Abstract
Burkholderia pseudomallei soil isolates from northeast Thailand were genotyped using multiple-locus variable-number tandem repeat (VNTR) analysis (MLVA) and multilocus sequence typing (MLST). MLVA identified 19 genotypes within three clades, while MLST revealed two genotypes. These close genetic relationships imply a recent colonization followed by localized expansion, similar to what occurs in an outbreak situation.
Burkholderia pseudomallei is the causative agent of melioidosis, a potentially fatal disease endemic in areas of northern Australia and southeast Asia (10, 19). In the northeast of Thailand, melioidosis is accountable for 20% of community-acquired septicemias and is associated with a 40% mortality rate (2). Melioidosis is acquired by skin exposure, inhalation, or ingestion of the pathogen (19). Thus, commonly infected Thai individuals are those that encounter soil and surface water contaminated with B. pseudomallei, especially rice farmers (13). While this disease is exotic and rare in much of the world, it is a major public health problem in Thailand.
As expected from such high incidences of melioidosis, B. pseudomallei is widely distributed and commonly isolated from soil and water throughout areas of endemicity (6, 17, 18). Molecular analyses of the genetic diversity of clinical and environmental B. pseudomallei isolates have revealed that at various geographic and temporal scales, B. pseudomallei has substantial genetic diversity (7, 9, 16, 17). However, few studies have examined the soil reservoir at defined geographic and temporal scales to assess whether B. pseudomallei genetic diversity is representative of regional diversity or alternatively demonstrates lower levels of diversity. To examine this question, we assessed the genetic diversity and spatial distribution of B. pseudomallei in the soil from a 50-km2 region of Khon Kaen Province, Thailand, during a 6-month time span in 2004.
In this study, soils were collected from 50 sites along both sides of intervillage roads. Sampling locations were recorded using a Global Positioning System receiver and mapped using ArcGIS 9.1 software. At each site, three holes, positioned in a triangle, were dug using a 5-inch-diameter hand auger. Three grams of soil was collected from three depths in each hole (15, 30, and 45 cm). Soil was vortexed with 3 ml distilled water for 30 s, followed by a settling time of 30 min. Five hundred microliters of supernatant was transferred into 3 ml selective enrichment broth (threonine-basal salt solution with 20 μg/liter of colistin) (20) and incubated at 42°C for 48 h with shaking. One hundred microliters of broth was 10-fold serially diluted to concentrations of 10−2, 10−3, and 10−4, and 10 μl of each dilution was cultured on Ashdown's agar (1), incubated at 42°C for 4 to 7 days, and examined daily for emerging colonies. Presumptive B. pseudomallei colonies were confirmed using biochemical tests, including l-arabinose assimilation.
Nineteen of the 50 sites were culture positive for B. pseudomallei. From those 19 sites, 68 B. pseudomallei colonies were cultured, and total genomic DNA was extracted by a phenol-chloroform technique (11) from single colonies. In addition, from five culture-positive sites, eight DNA samples (Table 1) were extracted directly from 2 g of soil using a bead beating (5) and phenol-chloroform protocol (21). DNA was then purified to remove inhibitors by agarose gel extraction (22).
TABLE 1.
