Abstract
NrpR is an euryarchaeal transcriptional repressor of nitrogen assimilation genes. Previous studies with Methanococcus maripaludis demonstrated that NrpR binds to palindromic operator sequences, blocking transcription initiation. The metabolite 2-oxoglutarate, an indicator of cellular nitrogen deficiency, induces transcription by lowering the affinity of NrpR for operator DNA. In this report we build on existing genetic tools for M. maripaludis to develop a screen for change-of-function mutations in a transcriptional regulator and demonstrate the use of an X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) screen for strict anaerobes. We use the approach to address the primary structural requirements for the response of NrpR to 2-oxoglutarate. nrpR genes from the mesophilic M. maripaludis and the hyperthermophilic Methanopyrus kandleri were targeted for mutagenesis. M. maripaludis nrpR encodes a protein with two homologous NrpR domains while the M. kandleri nrpR homolog encodes a single NrpR domain. Random point mutagenesis and alanine replacement mutagenesis identified two amino acid residues of M. kandleri NrpR involved in induction of gene expression under nitrogen-deficient conditions and thus in the response to 2-oxoglutarate. Mutagenesis of the corresponding regions in either domain of M. maripaludis NrpR resulted in a similar effect, demonstrating a conserved structure-function relationship between the two repressors. The results indicate that in M. maripaludis, both NrpR domains participate in the 2-oxoglutarate response. The approach used here has wide adaptability to other regulatory systems in methanogenic Archaea and other strict anaerobes.
Reporter gene-based genetic screens for regulatory mutations have enjoyed wide utility in the Bacteria but have had limited use in the Archaea, due in part to the extreme growth conditions typically required by these organisms. However, effective tools for genetic manipulation are now available in several species of Archaea (21, 23), and the time is ripe for the development of genetic screens for mutations that affect the regulation of gene expression. Methanococcus maripaludis, like all methanogenic archaea, requires strict anaerobic conditions for growth. However, genetic screens involving the use of β-galactosidase and its artificial substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) require oxidized conditions for color development (8). Here, we describe for M. maripaludis a procedure involving appropriate genetic manipulations and an X-Gal screen and its use in the identification of induction-deficient mutant alleles of the nitrogen repressor NrpR. The approach has wide adaptability to other regulatory systems in methanogenic Archaea and other strict anaerobes.
Most organisms regulate their nitrogen assimilation functions. Among the Bacteria and Archaea, a variety of nitrogen sensors and regulators are known, and most respond to cellular 2-oxoglutarate (2OG) as an indicator of nitrogen deficiency (11, 20). In the Bacteria and Euryarchaeota the most widespread nitrogen sensors belong to the PII superfamily of 2OG sensor proteins, which consists of the GlnB-GlnK family and the NifI family of nitrogenase regulators. In Proteobacteria, GlnB and GlnK proteins sense 2OG and also indirectly sense glutamine as an indicator of nitrogen sufficiency (5, 19). However, it is 2OG sensing that appears to be nearly universal in nitrogen regulation among the Bacteria and many Euryarchaeota (11).
In addition to the PII proteins, many 2OG-responsive nitrogen regulators and enzymes are known. These include the transcriptional activators NifA of Gammaproteobacteria (17) and NtcA of unicellular cyanobacteria (7), the glutamine synthetase modifying adenylyltransferase of Gammaproteobacteria (9), and glutamine synthetase itself in the archaeal genus Methanosarcina (4). A recently characterized 2OG-responsive regulator that operates at the transcriptional level in a wide range of Euryarchaeota is the repressor NrpR (13).
Detailed studies carried out in M. maripaludis have shown that NrpR binds as a dimer to palindromic operator sequences in promoter regions (15). 2OG acts as an inducer, directly interacting with NrpR and lowering its affinity for operators, resulting in derepression (induction) under nitrogen-deficient conditions (15). In M. maripaludis the regulation by NrpR of glnA encoding glutamine synthetase, the nif operon containing genes for nitrogen fixation (13), and a glnK-amtB operon (E. L. H. Hendrickson et al., unpublished data) containing genes for a PII protein and an ammonia transporter, has been studied. The presence of consensus NrpR binding sequences suggests that NrpR regulates transcription of other genes related to nitrogen assimilation as well (10).
