Abstract
The current study focuses on a symbiotic bacterium found in the slender pigeon louse, Columbicola columbae (Insecta: Phthiraptera). Molecular phylogenetic analyses indicated that the symbiont belongs to the gamma subdivision of the class Proteobacteria and is allied to Sodalis glossinidius, the secondary symbiont of tsetse flies (Glossina spp.) and also to the primary symbiont of grain weevils (Sitophilus spp.). Relative-rate tests revealed that the symbiont of C. columbae exhibits accelerated molecular evolution in comparison with the tsetse fly symbiont and the weevil symbiont. Whole-mount in situ hybridization was used to localize the symbiont and determine infection dynamics during host development. In first- and second-instar nymphs, the symbionts were localized in the cytoplasm of oval bacteriocytes that formed small aggregates on both sides of the body cavity. In third-instar nymphs, the bacteriocytes migrated to the central body and were finally located in the anterior region of the lateral oviducts, forming conspicuous tissue formations called ovarial ampullae. In adult females, the symbionts were transmitted from the ovarial ampullae to developing oocytes in the ovarioles. In adult males, the bacteriocytes often disappeared without migration. A diagnostic PCR survey of insects collected from Japan, the United States, Australia, and Argentina detected 96.5% (109/113) infection, with a few uninfected male insects. This study provides the first microbial characterization of a bacteriocyte-associated symbiont from a chewing louse. Possible biological roles of the symbiont are discussed in relation to the host nutritional physiology associated with the feather-feeding lifestyle.
Symbiotic associations with microorganisms are ubiquitous among a diverse array of insects. Some obligate symbionts are mutualistic in nature and contribute to the fitness of their hosts, while others are facultative and may have negative impacts upon host fitness (4, 5, 8). In many of these intimate associations, the symbionts are housed in specialized cells known as bacteriocytes or mycetocytes. Regardless of their obligate or facultative nature, most of these symbiotic microorganisms are vertically transmitted at early stages of oogenesis or embryogenesis, wherein the transmission process is integrated into the life cycle of the host insects (6, 14, 42).
A number of insects live solely on diets that are nutritionally incomplete or difficult to utilize, such as woody materials (hard to digest, low nitrogen), plant sap (few proteins and lipids), vertebrate blood (deficient in B vitamins), and others. In many of these cases, symbiotic microorganisms have been shown to play crucial roles in compensating for these nutritional deficiencies. In termites, for example, gut protozoans and bacteria enable the host to digest cellulose. In addition, some of these bacterial symbionts are involved in nitrogen fixation for the termite host (7, 30). In aphids, the endocellular bacterial symbiont Buchnera aphidicola efficiently synthesizes essential amino acids that are lacking in plant phloem sap (12, 39). In tsetse flies, the endocellular bacterial symbiont Wigglesworthia glossinidia provides the host with B vitamins that are lacking in vertebrate blood (1, 28).
Chewing lice (Insecta: Phthiraptera), embracing over 4,400 described species in the world, are ectoparasitic insects feeding on avian feather or mammalian skin and skin products (32). The main component of feather and hair is keratin, a protein constituting the intermediate filament of eukaryotic cells, concentrated in hard animal tissues such as feather, hair, nail, scale, beak, and horn and resistant to solubilization, proteolysis, and digestion (18, 27). Although protein rich and potentially nutritious, these hard tissues are difficult to utilize for most animals, with only a few insect groups such as chewing lice (Phthiraptera: Ischnocera, Amblycera), carpet beetles (Coleoptera: Dermestidae), keratin beetles (Coleoptera: Trogidae), and clothes moths (Lepidoptera: Tineidae) having evolved the ability to live on this difficult diet (21, 43). Possibly relevant to the nutritional difficulty, some chewing lice possess a well-developed endosymbiotic association, wherein bacteriocyte-associated symbiotic bacteria migrate to the ovary in a peculiar passage and are vertically transmitted to the oocytes in the maternal body (8, 34). Although these bacteria were visualized in early histological studies, no formal identification has yet been provided by molecular phylogenetic analyses. Previous studies have however demonstrated the presence of facultative endosymbiotic bacteria of the genus Wolbachia in many chewing and sucking lice (25) and a diverse assemblage of putative gut bacteria in the chewing lice of pocket gophers (33).
In this study, we present the first microbiological characterization of the bacteriocyte-associated symbiotic bacterium in the slender pigeon louse, Columbicola columbae, using molecular phylogenetic analyses and histological techniques. Although chewing lice are phylogenetically close to sucking lice (3, 22), the symbiont of C. columbae was not related to the symbionts of primate lice (Riesia spp.) (2, 37) but, unexpectedly, was allied to the symbionts of tsetse flies and grain weevils.
MATERIALS AND METHODS
Insect materials.
Samples of the slender pigeon louse, C. columbae, used in this study are listed in Table 1. The insects were collected from the rock pigeon, Columba livia, and immediately preserved in acetone (15).
TABLE 1.
