | ND |
| d-Xylose | ND |
| d-Ribose | ND |
| l-Arabinose | ND |
| l-Fucose | ND |
ND, not detected.
TABLE 3.
Substrate donor specificity of NahK
| Donor | Sp act (μmol·min−1·mg protein−1) |
|---|---|
| ATP | 1.48 |
| GTP | 0.65 |
| ITP | 0.48 |
| TTP | 0.02 |
| UTP | 0.02 |
| CTP | 0.005 |
| Polyphosphate | NDa |
ND, not detected.
TABLE 4.
Effects of metal ions on NahK activity
| Metal ion (1 mM) | Relative activity (%) |
|---|---|
| None | 0 |
| Mg2+ | 100 |
| Co2+ | 21 |
| Zn2+ | 8 |
| Mn2+ | 7 |
| Ni2+ | 4 |
| Ca2+ | 1 |
FIG. 3.
Lineweaver-Burk plot of the NahK reaction with GlcNAc at ATP concentrations of 0.2 mM (closed circles), 0.4 mM (open squares), 0.6 mM (closed squares), and 1.0 mM (triangles). The kinetic parameters in the equation v = V[GlcNAc][ATP]/(KmATPKiGlcNAc + KmATP[GlcNAc] + KmGlcNAc[ATP] + [GlcNAc][ATP]) were calculated to be as follows: V = 1.90 μmol/min·mg protein, KmGlcNAc = 0.10 mM, KmATP = 0.13 mM, and KiGlcNAc = 0.40 mM. v, initial reaction rate at the concentrations of the substrates; V, maximum velocity.
The activities of LnpC and LnpD (BL1643 and BL1644 homologs).
BL1643 was annotated to be a GalT2 (EC 2.7.7.10), transferring the UMP unit of UTP to Gal1P. Although the protein encoded by the lnpC gene did not have this activity, it showed GalT (EC 2.7.7.12) activity, transferring the UMP unit from UDP-Glc to Gal1P. Unusually, the enzyme also transferred the UMP unit to GlcNAc1P and GalNAc1P. The UMP-transferring activities from UDP-Glc to GlcNAc1P (2.24 U/mg protein) and GalNAc1P (2.30 U/mg protein) were found to be twofold larger (2.24 and 2.30 U/mg protein, respectively) than that to Gal1P (1.08 U/mg protein).
LnpD had GalE activity, as well as epimerizing UDP-GalNAc into UDP-GlcNAc, with an activity of 147 U/mg protein, the same level of activity as on UDP-Gal (158 U/mg). We therefore describe the catalytic activities of the LnpC and LnpD proteins as GalTBl and GalEBl (GalT and GalE of B. longum), respectively.
DISCUSSION
NahK.
Our results indicate that the protein encoded by the lnpB gene is not a “desulfatase for mucin” but a novel enzyme. Generally, most hexose kinases phosphorylate at the 6 position, with the exception of galactokinase (EC 2.7.1.6) (26) and N-acetylgalactosamine kinase (EC 2.7.1.157) (25), both of which recognize only galacto-type substrates. In contrast, NahK phosphorylates both GlcNAc and GalNAc at similar rates and possesses weak activities on various substrates. To our knowledge, NahK is a unique enzyme, as no other kinases at the anomeric position of a gluco-type substrate have yet been described.
GalTBl and GalEBl.
Our results also indicate that LnpC and LnpD are involved not only in general glucose/galactose metabolism, but also in GlcNAc/GalNAc metabolism. GalTBl is a novel type of GalT, having wide substrate specificity; no other GalT has been found to recognize GlcNAc1P or GalNAc1P. Based on their amino acid sequences, GalT enzymes have been classified into two groups: class I, which is present in most species, and class II, which is present only in bacteria (InterPro database; http://www.ebi.ac.uk/interpro/) (17). The amino acid sequence of GalTBl showed weak homology with class II enzymes. However, it is not clear if the substrate specificity of GalTBl is the same as those of other class II enzymes, because the difference between classes I and II has not been completely determined.
GalE from E. coli catalyzes only the conversion between UDP-Glc and UDP-Gal, whereas GalE from humans converts both UDP-Glc/UDP-Gal and UDP-GlcNAc/UDP-GalNAc. Replacing the Tyr299 residue of E. coli GalE with the cysteine residue from human GalE enables the enzyme to convert between UDP-GlcNAc and UDP-GalNAc (24). The tyrosine side chain has been shown to cause steric hindrance for the 2-acetoamide group during the rotation of GlcNAc to GalNAc through the 4-ketose intermediate. The amino acid residue at this position of the GalEBl protein is cysteine, explaining its ability to epimerize both UDP-Glc and UDP-GlcNAc.
Interpretation of the LNB/GNB pathway.