Location, depth, and genotypes for the 68 isolates of B. pseudomallei
| Isolate name | Sample site | Depth (cm) | Ribotype | ST by MLST | MLVA genotype |
|---|---|---|---|---|---|
| 001a | 2 | 15 | 23 | 70 | A1 |
| 002a | 2 | 15 | 23 | A1 | |
| 003 | 2 | 30 | 23 | A1 | |
| 005 | 2 | 30 | 23 | A1 | |
| 009 | 8 | 30 | 23 | A1 | |
| 010 | 8 | 30 | 23 | A1 | |
| 011 | 10 | 15 | 16 | A1 | |
| 014 | 11 | 30 | 23 | A1 | |
| 016 | 11 | 30 | 23 | A1 | |
| 018a | 13 | 30 | 23 | 70 | A2 |
| 004a | 2 | 30 | 23 | 70 | A3 |
| 008 | 8 | 30 | 23 | A4 | |
| 012 | 10 | 30 | 23 | A4 | |
| 013 | 11 | 30 | 23 | A4 | |
| 19 | 13 | 30 | 23 | A4 | |
| 020a | 15 | 15 | NAc | A4 | |
| 024 | 22 | 15 | 16 | A4 | |
| 025 | 22 | 30 | 23 | A4 | |
| 026 | 22 | 30 | 23 | A4 | |
| 028 | 22 | 30 | 23 | A4 | |
| 029 | 24 | 15 | 23 | A4 | |
| 030a | 24 | 30 | 23 | A4 | |
| 031 | 24 | 30 | 23 | A4 | |
| 032a | 24 | 30 | 23 | A4 | |
| 033 | 50 | 15 | 23 | A4 | |
| 034 | 50 | 30 | 23 | A4 | |
| 036 | 50 | 45 | 23 | A4 | |
| 037a | 26 | 30 | NA | A4 | |
| 038 | 26 | 30 | 23 | 70 | A4 |
| 039 | 26 | 45 | 23 | A4 | |
| 040 | 27 | 15 | 23 | A4 | |
| 041 | 27 | 15 | 23 | A4 | |
| 042 | 27 | 30 | 23 | A4 | |
| 046 | 29 | 30 | 23 | A4 | |
| 047 | 29 | 30 | 23 | A4 | |
| 048 | 29 | 30 | 23 | A4 | |
| 049 | 30 | 15 | 23 | A4 | |
| 052 | 30 | 30 | 23 | A4 | |
| 053 | 31 | 30 | 23 | A4 | |
| 054 | 31 | 30 | 23 | A4 | |
| 055 | 36 | 15 | 23 | A4 | |
| 056 | 36 | 30 | 23 | A4 | |
| 058 | 36 | 30 | 23 | A4 | |
| 059 | 36 | 30 | 23 | A4 | |
| 060 | 38 | 30 | 23 | A4 | |
| 061 | 38 | 45 | 23 | A4 | |
| 062 | 41 | 15 | 23 | A4 | |
| 063 | 41 | 15 | 23 | A4 | |
| 066 | 38 | 30 | 23 | A4 | |
| 035 | 50 | 30 | 23 | 70 | A5 |
| 023 | 15 | 30 | 23 | 70 | A6 |
| 015 | 11 | 30 | 23 | 70 | A7 |
| 045 | 29 | 15 | 23 | 70 | A8 |
| 050 | 30 | 15 | 23 | 70 | A9 |
| 064 | 45 | 30 | 23 | NA | A10 |
| 017 | 13 | 15 | 23 | 70 | A11 |
| 068 | 45 | 15 | 23 | A11 | |
| 057 | 36 | 30 | 23 | 70 | A12 |
| 027 | 22 | 30 | 23 | 70 | A13 |
| 051 | 30 | 30 | 23 | 70 | A14 |
| 067 | 44 | 30 | 23 | 70 | A15 |
| 007 | 8 | 15 | 13 | 70 | B16 |
| 021 | 15 | 30 | 13 | B16 | |
| 022 | 15 | 30 | 13 | B16 | |
| 006 | 8 | 15 | 21 | C17 | |
| 065 | 38 | 15 | 21 | NAb | C17 |
| 043 | 27 | 30 | 21 | NAb | C18 |
| 044 | 29 | 15 | 21 | NAb | C19 |
DNA also extracted directly from soil.
Novel ST (alleles 3, 1, 3, 2, 5, 2, 1).
NA, not available.
Before beginning genetic characterization, the 68 isolates were confirmed to be B. pseudomallei with a species-specific real-time PCR assay (15). For genetic characterization, 26 loci from a previously described multiple-locus variable-number tandem repeat (VNTR) analysis (MLVA) system were used to genotype the 68 cultured isolates as well as the eight DNAs extracted directly from the soil (16). DNA was also genotyped by a ribotyping method using HindIII enzyme for digestion and 16S and 23S rRNA genes of Escherichia coli as a probe (12). Further genetic analysis of isolates that were representative of each MLVA genotype was accomplished using multilocus sequence typing (MLST) (Table 1) (7), with the exception of genotype 10, which could not be sequenced. Sequences for each locus were queried against the online MLST database (http://bpseudomallei.mlst.net) to determine allelic designations and a subsequent allelic profile query was used to determine the sequence type (ST).
The degree of VNTR variability was assessed by the number of alleles observed and by Nei's diversity index: D = 1 − Σ(allele frequency)2. A Mantel test was performed to test the correlation between genetic distance and geographic distance (8). An unrooted distance-based phylogenetic tree and mean pairwise genetic diversity were calculated for representatives of each genotype from 26 VNTR loci using the neighbor-joining algorithm in PAUP 4.0 b10 (14).