Samples of C. columbae examined in this study, results of diagnostic PCR detection, and sequence accession numbers
| Sample code | Collection locality | Collection date, collectora | Infection frequencyb
|
Accession no. for:
|
||||
|---|---|---|---|---|---|---|---|---|
| No sexingc | Female | Male | Total | 16S rRNA gene | fusA gene | |||
| FKK99 | Ropponmatsu, Fukuoka, Japan | 27 August 1999, KY | AB303383 | |||||
| SPR06 | Sapporo, Hokkaido, Japan | 16 August 2006, KY | 100% (10/10) | 100% (10/10) | 100% (10/10) | 100% (30/30) | AB303382 | |
| SMY06 | Sumiyoshi, Osaka, Japan | 19 October 2006, TW | 90% (9/10) | 90% (9/10) | AB303384 | |||
| TTR06 | Tsuchiura, Ibaraki, Japan | 10 November 2006, KSF and TF | 100% (10/10) | 100% (10/10) | AB303385 | |||
| NNW07 | Naniwa, Osaka, Japan | 5 March 2007, NU | 100% (20/20) | 95% (19/20) | 97.5% (39/40) | |||
| BNS06 | Buenos Aires, Argentina | 19 October 2006, TF | 100% (10/10) | 100% (10/10) | AB303386 | |||
| UTH98 | Utah, United States | 29 June 1998, DC | 100% (3/3) | 100% (3/3) | EU021695 | |||
| UTH99 | Utah, United States | 1999, DC | EU021696 | |||||
| BRB02 | Brisbane, Australia | 2002, DC | EU021697 | |||||
| BRB07 | Brisbane, Australia | 16 February 2007, TF | 80% (8/10) | 80% (8/10) | AB303387 | |||
| Total | 94.3% (50/53) | 100% (30/30) | 96.7% (29/30) | 96.5% (109/113) | ||||
DC, Dale Clayton; KSF, Kayoko Sasaki-Fukatsu; KY, Kazunori Yoshizawa; NU, Nobutaka Urano; TF, Takema Fukatsu; TW, Takeshi Wada.
Percentage of infected insects; numbers in parentheses are numbers of insects infected/number examined by diagnostic PCR.
Including female adults, male adults, and nymphs.
DNA extraction and morphological inspection.
Each of the acetone-preserved insects was briefly dried in air, cut into two parts using a razor, and digested in 200 μl of lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 0.5% sodium dodecyl sulfate, 0.8 mg/ml proteinase K) at 55°C overnight. The exoskeleton of the insect was recovered, mounted on a microscope slide, and observed under a light microscope for morphological identification. The lysate was extracted with phenol-chloroform and subjected to ethanol precipitation, and the precipitated DNA was dried and dissolved in 50 μl of TE buffer (20 mM Tris-HCl [pH 8.0], 0.1 mM EDTA).
DNA cloning and sequencing.
The DNA samples from individual insects were subjected to PCR amplification of a 1.5-kb segment of the eubacterial 16S rRNA gene using the primers 16SA1 (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 16SB1 (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) (17) and a 0.76-kb segment of the fusA gene using the primers FusAF (5′-CAT CGG CAT CAT GGC NCA YAT HGA-3′) and FusAR (5′-CAG CAT CGG CTG CAY NCC YTT RTT-3′) (11). The PCR products were subjected to cloning, restriction fragment length polymorphism genotyping, and DNA sequencing as previously described (17).
Molecular phylogenetic analysis.
The DNA sequences were subjected to molecular phylogenetic analysis together with the sequences of related gammaproteobacteria that exhibited high BLAST scores in the DNA database search. A multiple alignment of the sequences was generated using the program Clustal W (40). Aligned nucleotide sites containing a gap were removed from the data set, and the final alignment was inspected and corrected manually. Neighbor joining (NJ) trees, with 1,000 bootstrap resamplings, were constructed using Clustal W (40). Kimura's two-parameter model was used for correction of multiple substitutions (23). Maximum parsimony (MP) trees, with 1,000 bootstrap resamplings, were generated by the program MEGA 3.1 (24). For finding the MP trees, the close-neighbor-interchange algorithm was used. An initial tree was generated by random addition of the sequences. Maximum likelihood (ML) trees were constructed by the program TREE-PUZZLE 5.2 (38), wherein supporting values for internal nodes were inferred by 1,000 puzzling steps. As a nucleotide substitution model, the HKY+Γ+Inv model was used. We tested several different substitution models and confirmed that the differences of the substitution models did not lead to any discrepancies in the tree topologies supported with high bootstrap values.
Relative rate test.
A relative rate test, based on genetic distances estimated under the Kimura's two-parameter model (23), was performed using the program RRTree (35). For 16S rRNA gene sequences, 1,444 unambiguously aligned nucleotide sites were subjected to the analysis. For fusA gene sequences, 499 unambiguously aligned nucleotide sites at the first- and second-codon positions were analyzed, while nucleotide sites at the third-codon positions were omitted from the analysis due to saturated nucleotide substitutions.
wFISH.
An oligonucleotide probe specific to the 16S rRNA sequence from C. columbae, AlexaFluor555-CcolSol427R (Al555-5′-CAT CGC CTT CCT CCC AGT CG-3′), was used for whole-mount fluorescent in situ hybridization (wFISH). After being decapitated to facilitate infiltration of reagents, the acetone-preserved insects were fixed in Carnoy's solution (chloroform-ethanol-acetic acid [6:3:1]) for 2 days. Subsequently the insects were incubated with 6% H2O2 in ethanol for 2 weeks to quench the autofluorescence of insect tissues. The insects were thoroughly washed and equilibrated with a hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate, 30% formamide), and the probe and SYTOX green were added at final concentrations of 10 nM and 5 μM, respectively. After an overnight incubation, the samples were thoroughly washed in 20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate and observed under an epifluorescence microscope (Axiophoto; Carl Zeiss) and a laser confocal microscope (PASCAL5; Carl Zeiss). To confirm specific detection of the symbionts, a series of control experiments, namely, the no-probe control, RNase digestion control, and competitive-suppression control with excess unlabeled probe, were conducted as previously described (36).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession numbers AB303382 to AB303387 and EU021695 to EU021697 (Table 1; also see Fig. 1 and 2).
