Abstract
A multiplex PCR-based method, in which two small-subunit rRNA regions are simultaneously amplified in a single reaction, was designed for parallel detection of honeybee microsporidians (Nosema apis and Nosema ceranae). Each of two pairs of primers exclusively amplified the 16S rRNA targeted gene of a specific microsporidian. The multiplex PCR assay was useful for specific detection of the two species of microsporidians related to bee nosemosis, not only in purified spores but also in honeybee homogenates and in naturally infected bees. The multiplex PCR assay was also able to detect coinfections by the two species. Screening of bee samples from Spain, Switzerland, France, and Germany using the PCR technique revealed a greater presence of N. ceranae than of N. apis in Europe, although both species are widely distributed. From the year 2000 onward, statistically significant differences have been found in the proportions of Nosema spp. spore-positive samples collected between and within years. In the first period examined (1999 to 2002), the smallest number of samples diagnosed as Nosema positive was found during the summer months, showing clear seasonality in the diagnosis, which is characteristic of N. apis. From 2003 onward a change in the tendency resulted in an increase in Nosema-positive samples in all months until 2005, when a total absence of seasonality was detected. A significant causative association between the presence of N. ceranae and hive depopulation clearly indicates that the colonization of Apis mellifera by N. ceranae is related to bee losses.
Colonization is a transmission process involving the spread of a parasite into new geographical areas and leads to establishment of that species in a host population in which it was not previously present (5). Over the past few decades, many free-living animals (hosts) and their parasites have invaded new areas and have been identified as alien or exotic species. After a parasite invades a new area, it is unlikely that it will remain focally localized. Following establishment of a viable, self-sustaining population, the subsequent stage of a species' colonization is usually spread or dispersal throughout the area (host) or in both the distribution range of a susceptible native host and the novel range of a recently invaded exotic host (parasite) (29).
When Apis mellifera, a superior producer of honey, was introduced into Asia some 50 years ago, it came in contact with native bees (Apis cerana, for example) and therefore with their parasites (Varroa jacobsoni). Subsequent adaptation to this new and suitable host could have led to genetic changes that had even more severe effects on the new host; this resulted in description of the new species Varroa destructor in A. mellifera (2). In a similar way, contact between the two host species could have had repercussions for microsporidian infections. Transportation of A. mellifera throughout the world is associated with transportation of the microsporidian Nosema apis, a parasite of the bee's digestive tract considered to be not highly virulent and also described as a parasite of A. cerana (11, 27, 28). For this Asian host, a new species, Nosema ceranae, was described in 1996 on the basis of gene sequences and ultrastructural features, and this species was thought to be restricted to this host and geographically limited to Asia (12). Quite recently, this new parasite has also been detected infecting A. mellifera both in Asia (20) and in Europe (17), which implies that either a great spatial leap occurred or a great deal of ignorance exists about the real situation. In any case, the situation is worrisome as experimental infection by N. ceranae of this new host species has recently proved to be highly pathogenic (15).
During the last 10 years, an increase in infections by microsporidian parasites in the honeybee (A. mellifera) has been detected in several European countries (8, 18, 23), while increasing numbers of honeybee colony deaths and low production in the same areas have been reported by beekeepers. One of the principal hypotheses that might explain these problems is the recent entry and rapid dissemination of N. ceranae in Europe (16).
The spores of the two Nosema species are very similar and can hardly be distinguished by optical microscopy, so that in the absence of clear morphological characteristics for species recognition, other techniques using molecular markers may greatly assist in the diagnosis and identification of honeybee microsporidians. The PCR technique provides a very sensitive test for detecting microsporidian infection because it enables detection of the parasite at very low levels of infection and can reveal all the stages of its life cycle (35). Multiplex PCR provides a significant improvement to the conventional technique by incorporating multiple primers that amplify regions of DNA from two honeybee microsporidians simultaneously in a single reaction.
Using stored samples of microsporidian-infected honeybees, it may be possible to trace the infection back in time (13), but due to the unavailability of old samples for molecular biology testing, in this study some retrospective data are presented to support the hypothesis concerning the introduction of a new parasite species and its relationship with the depopulation syndrome. Also, the presence of high levels of this parasite in Europe was determined by means of a quick, specific, and sensitive method developed for differential diagnosis of N. apis or N. ceranae in honeybee samples in just one step and establishment of the relationship between the presence of N. ceranae and bee colony depopulation.
MATERIALS AND METHODS
Samples.
The retrospective study of the prevalence of Nosema infection included all the results for Nosema spp. (5,776 data) registered for adult honeybee samples from different areas of Spain (which had been sent by beekeepers or veterinary services from hives with different pathological problems) in the Laboratory of Bee Pathology (Centro Apícola Regional [CAR]) from 1999 until 2005.
The diversity of Nosema species was studied using 290 honeybee samples sent to the CAR laboratory from different European countries (Spain, France, Switzerland, and Germany), as shown in Table 1. Data from the Spanish samples were also used to establish a relationship between clinical signs and Nosema detection.
TABLE 1.
Samples used to study the diversity of Nosema species
| Country | Source | No. of samplesa | Dates | Observations |
|---|---|---|---|---|
| Spain | Veterinary services and beekeepers | 149 | June 2005 to December 2006 | Sent to CAR laboratory for different pathological problems (depopulation, weakness, or asymptomatic); samples also used for the causative study |
| France | R. Borneck | 6 | February to May 2006 | Apiaries from Jura region, collected from dead hives |
| D. Fortini and J. F. Odoux | 30 | April to June 2006 | Apiary of Experimental Centre in Surgères; high bee colony mortality and weakness; seven controlled apiaries | |
| Switzerland | A. Imdorf | 36 | March 2006 | Collected from dead hives and colonies without abnormal findings |
| Germany | W. Ritter | 69 | 2003, 2005, and 2006 | Different German regions |
The total number of samples was 290.