NrpR proteins, defined as containing conserved NrpR domains, are present in all methanogens with known genome sequences as well as Archaeoglobus and a few bacterial species (12). Organisms containing NrpR proteins fall into three groups based on how many NrpR domains they contain and the configuration of the NrpR domains (12). M. maripaludis represents one group, containing two tandem NrpR domains within a single polypeptide with an N-terminal helix-turn-helix (HTH) motif. Methanosarcina species represent another group, where two NrpR domains are contained in two separately encoded polypeptides, one with an N-terminal HTH motif and the other without. Methanopyrus kandleri represents the third group, containing a single NrpR domain that is joined to an N-terminal HTH motif. Furthermore, the NrpR domains fall into five phylogenetic groups that correspond to their context: the two domains of the double-domain proteins make up two groups, the two separately encoded domains of Methanosarcina represent two more groups, and the single domain of M. kandleri represents the fifth group. NrpR domains from each of these five groups are aligned in Fig. 1. Despite this diversity of the NrpR family, each NrpR protein functions in nitrogen regulation, as evidenced by genetic complementation of an M. maripaludis nrpR null mutation (12). Hence, NrpR and its conserved mechanism of action appear to be widespread.
FIG. 1.
Alignment of NrpR domains of M. kandleri (Mk), M. maripaludis (Mm), and M. acetivorans (Ma). D1 and D2 differentiate between individual NrpR domains of M. maripaludis and M. acetivorans. Numbers at ends of rows indicate the amino acid positions based on the full-length NrpR sequences. Amino acids shown to be involved in 2OG sensing are underlined, and their mutations are indicated in brackets. Black, grey, and light grey represent 100%, 80%, and 60% conservation, respectively.
Although 2OG modulates a variety of nitrogen sensors and regulators, the structural basis for this response is not understood (11). Crystal structures of GlnB and GlnK-like PII proteins are known (see references 3, 6, 16, and 24), including recently a structure with bound 2OG (25). The tertiary structure of NrpR is unknown. Nevertheless, NrpR offers the opportunity to initiate the identification of structural elements involved in responding to 2OG. Specifically, mutant alleles of NrpR that result in repression of gene expression even under inducing conditions should be deficient in the response to 2OG. To this end, we initiated a genetic screen for such mutations in NrpR in M. maripaludis. We report here the identification of four amino acid residues in M. maripaludis NrpR and two in M. kandleri NrpR that are required for the full response to 2OG. The results identify two homologous positions in the conserved NrpR domain that participate in the induction of gene expression by 2OG.
MATERIALS AND METHODS
Strains and growth conditions.
Cultures of M. maripaludis were grown at 37°C in either McCas (McC medium with Casamino Acids in place of yeast extract) medium or nitrogen-free medium with NH4Cl or N2, as described previously (1, 18). For growth on solid medium, 15 g/liter of Noble agar (Difco) was used. Agar plates were incubated in sealed steel anaerobic vessels. For growth on agar medium with N2, anaerobic vessels were pressurized to 10 lb/in2 with 80% N2-20% CO2 gas mix and then to 20 lb/in2 with 80% H2-20% CO2. For growth with NH4Cl, 80% H2-20% CO2 (20 lb/in2) was used. Puromycin was added as needed (1). Escherichia coli strains were grown in LB broth with ampicillin (0.1 mg/ml).
Alanine replacement mutagenesis.