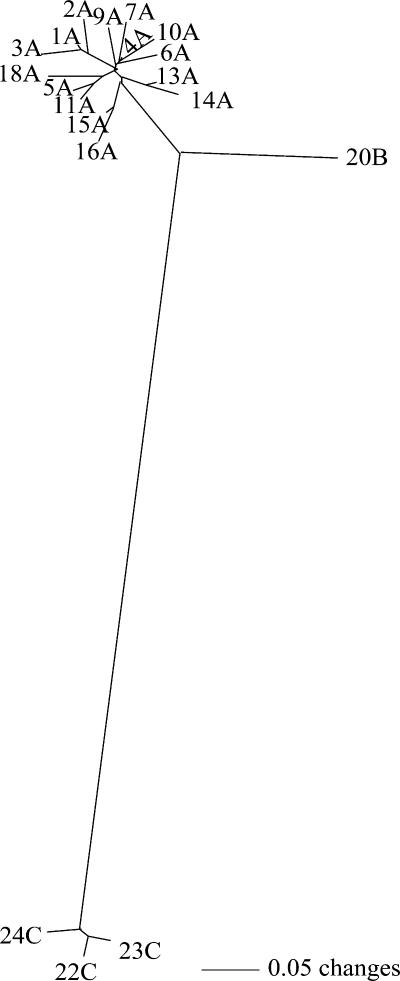
In the collection of B. pseudomallei isolates, 23 VNTR loci were polymorphic while 3 were monomorphic. For polymorphic loci, the number of alleles ranged from one to five, with Nei's diversity values ranging from 0.11 to 0.29. Phylogenetic analysis of MLVA data identified 19 genotypes within three clades (A, B, and C) (Fig. 1). Sixty-one of the isolates belonged to clade A, which was dominated by a single genotype (A4) (n = 38; 56%). Clade B contained three isolates of the same genotype. Clade C contained four isolates and three genotypes (Fig. 1). The mean pairwise genetic distance for the 19 genotypes was 0.16, which is lower than the calculated distance for a similarly localized B. pseudomallei population in northern Australia, even when considering only isolates from the same year (distance = 0.289) (9).
FIG. 1.
Unrooted neighbor-joining phylogram of 68 B. pseudomallei isolates. Numbers on the branches indicate the genotype, and A, B and C indicate the three genetic clades.
The genetic groups defined by MLVA were also confirmed by other typing methods (Table 1). For example, MLST revealed two STs for representatives of the 19 MLVA genotypes (ST70 for clades A and B and a novel ST for clade C). The novel ST is a single-locus variant to ST173 where the narK locus contains allele 2 instead of allele 4. Ribotyping identified four types (R16 and R23 for clade A, R13 for clade B, and R21 for clade C) which mirror the genetic relationship revealed by MLVA (Table 2). Additionally, a phylogenetic comparison of all 212 Thai MLST types (obtained from the online database www.bpseudomalleimlst.net) reveals that the STs found during this study are distantly related, therefore, supporting the genetic distance of MLVA clades A and B from clade C. Thus, the overall pattern of three typing methods demonstrates limited genetic diversity across the sampled region and the separation of isolates into a small number of phylogenetic groups.
TABLE 2.
Ribotype patterns of B. pseudomallei isolates
| MLVA clade | Ribotype | Presence of ribotype fragmenta:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | ||
| A | 16 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 |
| A | 23 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| B | 13 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| C | 21 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 |
Differences in the ribotype pattern are highlighted in boldface. 1, present; 0, absent.
The effects of culture bias could potentially influence which genotypes were observed; hence, a preliminary analysis of the eight DNA isolates extracted directly from soil, which represent five culture-positive soil sites, was used to confirm the presence of genotypes found through culturing. Three of the soil isolates (all from site 2) failed to amplify across all loci, most likely due to the crude purification method used. Additionally, multiple minor peaks were observed for some loci, suggesting the presence of mixed genotypes. However the dominant allele peaks matched clade A alleles from cultures obtained at the same sites. This preliminary evidence suggests that culture-dependent techniques recover the dominant B. pseudomallei genotypes, although the presence of other low-frequency genotypes cannot be excluded.
The soil and associated environment represent the pathogen reservoir for B. pseudomallei and the primary source of melioidosis (3). Previous studies have demonstrated the link between environmental isolates and clinical isolates associated with disease in humans and animals (12). For example, a study in the nearby Thai province of Ubon Ratchathani found that clinical and environmental samples shared the same MLST STs, although genotype frequencies differed between the two categories (17). The most prevalent genotype in both categories was ST70 (17), which is associated with our MLVA genetic clades A and B in this study. In contrast, the ST associated with MLVA clade C is novel, although it is closely related to ST173 which is also linked with a human case of melioidosis. These results reinforce the perceived association between soil isolates and melioidosis infection in northeast Thailand.