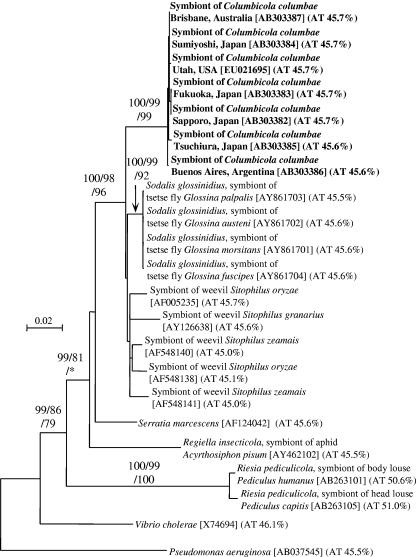
FIG. 1.
Molecular phylogenetic analysis on the basis of 16S rRNA gene sequences of the symbiont of C. columbae and allied gammaproteobacteria. An NJ tree inferred from 1,363 unambiguously aligned nucleotide sites is shown; MP and ML analyses gave essentially the same results (data not shown). Statistical support values higher than 70% are indicated at the nodes in the order of NJ/MP/ML. Sequence accession numbers are shown in brackets. AT contents of the sequences are shown in parentheses.
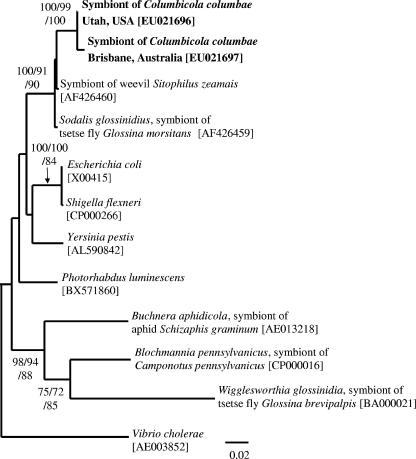
FIG. 2.
Molecular phylogenetic analysis on the basis of fusA gene sequences of the symbiont of C. columbae and allied gammaproteobacteria. An NJ tree inferred from 499 unambiguously aligned nucleotide sites is shown; MP and ML analyses gave essentially the same results (data not shown). Statistical support values higher than 70% are indicated at the nodes in the order of NJ/MP/ML. Sequence accession numbers are shown in brackets.
RESULTS
Bacterial 16S rRNA gene sequences from C. columbae.
From all the insect samples collected in Japan, the United States, Australia, and Argentina, 1,478-bp 16S rRNA gene sequences exhibiting 99.9 to 100% sequence identities to each other were identified. For each of the samples, more than 10 clones of the 16S rRNA gene segment showed identical restriction fragment length polymorphism patterns, indicating a single bacterial species dominant in the insects. A BLAST search clearly showed that the sequence belongs to the Enterobacteriaceae in the Gammaproteobacteria. In the DNA databases, we found several high-score hits including the secondary symbiotic bacterium Sodalis glossinidius from tsetse flies (Glossina spp.; GenBank accession no. AY861701; 96.5% sequence identity) and the primary symbiotic bacteria from grain weevils (Sitophilus spp.; GenBank accession no. AF005235; 96.0% sequence identity).
Phylogenetic placement of the symbiont of C. columbae based on 16S rRNA gene sequences.
These 16S rRNA gene sequences were subjected to molecular phylogenetic analysis together with the sequences of related gammaproteobacteria that exhibited high BLAST scores in the DNA database search (Fig. 1). The bacterial sequences from different C. columbae populations formed a monophyletic group with nearly 100% statistical support, constituting a distinct lineage in the gamma subclass of the Proteobacteria. The sequences also formed a monophyletic group together with the sequences of the tsetse fly symbionts and the weevil symbionts, which also garnered close to 100% statistical support.
Phylogenetic placement of the symbiont of C. columbae based on a protein-coding gene.
From DNA samples from insects collected in Australia and the United States, we cloned and sequenced a 760-bp segment of the fusA gene, encoding elongation factor G, a bacterial ribosomal translocase (11). Molecular phylogenetic analysis also showed that the fusA sequences from C. columbae formed a clade with the sequences from the tsetse fly symbiont and the weevil symbiont (Fig. 2).
Accelerated molecular evolution in the symbiont of C. columbae.
On the phylogenetic trees (Fig. 1 and 2), the lineage of the symbionts of C. columbae exhibited remarkably elongated branches in comparison with the lineages of the tsetse fly symbionts and the weevil symbionts. Thus, we performed relative-rate tests based on genetic distances between the gene sequences. The evolutionary rate of the 16S rRNA gene sequence in the lineage of the symbionts of C. columbae was 3.1 times and 2.7 times faster than those in the lineages of the tsetse fly symbionts and the weevil symbionts, respectively. In both cases, the differences were highly significant (P < 0.001) (Table 2). The evolutionary rate of the fusA gene sequence in the lineage of the symbionts of C. columbae was 25 times and 22 times faster than those in the lineages of the tsetse fly symbionts and the weevil symbionts, respectively (Table 2; P < 0.01).