Spore detection and DNA extraction.
The abdomens of at least 10 to 20 adult honeybees from each sample were macerated in 5 ml of distilled water. A further 5 ml was added, and the suspension was filtered and centrifuged for 6 min at 800 × g. Pellets were analyzed by phase-contrast microscopy (magnification, ×400) to verify the presence of spores (26). This methodology was employed for determination of the presence of Nosema spores in all the samples used in this study.
For DNA extraction, spore germination was induced with 200 μl of freshly prepared germination buffer (6), and preparations were incubated at 37°C for 15 min (27). DNA extraction was conducted using a High Pure PCR template preparation kit (catalog no. 1796828; Roche Diagnostic), as previously described (15). Negative controls were processed in parallel to detect possible contamination.
Previously characterized and sequenced Nosema samples (17) from different sources were used for testing the new molecular tools.
Duplex PCR design methodology.
The 16S rRNA locus was selected to perform N. apis-N. ceranae duplex PCR. Published sequences in the GenBank database (http://www.ncbi.nlm.nih.gov/) from N. apis (accession numbers DQ235446, U76706, U97150, U26534, X73894, and X74112), N. ceranae (accession numbers DQ329034, U26533, DQ078785, and DQ286728), Nosema bombi (accession numbers AY008373, AY741108, and AY741110), Nosema portugal (accession number AF033316), and Nosema trichoplupsiae (accession number U09282) were compiled. Sequences from the species were aligned using ClustalW (http://www.ebi.ac.uk/clustalw/) in order to identify individual polymorphic nucleotide positions and to avoid their becoming primer binding zones. A consensus sequence (with variable sites, named by using IUB code) was obtained for each species.
Specific primers for both N. apis and N. ceranae were visually selected taking into account that primer sequences were specific to each of the two species. The second requirement was to obtain two amplicons that were different lengths so that they could be separated using agarose gel electrophoresis. The selected primers are shown in Table 2 (321APIS primers for N. apis and 218MITOC primers for N. ceranae). The expected number of amplified bases in N. ceranae using the 218MITOC primers can be either 218 or 219 depending on the sequences for N. ceranae available in the GenBank database (http://www.ncbi.nlm.nih.gov/). In the case of N. apis, the expected size of the amplicon using the 321APIS primers was 321 bp.
TABLE 2.
Primers selected for detection of N. ceranae and N. apis in multiplex PCR
| Primer | Sequencea | PCR product size (bp) | Specificity |
|---|---|---|---|
| 218MITOC-FOR | 5′-CGGCGACGATGTGATATGAAAATATTAA-3′ | 218-219b | N. ceranae |
| 218MITOC-REV | 5′-CCCGGTCATTCTCAAACAAAAAACCG-3′ | ||
| 321APIS-FOR | 5′-GGGGGCATGTCTTTGACGTACTATGTA-3′ | 321 | N. apis |
| 321APIS-REV | 5′-GGGGGGCGTTTAAAATGTGAAACAACTATG-3′ |
CG tails added to primers are underlined.
There is a 1-bp difference in the N. ceranae amplicon size depending on the sequences for N. ceranae available in GenBank (http://www.ncbi.nlm.nih.gov).
Primer suitability (G+C content and melting temperature) was checked using the IDT OligoAnalyzer program (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/). G or GC tails were added to the 5′ end of all primers in order to make the melting temperatures of each primer set equal. Potential primer interactions (hairpin, homodimer, and heterodimer structures for the four primers) were tested using the AutoDimer program (http://www.cstl.nist.gov/div831/strbase//AutoDimerHomepage/AutoDimerProgramHomepage.htm). Species specificity was determined by conducting a search for nearly exact matches with BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Once the specific annealing of primers was verified, single PCRs and sequencing of six samples were performed for each fragment before the primers were combined for the duplex assay in order to ensure that products that were the correct sequence and size were generated. A gradient PCR (60 ± 5°C) was performed to empirically determine the annealing temperatures of the two primer pairs. The best amplicons were obtained when the annealing temperature was 61.8°C.
In order to verify that the resulting amplicons in fact corresponded to the expected sequences, subsequent single PCRs were carried out at this annealing temperature with six N. ceranae and five N. apis extracted samples, which were previously characterized by sequencing using the methodology described by Higes et al. (17). The PCR products were purified with a QIAquick PCR purification kit (catalog no. 28104; QIAGEN) as previously described (17) and were fully sequenced in both directions (3730 DNA analyzer; Applied Biosystems). These sequences were aligned and compared to the N. ceranae and N. apis consensus reference sequences using Sequencher (version 4.1.4Fb4; GeneCodes, Ann Arbor, MI).
Once the effectiveness and accuracy of the single PCRs were verified, a duplex PCR was carried out by using equimolar amounts of the two primer pairs. The initial equimolar primer mixture was tested, and the PCR products obtained showed that empirically balanced primer mixtures were not necessary.
PCR conditions.
