Abstract
Whole-genome microarray experiments were performed to define the Listeria monocytogenes cold growth regulon and to identify genes differentially expressed during growth at 4 and 37°C. Microarray analysis using a stringent cutoff (adjusted P < 0.001; ≥2.0-fold change) revealed 105 and 170 genes that showed higher transcript levels in logarithmic- and stationary-phase cells, respectively, at 4°C than in cells grown at 37°C. A total of 74 and 102 genes showed lower transcript levels in logarithmic- and stationary-phase cells, respectively, grown at 4°C. Genes with higher transcript levels at 4°C in both stationary- and log-phase cells included genes encoding a two-component response regulator (lmo0287), a cold shock protein (cspL), and two RNA helicases (lmo0866 and lmo1722), whereas a number of genes encoding virulence factors and heat shock proteins showed lower transcript levels at 4°C. Selected genes that showed higher transcript levels at 4°C during both stationary and log phases were confirmed by quantitative reverse transcriptase PCR. Our data show that (i) a large number of L. monocytogenes genes are differentially expressed at 4 and 37°C, with more genes showing higher transcript levels than lower transcript levels at 4°C, (ii) L. monocytogenes genes with higher transcript levels at 4°C include a number of genes and operons with previously reported or plausible roles in cold adaptation, and (iii) L. monocytogenes genes with lower transcript levels at 4°C include a number of virulence and virulence-associated genes as well as some heat shock genes.
Listeria monocytogenes is a gram-positive, psychrotolerant, food-borne pathogen that has the ability to grow at temperatures as low as −0.4°C (48, 93). L. monocytogenes ' ability to grow at low temperatures is a concern for refrigerated ready-to-eat food products in which L. monocytogenes can grow to high levels that may cause human disease, particularly if products are stored at refrigeration temperatures for a prolonged time (43). Previous studies have identified some L. monocytogenes genes and proteins that are involved in cold shock and cold adaptation or that are upregulated at low temperatures (2, 5, 13, 27, 55, 61, 62, 101), including ATP-binding cassette (ABC) transporters, such as the carnitine transport system (encoded by opuCABCD) and the glycine betaine porter II system (encoded by gbuABC), which allow for the uptake of compatible solutes that appear to facilitate L. monocytogenes growth at low temperatures (2, 27, 55). In addition, an oligopeptide binding protein (encoded by oppA), which is part of an ATP-dependent oligopeptide permease, has been shown to be required for L. monocytogenes growth at 5°C in brain heart infusion (BHI) broth (13). Other genes with potential contributions to the growth of L. monocytogenes at low temperatures include the cold-inducible gene fri, which encodes ferritin (39), and ltrC, which encodes a low-temperature-requirement C protein (101).
Thermoregulation of several L. monocytogenes virulence genes has been well documented (45, 58, 64), including temperature-dependent translation of prfA (45), which encodes the positive regulatory factor PrfA. PrfA, which is preferentially translated at 37°C over 30°C (45), regulates the transcription of a number of virulence and virulence-associated genes (12, 59, 78), including genes located in the main L. monocytogenes pathogenicity island (e.g., plcA, hly, mpl, actA, and plcB) as well as genes located in other areas of the genome (e.g., inlA, inlB, and bsh) (21, 56).