Primers are listed in Table S2 in the supplemental material. All PCRs used Herculase polymerase (Stratagene) and the following thermocycler conditions: 94°C for 5 min; 30 cycles of 94°C for 45 s, 50°C for 30 s, and 68°C for 2 min 30 s; and a final extension of 68°C for 10 min. Site-specific mutagenesis of nrpR genes was accomplished in two stages. In the first stage, the 5′ and 3′ portions of the gene were amplified in two separate PCRs. For M. kandleri nrpR, pWLGMK2-3 (12) was used as a template, the 5′ portion of the gene was amplified using the forward primer nrprhom+pstIF2 and a reverse primer (see Table S2 in the supplemental material), and the 3′ portion of the gene was amplified using a forward primer (see Table S2 in the supplemental material) and the reverse primer MKnrprrevbglII2. The desired mutations were incorporated into the first three or six nucleotides of the forward primer for the 3′ portion of the gene. The first stage for site-specific mutagenesis of M. maripaludis nrpR was similar, except that the template was pWLG40RepHis (15), the forward primer for the 5′ portion of the gene was Rephispst, and the reverse primer for the 3′ portion of the gene was Rephisxba. All PCR products were gel purified. In the second stage of the procedure, the PCR products for the 5′ and 3′ portions of the gene were ligated and amplified using the primers nrprhom+pstIF2 and MKnrprrevbglII2 for M. kandleri nrpR and the primers Rephispst and Rephisxba for M. maripaludis nrpR. Where necessary (3′ portions obtained using a forward primer lacking 5′ phosphate), 5′ phosphate was added before ligation using polynucleotide kinase (New England Biolabs). The full-length PCR products were gel purified. M. kandleri nrpR was then digested with PstI and BglII and cloned into NsiI- and BglII-digested pWLG40+lacZ (13). M. maripaludis nrpR was digested with PstI and XbaI and cloned into NsiI- and XbaI-digested pWLG40+lacZ. Plasmid constructs were then transformed (22) into M. maripaludis strain Mm1043 (12). The control strains were Mm1171 (vector control; Mm1043 containing pWLG40ΔlacZ [12]), Mm1058 (wild-type M. kandleri nrpR; Mm1043 containing pWLGMK2-3 [12]), and Mm1048 (wild-type M. maripaludis nrpR-His6; Mm1043 containing pWLG40RepHis [15]).
Random mutagenesis.
Error-prone PCR was conducted with Taq polymerase (Promega) using reaction buffer supplied by the manufacturer and MgCl2 at a final concentration of 1.5 mM. Templates were pWLG40MK2-3 for M. kandleri nrpR and pWLG40RepHis for M. maripaludis nrpR. For each nrpR gene, four separate PCRs were carried out with one of the deoxynucleoside triphosphates at a final concentration of 40 μM while each of the other three was used at a concentration of 200 μM. Thermocycler conditions were as above (see “Alanine replacement mutagenesis”). PCR products were gel purified, digested, and cloned into pWLG40+lacZ as described above. The plasmids were then transformed into E. coli (XL10Gold; Stratagene), and the cells were grown for 2 days. Plasmids were then extracted using a Promega Wizard Plus SV Miniprep kit and transformed into M. maripaludis strain Mm1043 as described above. M. maripaludis transformants were plated onto nitrogen-free agar plates with puromycin and grown with N2 as the sole nitrogen source. Plates were incubated for about 1 week or until a pressure drop of at least 5 lb/in2 occurred.
β-Galactosidase assay and X-Gal colony screen.
β-Galactosidase assay of M. maripaludis liquid cultures was performed as described previously (14). β-Galactosidase screening of M. maripaludis colonies was as follows. After anaerobic growth of colonies, agar plates were exposed to air for about 30 min until the resazurin in the medium turned pink (color formation from X-Gal requires oxidized conditions) (8). X-Gal (2 ml of 25 mg/ml X-Gal in 100% dimethylformamide) was mixed with 3 ml of nitrogen-free medium without sulfide. This mixture was sprayed onto plates until the agar surface was completely covered and was allowed to remain on the plates for at least 3 s before excess was decanted. The plates were then allowed to dry in a fume hood before they were placed on the benchtop. Color development was allowed to occur for 30 min to 1 h. Once targeted colonies were identified, plates were brought into an anaerobic chamber where colonies were picked and suspended in 0.1 ml of liquid McCas medium (reduced with sodium sulfide) with puromycin, inoculated into tubes with the same medium, and allowed to grow. A total of 0.1 ml of this culture was then transferred to another tube containing nitrogen-free medium with puromycin, grown under N2, and assayed for β-galactosidase activity. Low β-galactosidase activities were confirmed by comparing samples against controls expressing wild-type nrpR genes. Positive strains were streak purified on McCas medium with puromycin, after which liquid cultures were grown on McCas medium with puromycin. Plasmids were extracted using a Promega Wizard Plus SV Miniprep kit, and the nrpR gene was PCR amplified with high-fidelity Herculase polymerase (Stratagene) and sequenced at the University of Washington DNA sequencing facility. Alternatively, plasmids were transformed into E. coli XL10 Gold, extracted, amplified, and sequenced.