All PCRs were carried out with a Mastercycler ep gradient S (Eppendorf), and the PCR conditions were as follows: 50-μl reaction cocktail containing 25 μl of High Fidelity PCR Master Mixture (catalog no. 12140314001; Roche Diagnostic), 0.4 μM of each primer, 0.4 mM of each deoxynucleoside triphosphate, 3 mM Cl2Mg, 0.2 mg/ml bovine serum albumin, 0.1% Triton X-100, and 5 μl of N. apis or N. ceranae DNA template. The thermocycler program consisted of 94°C for 2 min, followed by 10 cycles of 15 s at 94°C, 30 s at 61.8°C, and 45 s at 72°C, 20 cycles of 15 s at 94°C, 30 s at 61.8°C, and 50 s at 72°C plus an additional 5 s of elongation for each successive cycle, and a final extension step at 72°C for 7 min. Negative controls (from DNA extraction) were included in all PCR experiments.
In order to evaluate the amount and quality of amplicons, single and duplex PCR amplicons were visualized by electrophoresis in 2% agarose gel (E-gels; Invitrogen) parallel to standard size electrophoresis.
Validation of duplex PCR: reproducibility, sensitivity and specificity. (i) Reproducibility.
Twenty samples from bees known to be infected with either N. apis or N. ceranae or both, as well as from noninfected bees, were prepared and analyzed to ensure that the specimens exhibited accurate, interpretable, and reproducible DNA types.
(ii) Sensitivity.
The sensitivity studies were performed with serial dilutions of N. apis or N. ceranae DNA templates. A first DNA template, extracted from a sample containing 3 × 106 spores of each microsporidian as determined with a hemocytometer, diluted either in 100 μl of double-distilled H2O or in uninfected honeybee macerate, was serially diluted to obtain DNA templates equivalent to 106, 105, 8 × 104, 6 × 104, 4 × 104, 2 × 104, 1.5 × 104, 1 × 104, 5 × 103, 2 ×103, 2 × 102, and 20 spores in 100 μl. Sensitivity was studied by testing N. apis or N. ceranae templates individually and then testing a mixture of the two species in order to investigate the ability of the system to detect the components of mixed specimens and define the limitations of the duplex PCR.
(iii) Specificity.
The primer pairs were tested for specificity by using them in PCRs with Nosema DNA isolated from different sources. We analyzed different European and Taiwanese N. ceranae (previously characterized and sequenced) isolates (supplied by W. F. Huang), N. bombi (supplied by R. Paxton), N. trichoplupsiae strain ATCC 30702, and three different N. apis samples isolated in our laboratory.
All the validation experiments were carried out using three PCRs performed on three different days with the parameters described above (an annealing temperature of 61.8°C and 0.4 μM of each primer). The amplicon products of each PCR were analyzed by electrophoresis twice. Negative controls were tested in parallel to detect possible contamination.
PCR analysis to determine the diversity of Nosema species.
Once the duplex PCR had been validated, a study of the presence of both Nosema species in samples having different origins was conducted.
After 290 samples from different European countries (Spain, France, Switzerland, and Germany) were processed, the 16S rRNA was amplified by using primer pairs 218MITOC-FOR/218MITOC-REV and 321APIS-FOR/321APIS-REV. The duplex PCR and electrophoresis conditions were the same as those described above. Extraction and PCR negative controls were included in all PCR experiments.
Additionally, samples from France, Switzerland, and Germany (three different samples from each country) were selected, and 16S rRNA was amplified using primer pairs MICRO-F/MICROCE-R and INTER-FOR/INTER-REV and sequenced as previously described (15).
Causative association between colony signs and the presence of Nosema spp.
The 149 Spanish samples used to determine Nosema species diversity (Table 1) were used to establish the causative association between colony signs and the presence of N. ceranae and/or N. apis. Each of these samples came from a different beekeeper. A total of 53% of the samples were from professional beekeepers (150 to 2,000 hives per beekeeper), 42.3% were from nonprofessional beekeepers (16 to 149 hives), and 4.7% were from self-consumption beekeepers (less than 16 hives). Together, all the beekeepers had 50,091 hives.
The term “depopulation” was used when samples came from dead colonies with depopulation or from apiaries with large numbers of dead colonies. “Weakness” was used to describe colonies whose beekeepers had observed low production without bee mortality, and the asymptomatic group included colonies without any clinical signs in routine or controlled surveys.
In order to avoid bias owing to possible geographically related differences, only Spanish samples were included in the analysis of causative association.
Statistical analysis.
In the retrospective study, different statistical analyses were carried out (all at the 95% confidence level). The Pearson chi-square test with Monte Carlo correction (to determine the exact probability) was used to evaluate the differences in frequency of Nosema-positive diagnosis in the different years studied (according to the distribution of the number of samples analyzed per month in each year or per year and the results of the Nosema diagnosis [positive samples versus negative samples]). The Fisher exact test was used to compare differences in the distribution of positive samples and negative samples for the total number of samples analyzed in the previously determined periods. Finally, differences in the number of Nosema-positive samples in the different years studied were compared using the nonparametric Kruskal-Wallis and median tests.
Causative associations between the presence of N. ceranae and/or N. apis and the signs observed in the samples (depopulation, weakness, or asymptomatic) were estimated by calculating the relative risk and its confidence interval (95% confidence level).
All calculations were performed with SPSS v. 13.
Nucleotide sequence accession numbers.
The consensus sequences of isolates from Switzerland, Germany, and France have been deposited in the GenBank database under accession numbers DQ673615, DQ374656, and DQ374655, respectively.
RESULTS
Duplex PCR: checking and optimizing.