TABLE 2.
Relative-rate tests for comparing the molecular evolutionary rates of the 16S rRNA gene and fusA gene between the symbionts of C. columbae, the symbionts of tsetse flies, and the symbionts of grain weevils
| Gene | Lineage
|
Outgroup | K1a | K2b | Difference of distance (K1 − K2) | Rate ratio (K1/K2) | Pc | |
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | |||||||
| 16S rRNA | Symbionts of C. columbaed | Symbionts of tsetse fliese | S. marcescensf | 0.027 | 0.009 | 0.018 | 3.1 | 0.00059 |
| gene | Symbionts of C. columbaed | Symbionts of grain weevilsg | S. marcescensf | 0.026 | 0.010 | 0.016 | 2.7 | 0.00092 |
| fusA | Symbiont of C. columbaeh | Symbiont of tsetse fliesi | E. colij | 0.025 | 0.001 | 0.024 | 25.0 | 0.0020 |
| Symbiont of C. columbaeh | Symbiont of grain weevilsk | E. colij | 0.022 | 0.001 | 0.021 | 22.0 | 0.0034 | |
K1, estimated mean distance between lineage 1 and the last common ancestor of lineages 1 and 2.
K2, estimated mean distance between lineage 2 and the last common ancestor of lineages 1 and 2.
P values were generated using the program RRTree (35).
Symbionts of C. columbae from Sapporo, Japan (GenBank accession no. AB303382), and Buenos Aires, Argentina (GenBank accession no. AB303386).
Sodalis glossinidius symbionts of tsetse flies Glossina morsitans (GenBank accession no. AY861701) and Glossina palpalis (GenBank accession no. AY861703).
Serratia marcescens (GenBank accession no. AF124042).
Symbionts of grain weevils Sitophilus oryzae (GenBank accession no. AF005235) and Sitophilus zeamais (GenBank accession no. AF005235).
Symbionts of C. columbae from Utah (GenBank accession no. EU021696) and from Brisbane, Australia (GenBank accession no. EU021697).
Sodalis glossinidius, symbiont of tsetse fly G. morsitans (GenBank accession no. AF426459).
Escherichia coli (GenBank accession no. X00415).
Symbiont of grain weevil S. zeamais (GenBank accession no. AF426460).
AT content of 16S rRNA gene sequences of the symbiont of C. columbae.
The 16S rRNA gene sequences derived from the symbiont of C. columbae were determined to be 45.6 to 45.7% AT, values which were not significantly different from those for the 16S rRNA sequences of the tsetse fly symbionts, the weevil symbionts, and other free-living gammaproteobacteria (Fig. 1).
wFISH of the symbiont in C. columbae.
In order to investigate in vivo localization and infection dynamics of the symbiont, nymphs and adults of C. columbae at different developmental stages were subjected to wFISH.
General localization in nymphal and adult insects.
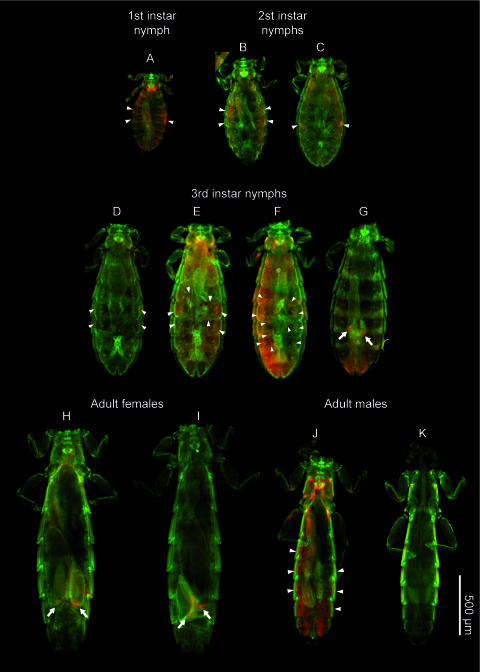
Figure 3 shows wFISH detection of the symbiont in the whole body of C. columbae at different developmental stages. In first-, second-, and third-instar nymphs, aggregates of bacteriocytes were located on the both sides of the body cavity (Fig. 3A to F). In a part of late third-instar nymphs and all adult females, the symbiont signals in the lateral body cavity disappeared and the symbiont cells were localized in the ovary (Fig. 3G to I). In adult males, some individuals possessed the symbiont-harboring bacteriocytes in the lateral body cavity (Fig. 3J) while other individuals exhibited few signals of the symbiont (Fig. 3K).
FIG. 3.
General localization of the symbiont in nymphs and adults of C. columbae. Red and green signals indicate symbiont cells and host nuclei, respectively, while cuticles often exhibit red signals due to autofluorescence. Arrowheads, hybridization signals of bacteriocytes in the body cavity; arrows, hybridization signals associated with lateral oviducts, ovarial ampullae, and/or oocytes.
Localization of the symbiont in lateral aggregates of bacteriocytes in first- and second-instar nymphs.