Electrophoretic mobility shift analysis.
Gel shift analysis was as described previously (15), except that cell extracts were used instead of purified NrpR. Cell extracts were made as described previously (14) and used in the following quantities: for wild-type NrpR, 0.26 μg of total protein; for mutant NrpR or vector control, 0.13 μg of total protein. 2OG (2 mM) was added as appropriate.
RESULTS AND DISCUSSION
Genetic screen for noninducing NrpR mutants.
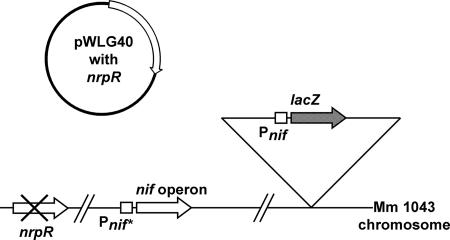
We used a genetic approach to identify amino acids of NrpR involved in responding to the inducer 2OG. Mutants of NrpR deficient in the 2OG response (induction-deficient mutants) should repress the expression of NrpR-regulated genes even under inducing conditions (i.e., growth with N2 as the nitrogen source). Hence, we made mutations in NrpR and screened for alleles in which expression from the nif promoter was decreased during growth on N2. For this purpose we used M. maripaludis strain Mm1043 (12), which has the following characteristics (Fig. 2): a nif promoter-lacZ reporter gene fusion, an operator mutation in the promoter region of the nif operon that renders expression constitutive (so that noninducing NrpR alleles will not prevent growth on N2, needed for genetic screening), and a deletion in the native nrpR gene. Mm1043 was the recipient for nrpR genes, which were mutagenized, introduced on a replicative plasmid, and screened.
FIG. 2.
Genetic arrangement for screening mutant nrpR alleles. M. maripaludis strain Mm1043 and the pWLG40-based replicative plasmid containing the nrpR gene are described in the text. Arrows represent genes or operons. Pnif represents wild-type nif promoter and Pnif* represents a mutated version to allow constitutive nif gene expression. The in-frame deletion of nrpR is indicated (×). The inverted triangle represents insertion of the Pnif-lacZ fusion into the chromosome at the upt site (12).
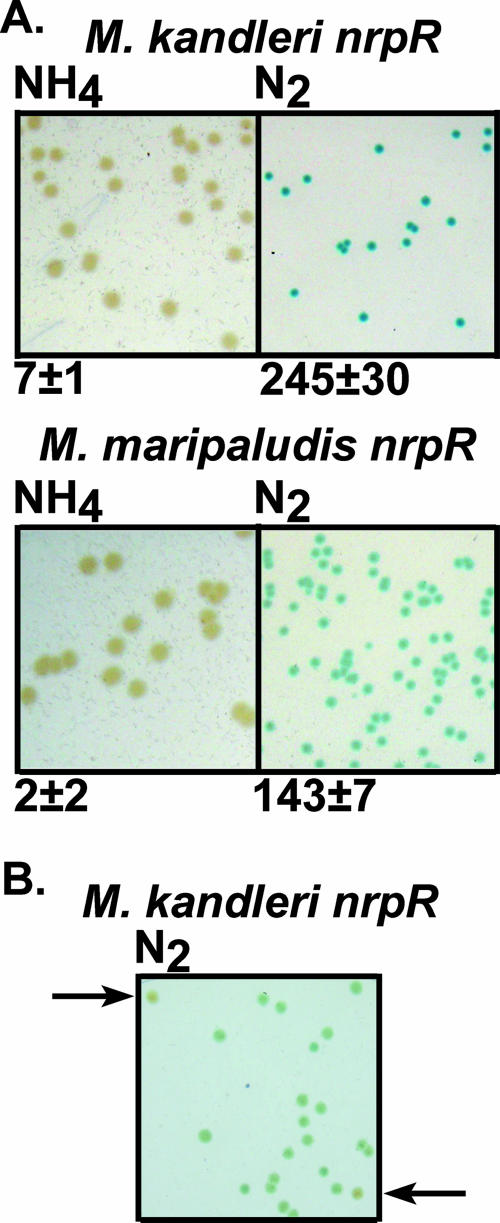
We devised a colony screen using X-Gal to determine expression from the nif promoter. X-Gal screens, which require oxygen for color development, are unusual with strict anaerobes and to our knowledge have not previously been carried out with methanogens. Previously, we determined that M. maripaludis NrpR (fused at the C terminus to an oligohistidine tag, NrpR-His6) conferred regulation of nif expression, as did several NrpR homologs from M. kandleri, Methanocaldococcus jannaschii, and Methanosarcina acetivorans (12). Colonies of M. maripaludis strains expressing either M. maripaludis nrpR-His6 or M. kandleri nrpR were grown anaerobically and then exposed to air and sprayed with X-Gal (See Materials and Methods). Expression of either M. maripaludis nrpR-His6 or M. kandleri nrpR in Mm1043 resulted in the expected pattern of color development: no blue developed in colonies grown with ammonia even 17 h