In the single-gradient PCR test of both primer pairs, the best PCR products were obtained when the annealing temperature was between 55.2 and 62.7°C. The temperature selected for the subsequent assays was 61.8°C because it was the highest temperature at which amplicons could clearly be seen; this, consequently, reduced the nonspecific amplification risk. After different primer concentrations in the PCR mixture (0.3 to 0.6 μM) (data not shown) were checked, an equimolar concentration of 0.4 μM was used to reduce primer dimerization.
Single PCR and sequencing analyses of both strands by using the designed primers (321APIS and 218MITOC primers) yielded the expected amplicon sizes (321 bp for N. apis and 218 bp for N. ceranae) and sequences. Neither primer interaction (hairpin, homodimer, and heterodimer structures among the four primers) nor undesirable amplicons were detected in agarose gels. Subsequent single PCRs and sequencing performed with the six N. ceranae and five N. apis known samples amplified the expected products.
Duplex PCR: reproducibility, sensitivity, and reliability.
All 20 samples and controls tested six times (three PCRs and two electrophoreses each) showed the same results, which demonstrated the reliability of the method.
The sensitivity of the duplex PCR was tested individually or using a mixture with DNA extracted from both microsporidians, and the minimum detection limit is shown in Table 3. N. apis-specific primers showed higher sensitivity, and N. apis was always detected in DNA extracted samples (macerated and diluted) equivalent to 750 N. apis spores per reaction mixture. In some cases, as few as 500 N. ceranae spores or 100 N. apis spores per reaction mixture yielded visible amplicons. N. ceranae spores diluted in water (PCR grade) yielded results similar to those obtained with spores diluted in macerates of naïve honeybees. In contrast, better results were obtained when N. apis spores were diluted in macerates of uninfected honeybees than when they were diluted in water. No differences were detected in the sensitivity of the duplex PCR when the two Nosema species were amplified either in a mixture or individually.
TABLE 3.
Sensitivity of the PCRs performed with the 218MITOC-FOR/218MITOC-REV and 321APIS-FOR/321APIS-REV primer pairs with spores diluted in water or uninfected honeybee maceratea
| Primers | Diluent | No. positive/no. tested with the following no. of spores/100 μl (no. of spores/reaction mixture):
|
No. positive/no. tested with:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 × 106 (15 × 104) | 106 (5 × 104) | 105 (5 × 103) | 8 × 104 (4 × 103) | 6 × 104 (3 × 103) | 4 × 104 (2 × 103) | 2 × 104 (103) | 1.5 × 104 (750) | 104 (500) | 5 × 103 (250) | 2 × 103 (100) | 2 × 102 (10) | 20 (1) | Uninfected honeybee macerate | Water | ||
| 218 MITOC | Water | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 5/6 | 5/6 | 3/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 |
| Uninfected honeybee macerate | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 3/6 | 1/6 | 1/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | |
| 321APIS | Water | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 2/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 |
| Uninfected honeybee macerate | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 5/6 | 4/6 | 2/6 | 0/6 | 0/6 | 0/6 | 0/6 | |
All samples were analyzed by three PCRs performed on three different days, and the amplicon products of each PCR were separated by electrophoresis twice.
To determine the primer specificity, a number of different species or isolates of Nosema (N. ceranae from Europe and Taiwan, N. bombi, N. trichoplupsiae, and N. apis) were tested by duplex PCR using the 218MITOC and 321APIS primer pairs. All reactions were found to be specific. The 218MITOC pair of primers amplified only N. ceranae, irrespective of the geographical area from which the samples originated (Taiwan or Europe). The specific primers for N. apis also showed a high degree of specificity and were positive only with previously characterized samples from different European countries. No amplification was obtained with N. bombi or N. trichoplupsiae microsporidia.
Retrospective analysis.
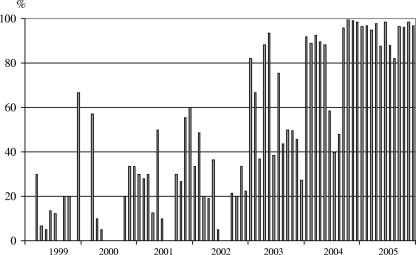
In the 3 years from 2003 to 2005 there was a surprising increase in Nosema spores detection in the CAR laboratory. Table 4 shows that the percentage of positive samples sent to the laboratory increased from 13 to 24.7% in the first 3 years to 95.6% in the last year (Fig. 1).
TABLE 4.
Numbers of samples positive for Nosema spores
| Year | No. of samples | % Positive |
|---|---|---|
| 1999 | 154 | 13.0 |
| 2000 | 124 | 9.7 |
| 2001 | 146 | 24.7 |
| 2002 | 443 | 23.5 |
| 2003 | 484 | 54.5 |
| 2004 | 3,002 | 89.0 |
| 2005 | 1,423 | 95.6 |
FIG. 1.
Percentages of samples positive for Nosema spores from 1999 to 2005 in the Laboratory of Bee Pathology.