There was no apparent spatial genetic differentiation, with members of all three clades distributed across the sampling area (Fig. 2). Genotypes within clade A were found at all sites, and the dominant genotype from this clade occurred in 17 of the 19 sites. Clade B genotypes were found at only two sites, and clade C genotypes were observed at four sites (Fig. 2). However, the Mantel test found no correlation between genetic distance and geographic distance for the isolates (R2 = 0.007). Additionally there was no association between soil depth and genotype (χ2 = 7.43, df = 4, P = 0.11).
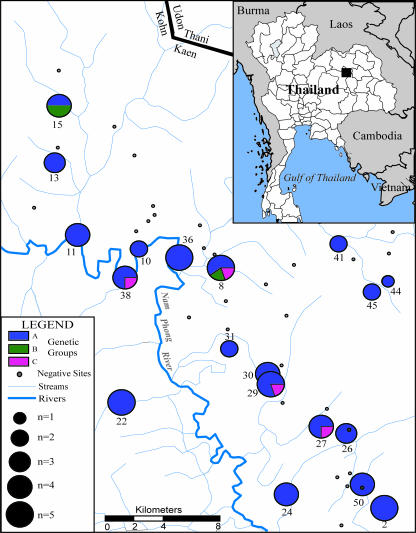
FIG. 2.
Map of regional study area within Khon Kaen Province. The figure illustrates the local watershed, as well as the locations of the 50 sites from which soil samples were collected. Each site is associated with a local village within the province. The center of each circle represents the location of a positive sampling area. Sites that were not positive for B. pseudomallei are shown as a black circle. Pie charts represent the percentages of isolates among three genetic clades (A, B, and C) for the B. pseudomallei DNA samples. The size of the pie chart is indicative of the number of samples from that site, ranging from one to five.
B. pseudomallei has been found to be both highly diverse even across very short geographic distances (9) and persistent in the environment for long periods of time (3). As such, genetic diversity at many geographic and temporal scales can be quite large (4, 7, 9, 16). In contrast, B. pseudomallei soil isolates collected for this study show lower levels of genetic diversity. This implies that there may have been a significant genetic bottleneck and subsequent population expansion in this region. The spatial homogeneity of genotypes supports this hypothesis of rapid dispersal and colonization, and given the sampling locations, the dispersal mechanism may likely be the flooding of the Nam Phong River during the rainy season.
The results of this study indicate that the overall diversity of B. pseudomallei in northeast Thailand may be a result of the cumulative effects of multiple locally differentiated subpopulations. However, further studies are needed to determine the population dynamics of B. pseudomallei, both over time and a wide geographic area. These studies, in combination with high-resolution subtyping methods, such as MLVA, will facilitate our understanding of the pathogen reservoir structure and how it contributes to melioidosis incidence.
Acknowledgments
Funding for this work was provided by the U.S. Departments of Energy and Homeland Security.
Footnotes
Published ahead of print on 24 August 2007.