after application of X-Gal, while colonies grown with N2 turned blue after approximately 20 min (Fig. 3A). The difference in lacZ expression with M. maripaludis NrpR versus M. kandleri NrpR under N2 growth conditions could also be readily discerned from the intensities of the blue. This difference correlated well with β-galactosidase assay results of liquid cultures (Fig. 3A). We also demonstrated that N2-grown colonies that contained lower β-galactosidase levels (below) could be easily differentiated (Fig. 3B), typically 45 min to 1 h after application of X-Gal, at which time plates were transferred to an anaerobic chamber and the colonies were picked. Viable cells could consistently be recovered from colonies exposed to air for as long as 2 h.
FIG. 3.
(A) M. maripaludis colonies expressing M. kandleri nrpR (strain Mm1058) or His-tagged M. maripaludis nrpR (strain Mm1048). Colonies were grown with ammonia or N2 as a nitrogen source. Numbers below images are β-galactosidase activities (Miller units) from the same strains grown in liquid medium with the same nitrogen sources. (B) Screen for random mutants of M. kandleri NrpR. Lighter-colored mutant colonies are indicated by arrows.
Alanine replacement and random mutagenesis of M. kandleri nrpR.
Our initial efforts at mutagenesis targeted M. kandleri NrpR, which contains a single NrpR domain and offered a smaller target than M. maripaludis NrpR, which contains two NrpR domains (12). In addition, mutagenesis of the single domain M. kandleri NrpR would avoid the possibility that a mutation in one domain might be masked by the other domain if the domains were functionally redundant. Truncation of the second M. maripaludis NrpR domain resulted in an inactive protein (unpublished data), so we had no opportunity to work with a single-domain variant of M. maripaludis NrpR.
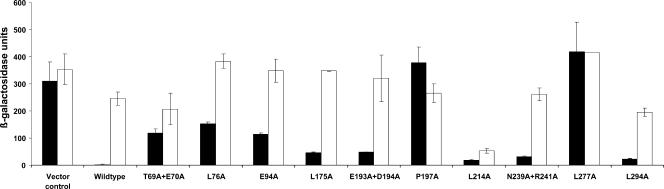
For alanine replacement mutagenesis of M. kandleri NrpR, we chose 13 amino acids that were identical in an alignment of the NrpR domain of M. kandleri with the first NrpR domains of M. maripaludis and M. jannaschii. Each of these amino acids was changed to alanine (see Table S1 in the supplemental material for details of all NrpR mutations made in this study.) All but one mutant retained high expression levels from the nif promoter during growth on N2. These mutants had various degrees of increased expression on ammonia, suggesting loss of repressor activity (Fig. 4). However, one mutation, L214A, had the desired effect: markedly lowered expression on N2, indicating that this mutant may be less responsive to 2OG.
FIG. 4.
nif regulation by wild-type M. kandleri NrpR and alanine replacement mutants. Vector control, strain Mm1171 (no NrpR); wild type, strain Mm1058 (M. kandleri NrpR). Amino acid changes are indicated on the x axis. Black bars, ammonia-grown cultures; white bars, dinitrogen-grown cultures.