A number of studies have used genomic tools, including microarrays, to study microbial gene expression during cold shock (i.e., cold exposure for <1 h) as well as during cold growth (31, 47, 49), including in Bacillus subtilis (7, 16, 31, 49), a close relative of L. monocytogenes. For example, Budde et al. (16) recently used genome-wide microarray-based transcriptome analyses to identify 279 B. subtilis genes that showed higher transcript levels during cold growth at 15°C than during growth at 37°C. While these data provided initial insights into cold adaptation in some gram-positive bacteria, no microarray studies of L. monocytogenes gene expression during cold growth or cold shock have been published to date. Studies using other techniques for the identification of differentially expressed genes or proteomic approaches have been performed with L. monocytogenes but have revealed only small numbers of cold-induced genes or proteins (5, 62, 74). A comprehensive, genome-wide understanding of L. monocytogenes gene expression during cold growth is critical to provide a better understanding of cold adaptation in this food-borne pathogen and to allow for the rational development of novel strategies to control the growth of this pathogen during refrigerated storage of foods. We thus used a whole-genome microarray strategy to identify L. monocytogenes genes differentially expressed at 4 and 37°C during log- or stationary-phase growth, with confirmation of selected cold-induced genes by quantitative reverse transcriptase PCR (qRT-PCR).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The L. monocytogenes serotype 1/2a strain 10403S (10) was used throughout this study. For each experiment, L. monocytogenes 10403S was streaked onto BHI (Difco, Sparks, MD) agar from glycerol stock cultures (stored at −80°C), followed by incubation at 37°C for 24 h. A single colony was subsequently inoculated into BHI broth and grown at 37°C overnight with shaking (250 rpm), and a 1-ml aliquot of this overnight culture was used to inoculate 99 ml of BHI broth. After growth to log phase (an optical density at 600 nm [OD600] of 0.4), the L. monocytogenes suspension was used to inoculate 75 ml of prechilled (4°C) or prewarmed (37°C) BHI broth (in a 300-ml Erlenmeyer flask) to a starting OD600 between 0.1 and 0.2. Cells were then incubated either at 4°C for up to 12 days or at 37°C for up to 8 h (without shaking). OD600 values were measured every day for cells grown at 4°C and every hour for cells grown at 37°C; in addition, bacterial cell numbers were determined during one experiment by spread plating on BHI agar plates at each time point.
Bacterial cells were collected for RNA isolation at (i) log phase (defined as an OD600 of 0.4 ± 0.05 for both temperatures) and (ii) stationary phase (defined as an OD600 of 1.0 ± 0.25; reached at day 9 and 4 h after inoculation for cells grown at 4 and 37°C, respectively) (Fig. 1); the L. monocytogenes cell densities at each of the two growth stages (log and stationary phases) were similar for both temperatures, indicating that observed differences in transcript levels are not likely due to different cell densities in L. monocytogenes cells grown at 4 and 37°C (Fig. 1), a consideration raised previously (73). Bacterial cultures were treated with RNAprotect bacterial reagent (QIAGEN, Valencia, CA), according to the manufacturer's protocol, to stabilize RNA prior to RNA isolation. Three biological replicates were performed for each growth experiment.
FIG. 1.
Growth of L. monocytogenes strain 10403S at 4 and 37°C in BHI broth. The OD600 of L. monocytogenes 10403S was measured once a day (for 12 days) for cells grown at 4°C (•) and once an hour (for 8 h) for cells grown at 37°C (▪). The data shown represent the average OD600 values for three independent experiments, and error bars indicate standard deviations. Cell counts were performed once a day (for 12 days) for cells grown at 4°C (○) and once an hour (for 5 h) for cells grown at 37°C (□); the cell count data shown represent the log CFU/ml determined for one growth experiment (average from duplicate platings). Numbers on the x axis represent days (for growth at 4°C) and hours (for growth at 37°C). White arrows indicate the time points used for the collection of log-phase cells (OD600 of 0.4 ± 0.05 for both temperatures) for microarray experiments; gray arrows indicate the time points used for the collection of stationary-phase cells (OD600 of 1.0 ± 0.25; reached 9 days and 4 h after inoculation for cells grown at 4 and 37°C, respectively).
Total RNA isolation.
Total RNA from L. monocytogenes cells grown to log or stationary phase at 4°C or 37°C was isolated using an RNeasy midi kit (QIAGEN) as described by Kazmierczak et al. (53) with a minor modification. Briefly, QIAGEN′s protocol for “enzymatic lysis with mechanical disruption” was used, with the exception that cells were lysed by sonication on ice three times with 30-s bursts at 18 to 21 W by using a Sonicator 3000 (Misonix, Farmingdale, NY). To remove contaminating DNA, total RNA was treated with RQ1 RNase-free DNase (Promega, Madison, WI) in the presence of RNase inhibitor (RNasin; Promega) as previously described (53). RNA concentration and purity were evaluated by gel electrophoresis and absorbance readings at 260 and 280 nm, using a Nanodrop ND-1000 UV spectrophotometer (Nanodrop Technologies, Wilmington, DE).
Microarray construction.