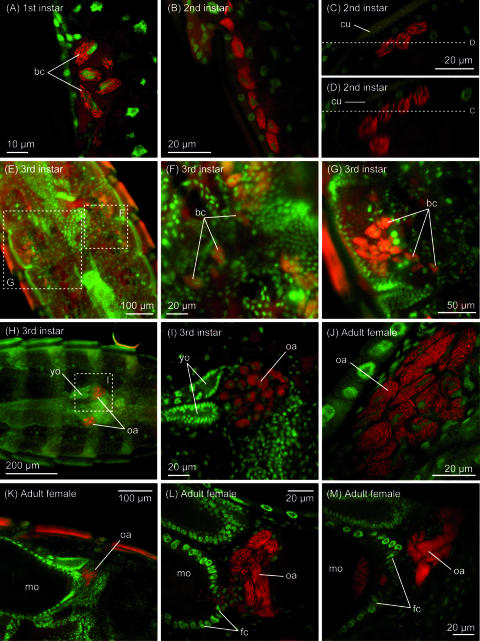
In first- and second-instar nymphs, oval bacteriocytes were 10 to 15 μm in the longer diameter and 5 to 7 μm in the shorter diameter and their cytoplasm was observed to be packed with symbiotic bacteria (Fig. 4A and B). These bacteriocytes were found in groups, usually on both sides of the third and fourth abdominal segments (Fig. 3A to C). The clustered bacteriocytes were arranged linearly just beneath the hypodermis of the abdominal segments (Fig. 4C and D).
FIG. 4.
Localization of the symbiont in nymphs and adults of C. columbae. (A) Aggregate of bacteriocytes in the lateral body cavity of a first-instar nymph. Cytoplasm of oval bacteriocytes is full of rod-shaped symbiont cells. (B) Aggregate of bacteriocytes in the lateral body cavity of a second-instar nymph. The number of bacteriocytes increases, and the area occupied by the aggregate stretches longitudinally. (C) Distribution of bacteriocytes beneath the hypodermis of the abdominal segment in a first-instar nymph. (D) z-axis reconstruction of the distribution of bacteriocytes through plane D shown in panel C. (E) Migration process of bacteriocytes in a third-instar female nymph. (F and G) Enlarged images of the migrating bacteriocytes. (H) Localization of bacteriocytes in lateral oviducts and formation of ovarial ampullae in a third-instar female nymph. (I) Enlarged image of an ovarial ampulla of the third-instar female nymph. (J) Enlarged image of an ovarial ampulla of a female adult. (K) Ovarial ampulla associated with a mature oocyte in an ovariole. (L) Infection process of symbiont cells from an ovarial ampulla to the posterior pole of an oocyte through follicle cells. (M) Symbiont cells localized in the posterior region of an oocyte. Abbreviations: bc, bacteriocytes; cu, cuticle; fc, follicle cells; mo, mature oocyte; oa, ovarial ampullae; yo, young oocyte.
Migration of bacteriocytes to lateral oviducts in third-instar nymphs.
In third-instar nymphs, localization of the bacteriocytes showed a drastic change. At the beginning, some of the bacteriocytes in the lateral body cavity were found outside the aggregates, apparently migrating toward the central body region (Fig. 3E and F and 4E). The more bacteriocytes participated in the migration, the further disintegration of the aggregates proceeded (Fig. 4E to G). Finally, all the bacteriocytes were located at the anterior region of the lateral oviducts and formed specialized tissue formations for symbiont transmission, so-called ovarial ampullae (8, 34) (Fig. 4H and I).
Vertical transmission of the symbiont from ovarial ampullae to oocytes in adult females.
In adult females, the symbiont cells were vertically transmitted from the well-developed ovarial ampullae (Fig. 3H and I and 4J) to the posterior pole of oocytes in the ovarioles (Fig. 4K to M). The symbionts from the ovarial ampullae passed through follicle cells and reached the posterior pole of oocytes (Fig. 4L), where a specific region was densely infected with the symbiont cells (Fig. 4M).
Prevalence of the symbiont in worldwide populations of C. columbae.
We examined individuals of C. columbae collected from Japan, the United States, Australia, and Argentina by diagnostic PCR for the symbiont infection. First, 53 insects from six populations were inspected without sexing, which revealed 94.3% (50/53) infection. Next, we inspected 30 adult females and 30 adult males from two populations and found 100% (30/30) infection in females and 96.7% (29/30) infection in males. In total, 109 of 113 insects examined were infected with the symbiont, indicating an infection frequency of 96.5% (Table 1).
DISCUSSION
Over 70 years ago, early investigators reported that some chewing lice harbor bacteriocyte-associated endosymbiotic bacteria (8, 34). Since then, however, the nature of the symbiosis has been elusive. To our knowledge, this study provides the first microbiological characterization of a bacteriocyte-associated symbiont from a chewing louse.
Morphologically, chewing lice, consisting mainly of bird lice such as C. columbae, are thought to be related to sucking lice, consisting exclusively of mammalian lice such as human lice (Pediculus humanus and Pediculus capitis) (46). Recent molecular phylogenetic analyses confirmed that the clade of sucking lice is actually nested in a clade of chewing lice (3, 22). However, we found that the endosymbiont of C. columbae was not closely related to the bacterial endosymbiont found in the human lice of Riesia spp. (Fig. 1). Hence, their symbiotic bacteria are likely of independent evolutionary origins, as reflected in the different types of a symbiotic organ, namely, the highly specialized organ called the stomach disk, in human lice (13, 34, 37) and the loosely associated bacteriocytes in C. columbae (Fig. 3 and 4) (34). The difference might be relevant to their distinct ecological niches and nutritional physiologies: sucking lice persist exclusively on a diet of vertebrate blood, whereas chewing lice feed on a keratin-rich diet composed primarily of feather or skin (32). Cytology, localization, and infection dynamics of the endosymbionts in not only sucking lice but also chewing lice are extremely diverse (8, 34), corroborating the idea that their symbiotic associations are of independent evolutionary origins. Interestingly, there are several peculiar features that are shared between the endosymbiotic systems of sucking lice and chewing lice, such as the symbiont migration to the ovary at the third instar and the specialized tissue formations for symbiont transmission called ovarial ampullae (Fig. 3 and 4) (8, 13, 34, 37). These shared features are perhaps indicative of some common developmental and evolutionary bases underlying their endosymbiotic systems. In order to better understand the complexities of endosymbiosis in these systems, more-intensive surveys are needed to characterize the host and symbiont diversity.