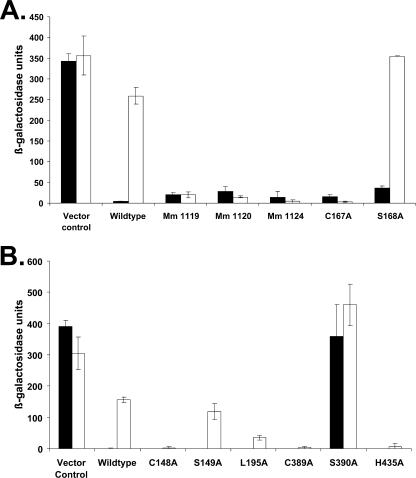
We next conducted random mutagenesis of M. kandleri nrpR, identifying candidate mutants with an X-Gal screen that we devised for this purpose (see above). We identified several white colonies grown on N2 (Fig. 3B), isolated the mutant strains, and assayed liquid cultures for β-galactosidase activity after growth with N2 or ammonia. This procedure yielded three induction-defective mutants, Mm1119, Mm1120, and Mm1124 (Fig. 5A). The nrpR genes in these strains were sequenced and found to contain multiple mutations resulting in more than one amino acid change (see Table S1 in the supplemental material). The NrpR proteins encoded in strains Mm1120 and Mm1124 contained four and three amino acid changes, respectively, including changes in one amino acid in common, S168P (Mm1120) and S168F (Mm1124). S168, and the adjacent C167 are highly conserved (Fig. 1). Hence, we constructed individual C167A and S168A mutations. Only C167A conferred an induction defect (Fig. 5A). The mutation of S168 to a bulky P or F residue could affect induction whereas a change to an A residue did not. In the remaining random mutant strain, Mm1119, the NrpR protein contained five amino acid changes, of which only three (R178G, E196G, and H215R) occurred in conserved regions of the NrpR domain. We recreated each of these changes as a single site-directed mutation. However, no single mutation resulted in a significant induction defect (data not shown). Evidently, only the cumulative effect of the mutations in Mm1119 produces an induction defect.
FIG. 5.
(A) nif regulation by mutants of M. kandleri NrpR resulting from random and site-specific mutagenesis. Vector and wild-type controls are as described in the legend of Fig. 4. Mm1119, Mm1120, and Mm1124 are mutant strains isolated after random mutagenesis. C167A and S168A are M. kandleri NrpR site-specific mutants. Black bars, ammonia-grown cultures; white bars, dinitrogen-grown cultures. (B) nif regulation by wild-type M. maripaludis NrpR and site-specific mutants. Vector control, strain Mm1171 (no NrpR); wild type, strain Mm1048 (His-tagged M. maripaludis NrpR); black bars, ammonia-grown cultures; white bars, dinitrogen-grown cultures.
Directed mutagenesis of M. maripaludis NrpR.
The above work led to the identification of two amino acid residues in M. kandleri NrpR whose mutation to alanine resulted in induction defects, C167 and L214 (Fig. 1). We made the corresponding mutations in each of the two M. maripaludis NrpR domains. Both C148A and L195A in the first domain and C389A and H435A in the second domain conferred defective induction (Fig. 5B). We also constructed mutations corresponding to S168A of M. kandleri NrpR. M. maripaludis NrpR mutant S149A retained normal regulation, similar to M. kandleri S168A. In the second domain of M. maripaludis nrpR, the corresponding mutation S390A resulted in complete loss of repression.
Electrophoretic mobility shift analysis of M. maripaludis NrpR mutants.