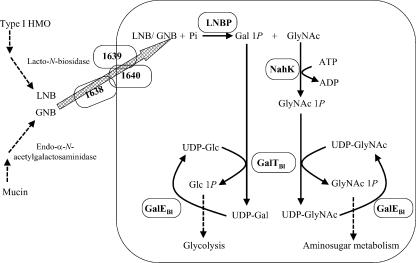
The enzyme activities of LnpA, LnpB, LnpC, and LnpD suggest that its gene cluster functions as an operon to metabolize whole molecules of LNB and GNB. LNB from HMO, liberated by the hydrolysis of lacto-N-tetraose by lacto-N-biosidase (EC 3.2.1.140), an enzyme that has not been isolated from bifidobacteria, and GNB from mucin, liberated by endo-α-N-acetylgalactosaminidase (EC 3.2.1.97) (10), are thought to be transported into the cell by a protein encoded by a putative ABC transporter gene located upstream of this cluster (the BL1638-BL1640 genes). Subsequently, LNB/GNB can be phosphorylated by LNBP to form Gal1P and GlcNAc/GalNAc. Gal1P is converted to Glc1P by the interaction of GalTBl and GalEBl and then joins the glycolytic pathway. In contrast, GlcNAc/GalNAc can be phosphorylated by NahK to form GlcNAc1P/GalNAc1P, which are converted by GalTBl and GalEBl, with GlcNAc1P entering an aminosugar metabolic pathway. Thus, the operon encodes a sufficient number of enzymes to metabolize the entire LNB and GNB molecules to substrates of a subsequent metabolic pathway. We call the metabolic pathway encoded by this operon the “LNB/GNB pathway” (Fig. 4).
FIG. 4.
Scheme of the LNB/GNB pathway in B. longum. Solid arrow, reaction catalyzed by an enzyme encoded in the operon; dashed arrow, reaction catalyzed by an enzyme not encoded in the operon.
Comparison with the Leloir pathway.
In most organisms, Gal is metabolized to Glc1P by the Leloir pathway (5, 9, 16), which consists of four enzymes (Fig. 2): galactose mutarotase (GalM), which converts β-Gal to α-Gal; galactokinase, which converts α-d-Gal to Gal1P; GalT, which converts Gal1P to UDP-Gal, coupled with the conversion of UDP-Glc to Glc1P; and GalE, which converts UDP-Gal to UDP-Glc, with the resultant UDP-Glc consumed in the reaction of GalT. Although the LNB/GNB pathway resembles the Leloir pathway, the two pathways differ. For example, in the Leloir pathway, α-Gal is phosphorylated by galactokinase, consuming one molecule of ATP, whereas in the LNB/GNB pathway, LNBP produces Gal1P without consuming ATP by the phosphorolysis of the galactosides. Utilization of N-acetylhexosamine, the other part of LNB/GNB, in the LNB/GNB pathway also differs from that in the Leloir pathway. GalNAc is transformed into GlcNAc1P with consumption of ATP, like GalK in the Leloir pathway. No gene cluster encoding the full set of enzymes involved in the Leloir pathway was found in B. longum NCC2705. Only a gene cluster containing genes for two of the Leloir enzymes, galK (BL1210) and galT (BL1211), was found in the genome, except for the lnpABCD cluster. In contrast, genes encoding the Leloir enzymes for galactose metabolism, galM, galK, galT, and galE, are often found in operons in other species (1, 4, 27). These results suggest that the LNB/GNB pathway is the main metabolic pathway for galactose as an energy source in B. longum. Moreover, LNB and GNB may be bifidus factors for intestinal colonization through the metabolism of milk oligosaccharides and mucin carbohydrates. Because colonization of bifidobacteria in the intestine is beneficial for human health, the LNB/GNB pathway is thought to be related to the symbiosis between humans and bifidobacteria.
Acknowledgments
This work was supported in part by a grant from the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) of Japan.
Footnotes
Published ahead of print on 24 August 2007.