In vivo localization and infection dynamics of the symbiont of C. columbae were described in detail in the pioneering work of Ries (34). Our wFISH results (Fig. 3 and 4) were totally concordant with these early histological descriptions. Here we point out an enigmatic phenomenon that Ries (34) and ourselves observed consistently in the migration of the bacteriocytes from the lateral body cavity into the ovary. In third-instar nymphs of C. columbae, individual bacteriocytes begin to migrate toward the central body region (Fig. 3E and F and 4E to G), arriving at the anterior region of the lateral oviducts (Fig. 3G and 4I), whereupon ovarial ampullae are formed for symbiont transmission to the oocytes (Fig. 3H and I and 4J to M). It should be noted that the bacteriocytes in the lateral body cavity (Fig. 4A to D), the migrating bacteriocytes in the central body region (Fig. 4F and G), and the bacteriocytes located inside the lateral oviducts (Fig. 4I) look very similar cytologically. In the migration process, neither disintegrating bacteriocytes nor extracellular symbiont cells are observed, and thus it seems that the whole bacteriocytes somehow pass through the wall of the oviducts and gain entry into the ovarial cavity (34). The mechanism of whole-cell penetration into the ovary is intriguing and should be pursued by more-detailed histological examinations in the future.
Molecular phylogenetic analyses revealed that the symbiont of C. columbae is closely related to the secondary symbiont of the tsetse flies, S. glossinidius, and also to the primary symbiont of the grain weevils (Fig. 1 and 2). The phylogenetic proximity of the symbionts is somewhat puzzling, since chewing lice, tsetse flies, and grain weevils represent different insect orders, Phthiraptera, Diptera, and Coleoptera, respectively. To account for the sporadic distribution of the Sodalis-allied endosymbionts, there may have been horizontal transfer between the distant insect lineages some time in the evolutionary past, although biological connections between these insects are difficult to imagine. Recently, a new member of Sodalis-allied symbiont was identified from hippoboscid fly Craterina melbae (29). It therefore seems likely that the host insect range of this symbiont clade is much broader than previously envisioned.
Recent molecular evolutionary analyses have suggested that the lifestyle of obligate insect symbionts has strongly affected their genome evolution, causing AT-biased nucleotide composition, accelerated rate of molecular evolution, and significant genome reduction. These peculiar genetic traits are hypothesized to be the consequence of attenuated purifying selection due to small population size and frequent transmission bottlenecks, which are associated with the lifestyle of vertically transmitted symbionts (20, 45). The symbiont of C. columbae exhibited significantly faster molecular evolutionary rates in 16S rRNA and fusA gene sequences than the tsetse fly symbiont and the weevil symbiont (Table 2), suggesting that these population genetic parameters might be strongly affected in the symbiont of C. columbae. To better understand this phenomenon, further studies are required to determine the age of the association between Columbicola spp. and their bacterial symbionts. It should be noted that the endosymbioses in the tsetse flies and the grain weevils are presumably of relatively recent origins. The eroded genome of S. glossinidius suggests recent transition of the bacterial lifestyle from free-living to endosymbiotic (41). While many of the weevils of the family Dryophthoridae are associated with an ancient symbiont lineage of the genus Nardonella, only the grain weevils (Sitophilus spp.) are associated with the Sodalis-allied symbiont, which suggests a later replacement of the endosymbiotic associates in the ancestor of the weevil genus (26). Interestingly the AT contents of 16S rRNA gene sequences were not different among the related louse, tsetse fly, and weevil symbionts (Fig. 1). The genome size of the tsetse fly symbiont S. glossinidius is known to be 4,171,146 bp (41), whereas the genome size of the weevil symbiont is estimated to be around 3.0 Mb (9). It will therefore be of interest to determine the genome size of the symbiont of C. columbae.
For chewing lice, Ries (34) made histological observations of bacterial endosymbionts from the genera Columbicola, Sturnidoecus, Goniocotes, Campanulotes, Colocerus, Goniodes, Anaticola, Turdinirmus, Kelerinirmus, and Brueelia, while no endosymbionts were detected from the family Trichodectidae. Our ongoing work will determine whether these chewing lice harbor the Sodalis-allied endosymbionts in common with C. columbae or other unrelated symbiotic bacteria.