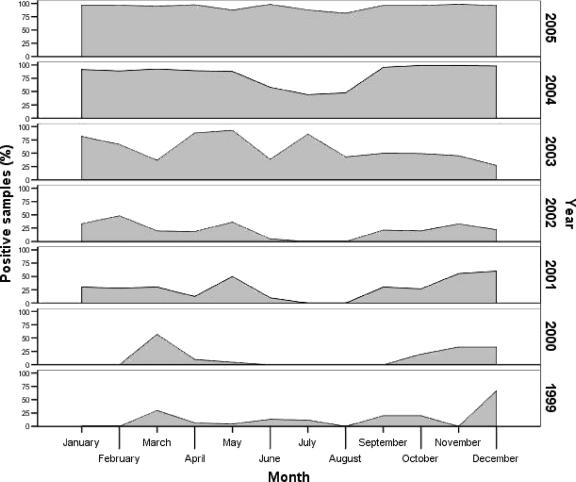
From the year 2000 onward, statistically significant differences have been found in the proportion of Nosema spp. spore-positive samples collected between and within years. The microsporidian species could not be identified for these samples. Two different periods for the frequency of Nosema-positive samples were established: 1999 to 2002 and 2003 to 2005. In the first period (1999 to 2002), the smallest number of samples diagnosed as Nosema positive was recorded during the summer months, showing the clear seasonality of N. apis. However, in 2003 there was a change in the tendency, with an increase in the proportion of Nosema-positive samples in all months independent of the increase in the number of samples received in the laboratory (as indicated by the sorting of the positive mean range and the position around the median). Finally, in 2005 no statistically significant differences were found in the frequency of Nosema-positive samples in the different months or seasons, indicating a total lack of seasonality in the diagnosis of microsporidiosis (Fig. 2).
FIG. 2.
Monthly distribution of Nosema-positive samples in every year from 1999 to 2005, obtained by comparing the number of samples positive for Nosema spores with the total number of samples received in the Laboratory of Bee Pathology.
Nosema species diversity.
A total of 290 samples from different European countries were investigated by multiplex PCR for the presence of N. apis and N. ceranae microsporidia. Eighty-eight samples (30.3%) were negative for both Nosema spp. N. ceranae was found to be the most frequent species; it was present alone in 53.8% of the samples and was detected in samples from every country studied (Table 5). N. apis alone was detected in 9.3% of the samples studied. Mixed infections were detected in 6.6% of the samples.
TABLE 5.
Results of Nosema species screening
| Group | No. (%) of samples screened
|
||||
|---|---|---|---|---|---|
| Total | Spain | France | Switzerland | Germany | |
| Negative | 88 (30.3) | 65 (43.9) | 3 (8.3) | 12 (33.3) | 8 (11.6) |
| N. ceranae positive alone | 156 (53.8) | 52 (34.9) | 27 (75) | 23 (63.9) | 54 (78.3) |
| N. apis positive alone | 27 (9.3) | 21 (14.1) | 0 (0) | 1 (2.8) | 5 (7.2) |
| N. apis and N. ceranae | 19 (6.6) | 11 (7.4) | 6 (16.7) | 0 (0) | 2 (2.9) |
| Total | 290 | 149 | 36 | 36 | 69 |
Both species were found in all the countries studied, with different levels of prevalence (Table 5). N. ceranae was the most prevalent species. Most French and German samples were positive for Nosema; no mixed infections were detected in Swiss samples, and in French samples, N. apis was not detected alone. N. ceranae was present in 75% of positive Spanish samples (in mixed infections with N. apis in 13.1% of the samples), while N. apis was present in 38.1%.
Causative association between colony signs and the presence of Nosema spp.
The 149 Spanish samples were used to study the association between the presence of microsporidian infection and the symptoms observed in hives. These samples were classified as shown in Table 6. Sixty-six samples were classified as members of the “depopulation” group, and 79 samples were classified as members of the asymptomatic group. “Weakness” was excluded from the causative association study due to the small number of colonies (four) classified in that group.
TABLE 6.
Association between the detection of microsporidian infection in Spanish honeybee samples and the signs observed in hives
| PCR result | No. in depopulation group | No. in asymptomatic group | Total | Relative risk | 95% confidence interval | P |
|---|---|---|---|---|---|---|
| N. apis + N. ceranae | 10 | 1 | 11 | 5.82 | 3.20-10.59 | 0.0000 |
| N. ceranae | 44 | 8 | 52 | 5.42 | 3.03-9.68 | 0.0000 |
| N. apis | 2 | 16 | 18 | 0.71 | 0.17-2.92 | 0.4869 |
| Negative | 10 | 54 | 64 | |||
| Total | 66 | 79 | 145 |
The relative risks determined are also shown in Table 6. The colonies in which either both species or only N. ceranae was detected exhibited a risk almost six times greater than the risk for colonies with negative PCR results. The confidence intervals (95% level of confidence) and P values (significance level) indicate the statistical significance of the causative association. In contrast, the colonies in which only N. apis was detected did not show any increased risk compared with the asymptomatic group (P > 0.05).
Data also showed that in 15% of hives belonging to the “depopulation” group N. ceranae was not detected and that 11.3% of the hives positive for N. ceranae were asymptomatic.
DISCUSSION
In this study, a change in the epidemiological pattern of nosemosis and a relationship between detection of N. ceranae and bee depopulation were established. To make differential diagnosis possible, it was necessary to develop a new rapid technique. The duplex PCR method developed in this study has been shown to be a sensitive and specific test for the detection of honeybee microsporidians. Among the many molecular techniques, PCR has been the most widely used for the diagnosis of microsporidian infections and epidemiological studies. The small-subunit (SSU) rRNA (16S rRNA) genes of many microsporidians have been sequenced and have been found to diverge greatly from those of other eukaryotes; the sequences are shorter and share little homology with the sequences of other eukaryotes (30, 35). Thus, the SSU rRNA genes of the microsporidians possess characteristics suitable for molecular detection (32).