REFERENCES
- 1.Ajdic, D., I. C. Sutcliffe, R. R. Russell, and J. J. Ferretti. 1996. Organization and nucleotide sequence of the Streptococcus mutans galactose operon. Gene 180:137-144. [DOI] [PubMed] [Google Scholar]
- 2.Benno, Y., and T. Mitsuoka. 1986. The development of gastrointestinal micro-flora in humans and animals. Bifidobact. Microflora 5:13-25. [Google Scholar]
- 3.Benno, Y., K. Sawada, and T. Mitsuoka. 1984. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol. Immunol. 28:975-986. [DOI] [PubMed] [Google Scholar]
- 4.Bettenbrock, K., and C. A. Alpert. 1998. The gal genes for the Leloir pathway of Lactobacillus casei 64H. Appl. Environ. Microbiol. 64:2013-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouffard, G. G., K. E. Rudd, and S. L. Adhya. 1994. Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J. Mol. Biol. 244:269-278. [DOI] [PubMed] [Google Scholar]
- 6.Chow, J. 2002. Probiotics and prebiotics: a brief overview. J. Ren. Nutr. 12:76-86. [DOI] [PubMed] [Google Scholar]
- 7.Collins, M. D., and G. R. Gibson. 1999. Probiotics, prebiotics, and synbiotics: approaches for modulating the microbial ecology of the gut. Am. J. Clin. Nutr. 69:1052S-1057S. [DOI] [PubMed] [Google Scholar]
- 8.Derensy-Dron, D., F. Krzewinski, C. Brassart, and S. Bouquelet. 1999. β-1,3-Galactosyl-N-acetylhexosamine phosphorylase from Bifidobacterium bifidum DSM 20082: characterization, partial purification and relation to mucin degradation. Biotechnol. Appl. Biochem. 29:3-10. [PubMed] [Google Scholar]
- 9.Frey, P. A. 1996. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 10:461-470. [PubMed] [Google Scholar]
- 10.Fujita, K., F. Oura, N. Nagamine, T. Katayama, J. Hiratake, K. Sakata, H. Kumagai, and K. Yamamoto. 2005. Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-α-N-acetylgalactosaminidase from Bifidobacterium longum. J. Biol. Chem. 280:37415-37422. [DOI] [PubMed] [Google Scholar]
- 11.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 12.Katayama, T., A. Sakuma, T. Kimura, Y. Makimura, J. Hiratake, K. Sakata, T. Yamanoi, H. Kumagai, and K. Yamamoto. 2004. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-l-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J. Bacteriol. 186:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitaoka, M., J. Tian, and M. Nishimoto. 2005. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl. Environ. Microbiol. 71:3158-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunz, C., S. Rudloff, W. Baier, N. Klein, and S. Strobel. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu. Rev. Nutr. 20:699-722. [DOI] [PubMed] [Google Scholar]
- 15.Leatherbarrow, R. J. 1990. Using linear and non-linear regression to fit biochemical data. Trends Biochem. Sci. 15:455-458. [DOI] [PubMed] [Google Scholar]
- 16.Leloir, L. F. 1951. The enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch. Biochem. 33:186-190. [DOI] [PubMed] [Google Scholar]
- 17.Mollet, B., and N. Pilloud. 1991. Galactose utilization in Lactobacillus helveticus: isolation and characterization of the galactokinase (galK) and galactose-1-phosphate uridyl transferase (galT) genes. J. Bacteriol. 173:4464-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newburg, D. S., and S. H. Neubauer. 1995. Carbohydrate in milks: analysis, quantities, and significance, p. 273-349. In R. G. Jensen (ed.), Handbook of milk composition. Academic Press, San Diego, CA.
- 19.Nishimoto, M., and M. Kitaoka. 2007. Identification of the putative proton donor residue of lacto-N-biose phosphorylase (EC 2.4.1.211). Biosci. Biotechnol. Biochem. 71:1587-1591. [DOI] [PubMed] [Google Scholar]
- 20.Petuely, F. 1957. Bifidusflora bei Fraschenkindern durch bifidogene Sbstanzen (Bifidusfaktor). Z. Kinderheilkd. 79:174-179. [PubMed] [Google Scholar]
- 21.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schrezenmeir, J., and M. de Vrese. 2001. Probiotics, prebiotics, and synbiotics—approaching a definition. Am. J. Clin. Nutr. 73:361S-364S. [DOI] [PubMed] [Google Scholar]
- 23.Sumiyoshi, W., T. Urashima, T. Nakamura, I. Arai, T. Saito, N. Tsumura, B. Wang, J. Brand-Miller, Y. Watanabe, and K. Kimura. 2003. Determination of each neutral oligosaccharide in the milk of Japanese women during the course of lactation. Br. J. Nutr. 89:61-69. [DOI] [PubMed] [Google Scholar]
- 24.Thoden, J. B., J. M. Henderson, J. L. Fridovich-Keil, and H. M. Holden. 2002. Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks. J. Biol. Chem. 277:27528-27534. [DOI] [PubMed] [Google Scholar]
- 25.Thoden, J. B., and H. M. Holden. 2005. The molecular architecture of human N-acetylgalactosamine kinase. J. Biol. Chem. 280:32784-32791. [DOI] [PubMed] [Google Scholar]
- 26.Thoden, J. B., and H. M. Holden. 2003. Molecular structure of galactokinase. J. Biol. Chem. 278:33305-33311. [DOI] [PubMed] [Google Scholar]
- 27.Vaughan, E. E., R. D. Pridmore, and B. Mollet. 1998. Transcriptional regulation and evolution of lactose genes in the galactose-lactose operon of Lactococcus lactis NCDO2054. J. Bacteriol. 180:4893-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]