A whole-genome microarray was constructed to include 70-mer oligonucleotides representing 2,857 open reading frames (ORFs), identified based on the annotated genome for L. monocytogenes EGD-e (28); in addition, an inlD probe was designed, using ArrayOligoSelector (http://arrayoligosel.sourceforge.net/), based on the inlD sequence for L. monocytogenes 10403S (as inlD is not present in strain EGD-e [28]). Probes targeting five Saccharomyces cerevisiae genes were used as nonhybridizing controls as previously described (64, 97). Salmon sperm DNA and serial dilutions of chromosomal L. monocytogenes 10403S DNA were also spotted on the glass array for quality control and signal normalization purposes, respectively. The Array-Ready Oligo sets for 2,857 ORFs from L. monocytogenes EGD-e and all other 70-mer oligonucleotides were purchased from Operon Technologies (Huntsville, AL). L. monocytogenes strains EGD-e and 10403S represent the same L. monocytogenes lineage (II), serotype (1/2a), and ribotype (DUP-1039C) (94), and probes designed based on the EGD-e genome were thus expected to hybridize well with 10403S genes. As an unfinished genome sequence for strain 10403S has recently (and after initiation of this project) become available (15), we were also able to verify cross-hybridization identities (CHI) between the EGD-e probes and the target genes in strain 10403S; 2,107, 2,578, and 2,695 of the probes targeting ORFs in the EGD-e genome showed CHI values of 100, ≥95, and ≥ 90, respectively; only 45 probes showed CHI values of <90. A total of 117 of the EGD-e-based microarray probes were not detected in the 10403S genome, likely reflecting sequence divergence, deletion of these genes in 10403S, or genes located in the gaps of the unfinished genome sequence for 10403S. We thus were confident that the array used here would allow for the comprehensive identification of differentially expressed genes in strain 10403S, with the possibility of some false negatives (i.e., for genes targeted by a probe with a low CHI or for genes present in the 10403S genome and absent in the EGD-e genome). Mismatches to the selected target genes are unlikely to yield false positives (i.e., genes identified as differentially regulated even if they are not) since the same mismatches will occur with both RNAs (i.e., the RNA from cells grown at 4 and 37°C).
The 70-mer oligonucleotides and controls were spotted in duplicate on Corning UltraGAPS slides (Corning, NY), using a custom-built XYZ arrayer at the Microarray Core Facility at Cornell University (Ithaca, NY). After spotting of the oligonucleotides, the slides were UV cross-linked (300 mJ for 1 min) to immobilize the oligonucleotides onto the slide. Slides were stored in a desiccator at room temperature until use.
cDNA labeling and competitive microarray hybridization.
Total RNA (representing about 10 μg RNA for each RNA sample) was reverse transcribed into cDNA and labeled with Alexa Fluor dyes (either Alexa Fluor 555 or Alexa Fluor 647) by using the SuperScript Plus indirect cDNA labeling system (Invitrogen Inc., Carlsbad, CA). Fluorescently labeled L. monocytogenes cDNA from cells grown to the same growth phase (log or stationary) at 4 or 37°C was used in the competitive microarray hybridization. Immediately prior to hybridization, microarray slides were blocked and washed essentially as previously described (64). For each array, the two fluorescently labeled cDNAs (from cells grown at 4 and 37°C) were combined into one tube, dried, and resuspended in 50 μl of 1× hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate, 0.1 mM dithiothreitol, 0.5× formamide, 600 μg/ml salmon sperm DNA), denatured at 95°C for 5 min (two times), and applied to the microarray slide by using mSeries LifterSlips (Erie Scientific, Portsmouth, NH). The competitive hybridization was then performed at 42°C overnight. After the hybridization, the microarray slide was washed to remove unbound fluorescently labeled cDNA in a series of low-, medium-, and high-stringency washes as previously described (64). Slides were then centrifuged briefly to dry and scanned using a GenePix 4000B scanner at the Microarray Core Facility at Cornell University (Ithaca, NY). Microarray images were analyzed using GenePix Pro 6.0 (Molecular Devices Corp., Sunnyvale, CA). Microarray experiments were performed using RNA from three biological replicates.
Microarray data analyses.
Microarray data were analyzed using LIMMA (linear models for microarray data) software (83) in R/Bioconductor (26), including the performance of background correction and the normalization and assessment of differential expression (DE). Background corrections were performed for each microarray slide by using the “normexp” method (83), resulting in strictly positive values and reducing variability in the log ratios for genes with low transcript levels. For within-array normalization, the “print-tip loess” method (84) was used to correct for spatial variation and intensity-dependent bias. Between-array normalization was performed to scale the normalization of log ratios to the same median absolute deviation across arrays in a given data set. Correlation between duplicate spots on each array was calculated, and a linear model was fitted to the normalized log ratios for each gene; moderated t statistics and P values were generated to identify genes with DE. Statistical significance of DE results was assessed based on adjusted P values (P values were adjusted for multiple comparisons by controlling for the false discovery rate).