Diagnostic PCR surveys demonstrated that the symbiont consistently exhibited high infection frequencies in natural populations of C. columbae worldwide (Table 1). Considering the prevalence of symbiont infection (Table 1) and the highly developed endosymbiotic devices such as bacteriocytes and ovarial ampullae (Fig. 3 and 4), it seems likely that the symbiont does play some important biological roles for the host insect. In grain weevils (Sitophilus spp.), the primary symbiont contributes to growth and fecundity of the host insects (19). In tsetse flies (Glossina spp.), the biological roles of the secondary symbiont, S. glossinidius, have been obscured by the presence of the primary symbiont, W. glossinidia (41, 44). In C. columbae, although the biological roles of the symbiont are currently unknown, the well-developed endosymbiotic system might be relevant to the feather-feeding lifestyle and physiology of the insect. The main component of feather is keratin, a hard protein resistant to solubilization, proteolysis, and digestion (21, 43). Hence, the symbiont might possibly be involved in keratin digestion, although the nonintestinal localization of the symbiont (Fig. 3 and 4) is not favorable to the hypothesis. Alternatively, the symbiont might contribute to the host insect nutritionally. The amino acid composition of keratin is conspicuously biased (18, 27), and the symbiont might compensate for this bias. Feather might also be devoid of vitamins and other trace nutrients, which could be supplied by the symbiont. We are currently carrying out physiological studies using symbiotic and aposymbiotic insects and genomic studies of the symbiont to provide insights into these biological aspects of the endosymbiosis in the chewing louse.
Of 113 individuals examined in this study, only 4 insects were diagnosed as negative for the symbiont (Table 1). The diagnostic PCR results of sexed individuals (Table 1) and the FISH results for adult males (Fig. 3) suggest that these symbiont-free insects are probably males. Male-specific absence of symbiont infection has been reported from aphids, coccids, and other insects (8, 16), which might be relevant to the fact that males do not contribute to vertical transmission of the symbiont to the next generation. It should be noted, however, that the samples of C. columbae are field collected and thus may contain old insects and unhealthy insects, from which the symbiont infection could be accidentally lost irrespective of their sex.
Exceptionally among insect endosymbionts, the tsetse fly symbiont S. glossinidius is culturable in cell-free media (10), making the bacterium a unique model for studies of insect-microbe symbiosis (31). For example, it was experimentally demonstrated that, like many pathogenic bacteria, the symbiont recruits the type III secretion system for invasion to the host cells (11). In addition, the ability to maintain S. glossinidius in pure culture greatly facilitated genome sequencing of the symbiont (41). Considering its phylogenetic affinity to the tsetse fly symbiont, the symbiont of C. columbae would, in addition to the primary symbiont of the grain weevils, provide further insights into how endosymbiotic associations could evolve from parasitism, through commensalism, and ultimately toward mutualism. At present, attempts are under way to culture the C. columbae endosymbiont using the techniques established previously for S. glossinidius.
Acknowledgments
We thank Takeshi Wada and Nobutaka Urano for their help with sampling of C. columbae.
We are supported by NSF awards DEB-0614565 (to D.H.C. and C.D.) and EF-58501127 (to C.D.).
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. [DOI] [PubMed] [Google Scholar]
- 2.Allen, J. M., D. L. Reed, M. A. Perotti, and H. R. Braig. 2007. Evolutionary relationships of “Candidatus Riesia spp.,” endosymbiotic Enterobacteriaceae living within hematophagous primate lice. Appl. Environ. Microbiol. 73:1659-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker, S. C., M. Whiting, K. P. Johnson, and A. Murrell. 2003. Phylogeny of the lice (Insecta, Phthiraptera) inferred from small subunit rRNA. Zool. Scripta 32:407-414. [Google Scholar]
- 4.Bourtzis, K., and T. A. Miller. 2003. Insect symbiosis. CRC Press, Boca Raton, FL.
- 5.Bourtzis, K., and T. A. Miller. 2006. Insect symbiosis II. CRC Press, Boca Raton, FL.
- 6.Braendle, C., T. Miura, R. Bickel, A. W. Shingleton, S. Kambhampati, and D. L. Stern. 2003. Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 1:70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breznak, J. A., and A. Brune. 1994. Role of microorganisms in the digestion of lignocellulose by termites. Annu. Rev. Entomol. 39:453-487. [Google Scholar]
- 8.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience, New York, NY.