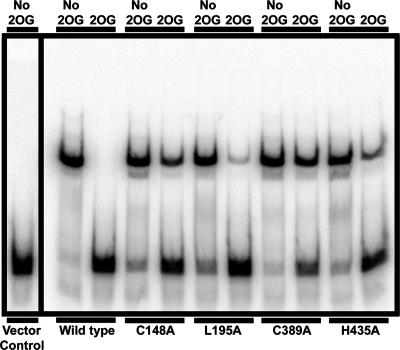
The effects of M. maripaludis NrpR mutations C148A, L195A, C389A, and H435A in vivo suggested a lowered response to 2OG. To determine whether this was the case, we tested the effect of 2OG on NrpR binding to nif operator DNA using electrophoretic mobility shift analysis with cell extracts. Previously, we demonstrated that the band shifted by the cell extract was wholly dependent on the presence of NrpR and the normal binding site sequence of the operator (2, 13). Using this assay, we adjusted the amount of cell extract from each nrpR-expressing strain to shift 70 to 85% of the DNA probe in the absence of 2OG (these amounts of cell extract varied no more than twofold in total protein). Then, adding 2 mM 2OG, we assessed the decrease in the amount of probe shifted. 2OG nearly abolished binding of wild-type NrpR to operator DNA. In contrast, each mutant NrpR retained some binding in the presence of 2OG (Fig. 6). Furthermore, the amount of binding retained in the presence of 2OG for each NrpR protein corresponded roughly to the degree of repression of gene expression (loss of induction) during growth on N2 that is shown in Fig. 5B. Thus, by both criteria, L195A was the least severely altered in the induction response, while the remaining three mutants were more markedly affected.
FIG. 6.
Electrophoretic mobility shift assay of binding of wild-type and mutant M. maripaludis NrpR in cell extracts to nif operator DNA. Vector control, Mm1171 (no NrpR); wild type, strain Mm1048 (His-tagged M. maripaludis NrpR).
Conclusions.
The work presented here establishes and validates a genetic approach in methanogenic Archaea to determine the structural basis for the induction of NrpR-regulated genes. The approach, which can be adapted to the analysis of other regulators and other strict anaerobes, involves genetic complementation in an appropriate background and the implementation of an X-Gal screen for anaerobes.
Two sites were identified in each NrpR domain that participate in induction through their role in responding to the inducer 2OG (Fig. 1). The results indicate that each NrpR domain of M. maripaludis NrpR functions similarly and that the two domains are both required for a full 2OG response; that is, they are not redundant. Furthermore, the homolog from the distantly related species M. kandleri conserves the same structure-function relationship. This observation is consistent with our previous finding that NrpR homologs, which are widely distributed in the Euryarchaeota, functionally replaced M. maripaludis NrpR in a genetic complementation assay (12). Possible roles that each amino acid site might play in the induction response include binding of 2OG or responding to it with a conformational change that decreases DNA binding. Future work, which will include the determination of the crystal structure of NrpR as well as the further characterization of these and additional mutant NrpR proteins, will distinguish between these possibilities.
Supplementary Material
Acknowledgments
This work was supported by grants GM55255 and GM74783 from the National Institutes of Health.
We thank Jeremy Dodsworth for helpful discussions.
Footnotes
Published ahead of print on 24 August 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Blank, C. E., P. S. Kessler, and J. A. Leigh. 1995. Genetics in methanogens: transposon insertion mutagenesis of a Methanococcus maripaludis nifH gene. J. Bacteriol. 177:5773-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen-Kupiec, R., C. Blank, and J. A. Leigh. 1997. Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen. Proc. Natl. Acad. Sci. USA 94:1316-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conroy, M. J., A. Durand, D. Lupo, X. D. Li, P. A. Bullough, F. K. Winkler, and M. Merrick. 2007. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. USA 104:1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehlers, C., K. Weidenbach, K. Veit, K. Forchhammer, and R. A. Schmitz. 2005. Unique mechanistic features of post-translational regulation of glutamine synthetase activity in Methanosarcina mazei strain Go1 in response to nitrogen availability. Mol. Microbiol. 55:1841-1854. [DOI] [PubMed] [Google Scholar]