GSEA.
Gene Set Enrichment Analysis (GSEA) (87) was conducted using GSEA software v2.0.1 (http://broad.mit.edu/GSEA) and the M values (log2 changes [n = fold]) obtained from the fitted normalized data in LIMMA. Data for log- and stationary-phase cultures were analyzed separately. For analysis of biological function, genes were classified into “main role category: subrole category” (e.g., “cellular processes: chemotaxis and motility”) based on The Institute for Genomic Research Comprehensive Microbial Resource (http://cmr.tigr.org) role categories for L. monocytogenes EGD-e. For analysis of enrichment of genes in the σB, CodY, and PrfA regulons among genes with high or low transcript levels at 4°C, genes were classified as follows: all 168 genes found by microarray analyses to be positively regulated by σB in L. monocytogenes cells grown under at least one of two σB-activating conditions (adjusted P < 0.05; >2.0-fold change) (S. Raengpradub, M. Wiedmann, and K. J. Boor, submitted for publication) were considered members of the σB regulon, all 85 CodY-dependent genes identified by Bennett et al. (8) were considered members of the CodY regulon, and all 12 positively regulated PrfA-dependent genes with upstream PrfA boxes (i.e., group I genes), as reported by Milohanic et al. (65), were considered members of the PrfA regulon. GSEA parameters were set to calculate 1,000 permutations, and the gene set size parameters were adjusted to include gene sets of all sizes, except gene sets with fewer than five members. For each data set (log and stationary phases), we report those gene sets found to be significant at a false discovery rate of <5% (i.e., q < 0.05 [q represents the estimated probability of a false positive]).
qRT-PCR.
Real-time qRT-PCR was used to confirm selected genes that showed higher transcript levels in cells grown at 4°C than in cells grown at 37°C in the microarray analyses (see Results). Primers and TaqMan minor groove binder probes were designed using Primer Express 1.0 (Applied Biosystems, Foster City, CA) (Table 1); primers and probes for the housekeeping genes rpoB and gap have previously been reported (82, 88).
TABLE 1.
Sequences of primers and TaqMan probes used in this studya
| Gene | Primer sequenceb | TaqMan probe sequencec |
|---|---|---|
| lmo0287 | 5′-GCTCGTGTGAAAGCCAACTTG-3′ | FAM-5′-CCGTCACAGCCAAGT-3′-MGB NFQ |
| 5′-TTCCTCGGCTGTGCTTGAG-3′ | ||
| lmo0866 | 5′-GCGGTTTACGGTGGTAGTGATAT-3′ | FAM-5′-CGTCAAATCCGTTCACT-3′-MGB NFQ |
| 5′-GGCGTACCAACTACGATTTGTG-3′ | ||
| lmo1364 (cspL) | 5′-CCAAGGCGACGGATTCAA-3′ | FAM-5′-CAAGCAGTAACTTTCG-3′-MGB NFQ |
| 5′-CGCGTTGGCCTTCTTCAA-3′ | ||
| lmo1722 | 5′-CAAAGACGGTGCAGATGTACTTG-3′ | FAM-5′-AGTATCACCAACTGGAAC-3′-MGB NFQ |
| 5′-GGCAGTGCATAAGCCACTGTT-3′ |
qRT-PCR was performed as previously described (54). All qRT-PCR experiments were performed using the three different RNA preparations (representing the three independent growth experiments) that were also used for the microarray experiments. qRT-PCR data were log10 transformed and normalized to the geometric means for the housekeeping genes rpoB and gap as also previously described (54). Statistical analyses were performed in SAS v 9.1 (SAS Institute, Inc., Cary, NC) using a general linear model (GLM), with multiple comparisons performed using Tukey's studentized range test. An α value of <0.05 was considered significant for qRT-PCR data.
Microarray data accession number.
Raw data and microarray files (MIAME format) are available at the Gene Expression Omnibus (GEO) database (4) under accession number GSE7465.
RESULTS
Microarray-based definition of L. monocytogenes cold stress genes.