Until recently, detection of honeybee microsporidians has been limited to microscopic examination, and to differentiate between the two Nosema spp., sequencing of PCR products is required (17). Microscopic detection of these microsporidians is expensive, laborious, and limited to known stages of development. The multiplex PCR method for detecting N. ceranae and N. apis offers a number of advantages over traditional microscopy, including increased sensitivity, specificity, and the ability to identify all developmental stages of the parasite (31). Thus, multiplex PCR decreases the risk of misidentification and should facilitate epizootiological studies related to these pathogens. Moreover, other molecular methods, such as sequencing or restriction analysis of PCR products, are expensive and time-consuming and involve a great deal of manipulation (which likely increases the risk of error). The duplex PCR described here is a reliable, rapid, cost-effective, and simple method for detecting N. apis and/or N. ceranae infections in a single step since PCR products can be easily separated and identified by agarose gel electrophoresis (with no additional treatment).
Multiplex PCR primer design and optimization are a greater challenge than designing single PCR primer pairs because multiple primer annealing events need to occur in the same annealing conditions without interfering with one another. So, since compatible primers are the key to successful multiplex PCR, careful primer design and appropriate quality control measure are essential in order to ensure that the PCR primers work under uniform PCR conditions and do not adversely interact with one another. Initially, single PCRs were tested using the 218MITOC and 321APIS primer sets for N. ceranae and N. apis, respectively, to ensure locus-specific amplification. After this, duplex PCR studies were carried out using annealing temperature adjustment and empirical performance tests in an attempt to generate sensitive, balanced, and specific signals for both Nosema species in a single PCR.
Subsequent PCR assays confirmed the suitability of our design. The N. apis and N. ceranae SSU rRNA gene-specific primers developed here successfully amplified a 218-bp fragment from N. ceranae and/or a 321-bp fragment from N. apis spores and even fragments from distant N. ceranae isolates, such as those from Taiwan and Europe. Conversely, these primers did not amplify DNA preparations from uninfected honeybees or other microsporidian species, such as N. trichoplupsiae or N. bombi, indicating that the 218MITOC and 321APIS primer pairs are specific for N. ceranae and N. apis, respectively. High specificity is frequently associated with low sensitivity, and to avoid this, nonspecific primers have been designed previously, as in the case of N. bombi (21). In our case, the degree of sensitivity achieved by using specific primers was satisfactory as the limits of detection for both Nosema species were consistent with those in other studies of invertebrate microsporidians (32, 36).
Although N. apis-specific primers have been previously described (34), this is the first study of specific and sensitive molecular diagnosis of N. ceranae and the first differential diagnosis of both honeybee microsporidians in just one reaction.
No notable differences were observed when the spores were diluted in water or in uninfected honeybee macerate, which differs from the results obtained with N. bombi (21). In our case, with N. apis-specific primers (321APIS), the degree of sensitivity was slightly higher when spores were diluted in honeybee macerate. The addition of bovine serum albumin and Triton X-100 to the PCR mixture probably prevents PCR-inhibiting substances (that might be present in honeybee macerates) from having any effect during the PCR (1, 22).
Data from the first period studied (1999 to 2002) seem to be consistent with the seasonal pattern previously described for Nosema infection (19). In the absence of molecular diagnosis, most studies performed in the last century considered N. apis the etiological agent of the disease while describing similar epidemiology and pathology of the disease all over the world. Reports of nosemosis were related to low levels of infection during the summer, a generally small peak in the autumn, and a usually slow rise during the winter. These seasonal trends related to N. apis also included a rapid and large increase in the spring as brood rearing started, while flight possibilities due to climatic conditions were still limited. It is generally agreed that colonies in northern climates are more seriously affected than colonies in southern regions because of the increased amount of time that bees are confined to the hive.
Our data are consistent with this pattern only for the first years of the study (1999 to 2002). However, in the period from 2003 to 2005 the increased percentage of positive samples in the summer was statistically different from the percentage in the previous period. In 2005 the absence of differences in the number of positive samples between months shows an evident lack of seasonality. Recent reports (8) indicate that the clinical and epidemiological pattern of nosemosis is changing.
Nosema seasonality has been related to precipitation (7), and in recent years high-precipitation regions have been thought to represent disease reservoirs from which Nosema can radiate each year, so that the epizootic years are closely related to higher precipitation in these areas (3). The relationship between precipitation and nosemosis is not clear at present. Even when an established pattern for nosemosis is detected (e.g., in the first years examined in this study), mean rainfall can be very different from one year to another. The high rainfall peak of 2000 had no direct effect on Nosema prevalence, and after 2004, which was the driest year in 60 years (25), in 2005 the highest prevalence of Nosema to date was recorded and there was no seasonal pattern. Our data strongly suggest that currently the prevalence of Nosema is not related to precipitation in Spain.
Nosema infection levels have also been related to stress due to management systems (4). Spanish manipulation of hives varies greatly according to region, but for the last period of the study there is no reason to believe that any significant changes occurred on a global level.
The lack of seasonality detected and the increasing number of pathological samples sent to the laboratory from 2003 to the present without any compatible Nosema signs of infection can be related to the higher N. ceranae prevalence in Spain in 2005. Some differences in the epidemiology of and pathology caused by the two microsporidians may explain this situation.