REFERENCES
- 1.Ashdown, L. R., and S. G. Clarke. 1992. Evaluation of culture techniques for isolation of Pseudomonas pseudomallei from soil. Appl. Environ. Microbiol. 58:4011-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaowagul, W., N. J. White, D. A. Dance, Y. Wattanagoon, P. Naigowit, T. M. Davis, S. Looareesuwan, and N. Pitakwatchara. 1989. Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand. J. Infect. Dis. 159:890-899. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, A. C., D. Godoy, M. Mayo, D. Gal, B. G. Spratt, and B. J. Currie. 2004. Isolates of Burkholderia pseudomallei from northern Australia are distinct by multilocus sequence typing, but strain types do not correlate with clinical presentation. J. Clin. Microbiol. 42:5477-5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Silva Costa, J. L., and V. C. de Oliveira. 2003. Enhancement of soil DNA extraction by the use of hand held mixer. Braz. J. Microbiol. 34:311-312. [Google Scholar]
- 6.Finkelstein, R. A., P. Atthasampunna, and M. Chulasamaya. 2000. Pseudomonas (Burkholderia) pseudomallei in Thailand, 1964-1967: geographic distribution of the organism, attempts to identify cases of active infection, and presence of antibody in representative sera. Am. J. Trop. Med. Hyg. 62:232-239. [DOI] [PubMed] [Google Scholar]
- 7.Godoy, D., G. Randle, A. J. Simpson, D. M. Aanensen, T. L. Pitt, R. Kinoshita, and B. G. Spratt. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peakall, R., and P. E. Smouse. 2001. genalex6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 8:288-295. [Google Scholar]
- 9.Pearson, T., M. U'Ren, J. J. M. Schupp, G. J. Allan, P. G. Foster, M. J. Mayo, D. Gal, J. L. Choy, R. L. Daugherty, S. Kachur, C. L. Friedman, B. Leadem, S. Georgia, H. Hornstra, A. J. Vogler, D. M. Wagner, P. Keim, and B. J. Currie. 2007. VNTR analysis of selected outbreaks of Burkholderia pseudomallei in Australia. Infect. Genet. Evol. 7:416-423. [DOI] [PubMed] [Google Scholar]
- 10.Rotz, L. D., A. S. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hughes. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Sermswan, R. W., S. Wongratanacheewin, S. Trakulsomboon, and V. Thamlikitkul. 2001. Ribotyping of Burkholderia pseudomallei from clinical and soil isolates in Thailand. Acta Trop. 80:237-244. [DOI] [PubMed] [Google Scholar]
- 13.Suputtamongkol, Y., A. J. Hall, D. A. Dance, W. Chaowagul, A. Rajchanuvong, M. D. Smith, and N. J. White. 1994. The epidemiology of melioidosis in Ubon Ratchatani, northeast Thailand. Int. J. Epidemiol. 23:1082-1090. [DOI] [PubMed] [Google Scholar]
- 14.Swofford, D. L. 1999. PAUP, phylogenetic analysis using parsimony (and other methods), version 4.0 Beta. Sinauer, Sunderland, MA.
- 15.U'Ren, J. M., M. N. Van Ert, J. M. Schupp, W. R. Easterday, T. S. Simonson, R. T. Okinaka, T. Pearson, and P. Keim. 2005. Use of a real-time PCR TaqMan assay for rapid identification and differentiation of Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 43:5771-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.U'Ren, J. M., J. Schupp, T. Pearson, H. Hornstra, C. Clark, K. Smith, R. Leadem Daugherty, S. Rhoton, B. Leadem, S. Georgia, M. Cardon, L. Huynh, D. Deschazer, S. P. Harvey, R. Robison, B. Currie, and P. Keim. 2007. Tandem repeat regions within the Burkholderia pseudomallei genome and their application for high resolution genotyping. BMC Microbiol. 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vesaratchavest, M., S. Tumapa, N. P. J. Day, V. Wuthiekanun, W. Chierakul, M. T. G. Holden, N. J. White, B. J. Currie, B. G. Spratt, E. J. Feil, and S. J. Peacock. 2006. Nonrandom distribution of Burkholderia pseudomallei clones in relation to geographical location and virulence. J. Clin. Microbiol. 44:2553-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vuddhakul, V., P. Tharavichitkul, N. Na-Ngam, S. Jitsurong, B. Kunthawa, P. Noimay, P. Noimay, A. Binla, and V. Thamlikitkul. 1999. Epidemiology of Burkholderia pseudomallei in Thailand. Am. J. Trop. Med. Hyg. 60:458-461. [DOI] [PubMed] [Google Scholar]
- 19.White, N. J. 2003. Melioidosis. Lancet 361:1715-1722. [DOI] [PubMed] [Google Scholar]
- 20.Wuthiekanun, V., D. A. Dance, Y. Wattanagoon, Y. Supputtamongkol, W. Chaowagul, and N. J. White. 1990. The use of selective media for the isolation of Pseudomonas pseudomallei in clinical practice. J. Med. Microbiol. 33:121-126. [DOI] [PubMed] [Google Scholar]
- 21.Yeates, C., M. R. Gillings, A. D. Davison, N. Altavilla, and D. A. Veal. 1998. Methods for microbial DNA extraction from soil for PCR amplification. Biol. Proced. Online 1:40-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young, C. C., R. L. Burghoff, L. G. Keim, V. Minak-Bernero, J. R. Lute, and S. M. Hinton. 1993. Polyvinylpyrrolidone-agarose gel electrophoresis purification of polymerase chain reaction-amplifiable DNA from soils. Appl. Environ. Microbiol. 59:1972-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]