- 5.Forchhammer, K. 2007. Glutamine signalling in bacteria. Front. Biosci. 12:358-370. [DOI] [PubMed] [Google Scholar]
- 6.Gruswitz, F., J. O'Connell III, and R. M. Stroud. 2007. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 Å. Proc. Natl. Acad. Sci. USA 104:42-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrero, A. 2004. New targets of the PII signal transduction protein identified in cyanobacteria. Mol. Microbiol. 52:1225-1228. [DOI] [PubMed] [Google Scholar]
- 8.Horwitz, J. P., J. Chua, R. J. Curby, A. J. Tomson, M. A. Darooge, B. E. Fisher, J. Mauricio, and I. Klundt. 1964. Substrates for cytochemical demonstration of enzyme sctivity. I. Some substituted 3-indolyl-beta-d-glycopyranosides. J. Med. Chem. 7:574-575. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, P., A. E. Mayo, and A. J. Ninfa. 2007. Escherichia coli glutamine synthetase adenylyltransferase (ATase, EC 2.7.7.49): kinetic characterization of regulation by PII, PII-UMP, glutamine, and alpha-ketoglutarate. Biochemistry 46:4133-4146. [DOI] [PubMed] [Google Scholar]
- 10.Kessler, P. S., and J. A. Leigh. 1999. Genetics of nitrogen regulation in Methanococcus maripaludis. Genetics 152:1343-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leigh, J. A., and J. A. Dodsworth. 2007. Nitrogen regulation in Bacteria and Archaea. Annu. Rev. Microbiol. 61:349-377. [DOI] [PubMed] [Google Scholar]
- 12.Lie, T. J., J. A. Dodsworth, D. C. Nickle, and J. A. Leigh. 2007. Diverse homologs of the nitrogen repressor NrpR function similarly in nitrogen regulation. FEMS Microbiol. Lett. 271:281-288. [DOI] [PubMed] [Google Scholar]
- 13.Lie, T. J., and J. A. Leigh. 2003. A novel repressor of nif and glnA expression in the methanogenic archaeon Methanococcus maripaludis. Mol. Microbiol. 47:235-246. [DOI] [PubMed] [Google Scholar]
- 14.Lie, T. J., and J. A. Leigh. 2002. Regulatory response of Methanococcus maripaludis to alanine, an intermediate nitrogen source. J. Bacteriol. 184:5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lie, T. J., G. E. Wood, and J. A. Leigh. 2005. Regulation of nif expression in Methanococcus maripaludis: roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators. J. Biol. Chem. 280:5236-5241. [DOI] [PubMed] [Google Scholar]
- 16.Machado Benelli, E., M. Buck, I. Polikarpov, E. Maltempi de Souza, L. M. Cruz, and F. O. Pedrosa. 2002. Herbaspirillum seropedicae signal transduction protein PII is structurally similar to the enteric GlnK. Eur. J. Biochem. 269:3296-3303. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Argudo, I., R. Little, N. Shearer, P. Johnson, and R. Dixon. 2005. Nitrogen fixation: key genetic regulatory mechanisms. Biochem. Soc. Trans. 33:152-156. [DOI] [PubMed] [Google Scholar]
- 18.Moore, B. C., and J. A. Leigh. 2005. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J. Bacteriol. 187:972-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ninfa, A. J., and M. R. Atkinson. 2000. PII signal transduction proteins. Trends Microbiol. 8:172-179. [DOI] [PubMed] [Google Scholar]
- 20.Ninfa, A. J., and P. Jiang. 2005. PII signal transduction proteins: sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 8:168-173. [DOI] [PubMed] [Google Scholar]
- 21.Rother, M., and W. W. Metcalf. 2005. Genetic technologies for Archaea. Curr. Opin. Microbiol. 8:745-751. [DOI] [PubMed] [Google Scholar]
- 22.Tumbula, D. L., R. A. Makula, and W. B. Whitman. 1994. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol. Lett. 121:309-314. [Google Scholar]
- 23.Tumbula, D. L., and W. B. Whitman. 1999. Genetics of Methanococcus: possibilities for functional genomics in Archaea. Mol. Microbiol. 33:1-7. [DOI] [PubMed] [Google Scholar]
- 24.Xu, Y., P. D. Carr, T. Huber, S. G. Vasudevan, and D. L. Ollis. 2001. The structure of the PII-ATP complex. Eur. J. Biochem. 268:2028-2037. [DOI] [PubMed] [Google Scholar]
- 25.Yildiz, O., C. Kalthoff, S. Raunser, and W. Kuhlbrandt. 2007. Structure of GlnK1 with bound effectors indicates regulatory mechanism for ammonia uptake. EMBO J. 26:589-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.