The introduction of N. ceranae from Southeast Asia into Spain in recent years may explain the lack of seasonality in the second period of our study. The dispersion of a parasite into a new habitat or geographical area is usually a chance event and does not guarantee success in colonizing a new host or in becoming established reproductively. The presence of N. ceranae in most European countries suggests that colonization has been successful, but there is still a lack of equilibrium between the new parasite and the new host. N. ceranae seems to be highly pathogenic to A. mellifera. High rates of mortality are tied to the presence of autoinfective spores that facilitate the rapid division and invasion of digestive tissue and affect the regenerative digestive cells (15). These differences can be related to the recently described higher colony losses during the last few years, especially at the end of winter (9, 24).
Direct evidence of the influence of the parasite on colonies is related to changes in the activity and longevity of the worker honeybee and queen (10, 14). Infection of workers by N. apis retards or inhibits development of the pharyngeal salivary glands (33), compromising the feeding of young larvae. A fairly high proportion of the eggs laid by the queen of a colony suffering from nosemosis fail to produce pupae, while artificially infected pupae are resistant to infection (14). The bee population begins to decrease as Nosema infection spreads. Due to changes in their physiological state, infected bees seem to become behaviorally older than healthy bees of the same age (33), starting foraging activities at an early stage (10), and appear to have a shorter life span. But somehow N. apis has reached an epidemiological equilibrium with the host that minimizes its effects on the total bee population. This equilibrium probably depends on climatic conditions and reduced transmission inside the hive in the summer months. N. ceranae does not seem to follow the seasonal pattern of N. apis, exhibiting similar rates of infection throughout the year, which indicates that the decreasing effect on transmission of the hottest months is not effective in controlling this infection in A. mellifera.
The relative risk of bee depopulation observed in colonies with either both species or only N. ceranae (almost six times greater than that in colonies with negative PCR results) indicates that there is a significant causative association between the presence of N. ceranae and the development of hive depopulation. The presence of N. ceranae in asymptomatic hives can be related to the incubation period, which is not known at present for this species, while the existence of a few negative hives in the depopulation group may be due to some other etiology or hypothesis associated with this symptom and not included in this study.
Acknowledgments
We thank D. Fortiny, A. Imdorf, J. F. Odoux, and W. Ritter for supplying the samples from France, Switzerland, and Germany, R. Paxton and W. F. Huang for providing the spores for N. bombi and N. ceranae controls, F. A. Rojo-Vázquez for constructive revision of the text, L. Barrios for statistical analysis, and N. Azpiazu and M. A. Chin for their linguistic input.
This study was supported by projects INIA/FEDER FOUNDS-RTA2005-00152, JCCM 05-280/PA-47, and API/FEGA-MAPYA FOUNDS-06-009.
Footnotes
Published ahead of print on 3 August 2007.
REFERENCES
- 1.Al-Soud, W. A., and P. M. Rådströ. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39:485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. L., and J. W. H. Trueman. 2000. Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp. Appl. Acarol. 24:165-189. [DOI] [PubMed] [Google Scholar]
- 3.Aydin L., I. Cakmak, E. Gulegen, and H. Wells. 2005. Honey bee Nosema disease in the Republic of Turkey. J. Apic. Res. 44:196-197. [Google Scholar]
- 4.Bailey, L., and B. V. Ball. 1991. Honey bee pathology. Academic Press, New York, NY.
- 5.Bush, A. O., J. C. Fernández, G. W. Esch, and J. R. Sheed. 2001. From reproduction and transmission to colonization, p. 384-399. In Parasitism. The diversity and ecology of animal parasites. Cambridge University Press, Cambridge, United Kingdom.
- 6.De Graaf, D. C., G. Masschelein, F. Vandergeynst, H. F. De Bravander, and F. J. Jacobs. 1993. In vitro germination of Nosema apis (Microspora: Nosematidae) spores and its effects on their αα-trehalose/d-glucose ratio. J. Invertebr. Pathol. 62:220-225. [Google Scholar]
- 7.Dyess, E. G., and C. A. Wilson. 1978. A study of the seasonal variations of Nosema apis Zander of honey bee in Mississippi. Am. Bee J. 118:33-35. [Google Scholar]
- 8.Faucon, J. P. 2005. La nosémose. Sante Abeille 209:343-367. [Google Scholar]
- 9.Faucon, J. P., L. Mathieu, M. Ribiere, A. C. Martel, P. Drajnudel, S. Zeggane, C. Aurieres, and M. F. A. Aubert. 2002. Honey bee winter mortality in France in 1999 and 2000. Bee World 83:14-23. [Google Scholar]
- 10.Fries, I. 1993. Nosema apis—a parasite in the honey bee colony. Bee World 74:5-19. [Google Scholar]
- 11.Fries, I., and F. Feng. 1995. Crossinfectivity of Nosema apis in Apis mellifera and Apis cerana, p. 151-155. In Proceedings of the 34th International Apicultural Congress. Apimondia, Bucharest, Romania.