- 9.Charles, H., G. Condemine, C. Nardon, and P. Nardon. 1997. Genome size characterization of the principal endocellular symbiotic bacteria of the weevil Sitophilus oryzae, using pulsed field gel electrophoresis. Insect Biochem. Mol. Biol. 27:345-350. [Google Scholar]
- 10.Dale, C., and I. Maudlin. 1999. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int. J. Syst. Bacteriol. 49:267-275. [DOI] [PubMed] [Google Scholar]
- 11.Dale, C., G. R. Plague, B. Wang, H. Ochman, and N. A. Moran. 2002. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc. Natl. Acad. Sci. USA 99:12397-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas, A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17-37. [DOI] [PubMed] [Google Scholar]
- 13.Eberle, M. W., and D. L. McLean. 1983. Observation of symbiote migration in human body lice with scanning and transmission electron microscopy. Can. J. Microbiol. 29:755-762. [DOI] [PubMed] [Google Scholar]
- 14.Frydman, H. M., J. M. Li, D. N. Robson, and E. Wieschaus. 2006. Somatic stem cell niche tropism in Wolbachia. Nature 441:509-512. [DOI] [PubMed] [Google Scholar]
- 15.Fukatsu, T. 1999. Acetone preservation: a practical technique for molecular analysis. Mol. Ecol. 8:1935-1945. [DOI] [PubMed] [Google Scholar]
- 16.Fukatsu, T., and H. Ishikawa. 1992. Soldier and male of an eusocial aphid Colophina arma lack endosymbiont: implications for physiological and evolutionary interaction between host and symbiont. J. Insect Physiol. 38:1033-1042. [Google Scholar]
- 17.Fukatsu, T., and N. Nikoh. 1998. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl. Environ. Microbiol. 64:3599-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillespie, J. M., and M. J. Frenkel. 1974. The diversity of keratins. Comp. Biochem. Physiol. 47B:339-346. [Google Scholar]
- 19.Heddi, A., and P. Nardon. 2005. Sitophilus oryzae L.: a model for intracellular symbiosis in the Dryophthoridae weevils (Coleoptera). Symbiosis 39:1-11. [Google Scholar]
- 20.Hosokawa, T., Y. Kikuchi, N. Nikoh, M. Shimada, and T. Fukatsu. 2006. Strict host-symbiont co-speciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4:e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes, J., and A. P. Vogler. 2006. Gene expression in the gut of keratin-feeding clothes moths (Tineola) and keratin beetles (Trox) revealed by subtracted cDNA libraries. Insect Biochem. Mol. Biol. 36:584-592. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, K. P., K. Yoshizawa, and V. S. Smith. 2004. Multiple origins of parasitism in lice. Proc. R. Soc. Lond. B 271:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 25.Kyei-Poku, G. K., D. D. Colwell, P. Coghlin, B. Benkel, and K. D. Floate. 2005. On the ubiquity and phylogeny of Wolbachia in lice. Mol. Ecol. 14:285-294. [DOI] [PubMed] [Google Scholar]
- 26.Lefevre, C., H. Charles, A. Vallier, B. Delobel, B. Farrell, and A. Heddi. 2004. Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Mol. Biol. Evol. 21:965-973. [DOI] [PubMed] [Google Scholar]
- 27.Marshall, R. C., D. F. G. Orwin, and J. M. Gillespie. 1991. Structure and biochemistry of mammalian hard keratin. Electron Microsc. Rev. 4:47-83. [DOI] [PubMed] [Google Scholar]
- 28.Nogge, G. 1982. Significance of symbionts for the maintenance of an optimal nutritional state for successful reproduction in hematophagus arthropods. Parasitology 82:299-304. [Google Scholar]
- 29.Novakova, E., and V. Hypsa. 2007. A new Sodalis lineage from bloodsucking fly Craterina melbae (Diptera, Hippoboscoidea) originated independently of the tsetse flies symbiont Sodalis glossinidius. FEMS Microbiol. Lett. 269:131-135. [DOI] [PubMed] [Google Scholar]
- 30.Ohkuma, M. 2003. Termite symbiotic systems: efficient bio-recycling of lignocellulose. Appl. Microbiol. Biotechnol. 61:1-9. [DOI] [PubMed] [Google Scholar]
- 31.Pontes, M. H., and C. Dale. 2006. Culture and manipulation of insect facultative symbionts. Trends Microbiol. 14:406-412. [DOI] [PubMed] [Google Scholar]
- 32.Price, R. D., R. A. Hellenthal, R. L. Palma, K. P. Johnson, and D. H. Clayton. 2003. The chewing lice: world checklist and biological overview. Illinois Natural History Survey Special Publication 24. Illinois Natural History Survey, Champaign, IL.
- 33.Reed, D. L., and M. S. Hafner. 2002. Phylogenetic analysis of bacterial communities associated with ectoparasitic chewing lice of pocket gophers: a culture-independent approach. Microb. Ecol. 44:78-93. [DOI] [PubMed] [Google Scholar]
- 34.Ries, E. 1931. Die Symbiose der Laüse und Federlinge. Z. Morphol. Oekol. Tiere 20:233-367. [Google Scholar]
- 35.Robinson-Rechavi, M., and D. Huchon. 2000. RRTree: relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics 16:296-297. [DOI] [PubMed] [Google Scholar]
- 36.Sakurai, M., R. Koga, T. Tsuchida, X.-Y. Meng, and T. Fukatsu. 2005. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl. Environ. Microbiol. 71:4069-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasaki-Fukatsu, K., R. Koga, N. Nikoh, K. Yoshizawa, S. Kasai, M. Mihara, M. Kobayashi, T. Tomita, and T. Fukatsu. 2006. Symbiotic bacteria associated with stomach discs of human lice. Appl. Environ. Microbiol. 72:7349-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt, H. A., K. Strimmer, M. Vingron, and A. Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 39.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and J. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toh, H., B. L. Weiss, S. A. H. Perkin, A. Yamashita, K. Oshima, M. Hattori, and S. Aksoy. 2006. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veneti, Z., M. E. Clark, T. L. Karr, C. Savakis, and K. Bourtzis. 2004. Heads or tails: host-parasite interactions in the Drosophila-Wolbachia system. Appl. Environ. Microbiol. 70:5366-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterhouse, D. F. 1957. Digestion in insects. Annu. Rev. Entomol. 2:1-18. [Google Scholar]
- 44.Weiss, B. L., R. Mouchotte, R. V. M. Rio, Y. N. Wu, Z. Y. Wu, A. Heddi, and S. Aksoy. 2006. Interspecific transfer of bacterial endosymbionts between tsetse fly species: infection establishment and effect on host fitness. Appl. Environ. Microbiol. 72:7013-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wernegreen, J. J. 2002. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3:850-861. [DOI] [PubMed] [Google Scholar]
- 46.Yoshizawa, K., and K. P. Johnson. 2006. Morphology of male genitalia in lice and their relatives and phylogenetic implications. Syst. Entomol. 31:350-361. [Google Scholar]