- 12.Fries, I., F. Feng, A. da Silva, S. B. Slemenda, and J. Pieniazek. 1996. Nosema ceranae n. sp. (Microspora, Nosematidae). Morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 32:356-365. [Google Scholar]
- 13.Fries, I., R. Martín, A. Meana, P. García-Palencia, and M. Higes. 2006. Natural infections of Nosema ceranae in European honey bees. J. Apic. Res. 45:230-233. [Google Scholar]
- 14.Hassanein, M. H. 1953. The influence of infection with Nosema apis on the activities and longevity of the worker honeybee. Ann. Appl. Biol. 40:418-423. [Google Scholar]
- 15.Higes, M., P. García-Palencia, R. Martín-Hernández, and A. Meana. 2007. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J. Invertebr. Pathol. 94:211-217. [DOI] [PubMed] [Google Scholar]
- 16.Higes, M., R. Martín, A. Sanz, N. Alvarez, A. Sanz, M. P Garcia, and A. Meana. 2005. El síndrome de despoblamiento de las colmenas en España. Consideraciones sobre su origen. Vida Apicola 133:15-21. [Google Scholar]
- 17.Higes, M., R. Martín, and A. Meana. 2006. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 92:93-95. [DOI] [PubMed] [Google Scholar]
- 18.Higes, M., R. Martín, and A. Sanz. 2004. Chronic nosemosis in Apis mellifera: an increasing problem, p. 637-638. In S. Mas-Coma, M. D. Bargues, J. G. Esteban, and M. A. Valero (ed.), Multidisciplinarity for parasites, vectors and parasitic diseases. Proceedings of the IX European Multicolloquium of Parasitology, Valencia, Spain.
- 19.Hornitzky, M. 11 January 2007, accession date. Nosema disease. Literature review and survey of beekeepers. Rural Industries Research and Development Corporation, Kingston, Australia. http://www.rirdc.gov.au/reports/HBE/05-055.pdf.
- 20.Huang, W. F., J. H. Jiang, Y. W. Chen, and C. H. Wang. 2007. A Nosema ceranae isolate from the honeybee Apis mellifera. Apidologie 38:1-8. [Google Scholar]
- 21.Klee, J., W. T. Tay, and R. Paxton. 2006. Specific and sensitive detection of Nosema bombi (Microsporidia: Nosematidae) in bumble bees (Bombus spp.; Hymenoptera: Apidae) by PCR of partial rRNA gene sequences. J. Invertebr. Pathol. 91:98-104. [DOI] [PubMed] [Google Scholar]
- 22.Kreader, C. A. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martín, R., A. Meana, and M. Higes. 2005. Increase of nosemosis in Spain, p. 49-50. In A. J. dos Santos Grácio (ed.), Acta Parasitológica Portuguesa. Socidedade Portuguesa de Parasitologia, Lisboa, Portugal.
- 24.Mussen, E. C. 2002. April 2002, posting date. Winter losses, again. Apiculture Newsletter. http://entomology.ucdavis.edu/faculty/mussen/03-04-02.pdf.
- 25.National Meteorology Institute. January 2007, accession date. Resumen del año hidrológico 2005-2006. Ministerio de Medio Ambiente, Madrid, Spain. http://www.inm.es/web/sup/tiempo/climat/res_cli/res_anual_clim_2005.pdf.
- 26.Office International des Epizooties. July 2004, posting date. Manual of standards for diagnostic test and vaccines. Office International des Epizooties, Paris, France. http//www.oie.int.
- 27.Rice, R. 2001. Nosema diseases in honeybees. Genetic variation and control. RIRDC 1/46. Rural Industries Research and Development Corporation, Kingston, Australia.
- 28.Singh, Y. 1975. Nosema in Indian honey bee (Apis cerana indica). Am. Bee J. 115:59. [Google Scholar]
- 29.Taraschewski, H. 2006. Hosts and parasites as aliens. J. Helminthol. 80:99-128. [DOI] [PubMed] [Google Scholar]
- 30.Tay, W. T., E. M. O'Mahony, and R. J. Paxton. 2005. Complete rRNA gene sequence reveals that the microsporidium Nosema bombi infects diverse bumble bee (Bombus spp.) hosts yet contains multiple polymorphic sites. J. Eukaryot. Microbiol. 52:505-513. [DOI] [PubMed] [Google Scholar]
- 31.Valles, S. M., D. H. Oi, J. A. Briano, and D. F. Williams. 2004. Simultaneous detection of Vairimorpha invictae (Microsporidia: Burenellidae) and Thelohania solenopsae (Microsporidia: Thelohaniidae) in fire ants by PCR. Fla. Entomol. 87:85-87. [Google Scholar]
- 32.Valles, S. M., D. H. Oi, O. P. Perera, and D. F. Williams. 2002. Detection of Thelohania solenopsae (Microsporidia: Thelohaniidae) in Solenopsis invicta (Hymenoptera: Formicidae) by multiplex PCR. J. Invertebr. Pathol. 81:196-201. [DOI] [PubMed] [Google Scholar]
- 33.Wang, D. I., and F. E. Moeller. 1971. Ultrastructural changes in the hypopharingeal glands of worker honey bee infected by Nosema apis. J. Invertebr. Pathol. 17:308-320. [Google Scholar]
- 34.Webster, T. C., K. W. Pomper, G. Hunt, E. M. Thacker, and S. C. Jones. 2004. Nosema apis infection in worker and queen Apis mellifera. Apidologie 35:49-54. [Google Scholar]
- 35.Weiss, L. M., and C. R. Vossbrinck. 1999. Molecular biology, molecular phylogeny and molecular diagnostic approaches to the microsporidia, p. 129-171. In M. Witner and L. M. Weiss (ed.), The microsporidia and microsporidiosis. American Society for Microbiology, Washington, DC.
- 36.Whipps, C. M., and M. Kent. 2006. Polymerase chain reaction detection of Pseudoloma neurophilia, a common microsporidian of zebrafish (Danio rerio) reared in research laboratories. J. Am. Assoc. Lab. Anim. Sci. 45:36-39. [PMC free article] [PubMed] [Google Scholar]