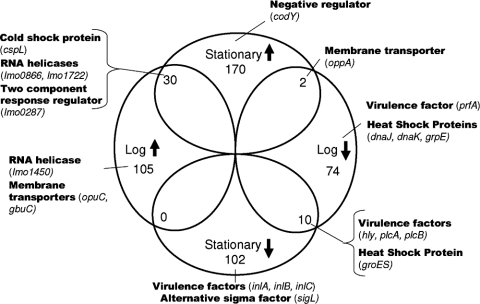
Competitive whole-genome microarray hybridizations were performed to experimentally define the L. monocytogenes cold stress regulon, using (i) cells grown to log phase at 4°C and 37°C and (ii) cells grown to stationary phase at 4°C and 37°C (Fig. 1). Statistical analysis was performed separately for each microarray data set. Initial analyses that used an adjusted P value of <0.05 and at least twofold DE to identify differentially expressed genes revealed (i) 110 and 237 genes, respectively, with significantly higher transcript levels in log- and stationary-phase cells grown at 4°C and (ii) 74 and 187 genes, respectively, with significantly lower transcript levels in log- and stationary-phase cells grown at 4°C (see Fig. S1 in the supplemental material); all genes identified as differentially transcribed with an adjusted P value of <0.05 are listed in Tables S1 through S4 in the supplemental material. Based on the large numbers of differentially expressed genes identified when an adjusted P value of <0.05 was used as the cutoff, and in order to include only genes identified with high confidence as differentially expressed in our analyses, we modified the P value cutoff to an adjusted P value of <0.001, consistent with other studies that have used P value cutoffs lower than 0.05 to define genes differentially expressed at low temperatures (16). Thus, the two criteria used to identify genes that showed either higher or lower transcript levels at 4°C (compared to 37°C) were (i) a conservatively adjusted P value of <0.001 and (ii) at least twofold DE in mRNA transcript levels between L. monocytogenes cells grown at 4°C and those grown at 37°C. Using these criteria, we identified a total of 245 genes that showed higher transcript levels in either log- or stationary-phase cells at 4°C, including (i) 105 genes that showed higher transcript levels in log-phase cells and (ii) 170 genes that showed higher transcript levels in stationary-phase cells; 30 of these genes showed higher transcript levels in both log- and stationary-phase cells grown at 4°C (Fig. 2; Table 2). In addition, 166 genes showed lower transcript levels in either log- or stationary-phase cells at 4°C (compared to 37°C), including 74 genes and 102 genes that showed lower transcript levels in log- and stationary-phase cells, respectively (Fig. 2).
FIG. 2.
Venn diagram of L. monocytogenes 10403S genes differentially transcribed at 4 and 37°C in log- and stationary-phase cells. Only genes that are differentially transcribed based on stringent cutoff criteria (≥2-fold change; adjusted P < 0.001) are shown (Fig. S1 in the supplemental material represents a similar Venn diagram but shows all genes that showed evidence of differential transcription based on less stringent criteria [≥2-fold change; adjusted P < 0.05]). In each circle, the total number of genes upregulated (↑) at 4°C or downregulated (↓) at 4°C under a given condition (log or stationary phase) is shown. Selected genes with relevant known or putative functions are also indicated.
TABLE 2.
Genes identified by microarray analysis to be upregulated at 4°C in both log- and stationary-phase cellsa
| Protein category and gene | Protein function(s)b | Change (n-fold) in cells grown toc:
|
|
|---|---|---|---|
| Log phase | Stationary phase | ||
| Cell membrane function | |||
| lmo0540 | Similar to penicillin binding protein | 2.3 | 3.1 |
| lmo0581 (met) | Putative S-adenosylmethionine-dependent methyltransferase | 4.7 | 2.2 |
| lmo1713 | Similar to cell-shape-determining proteins | 2.5 | 5.0 |
| lmo1864 | Similar to hemolysin III proteins, putative integral membrane protein | 4.1 | 8.3 |
| lmo2522 | Similar to hypothetical cell wall binding protein from B. subtilis | 10.6 | 4.5 |
| Lipid and/or carbohydrate metabolism | |||
| lmo0624 | Acetyltransferase, Gcn5-related N-acetyltransferase family | 3.6 | 2.1 |
| lmo0625 | Putative lipase/acylhydrolase | 2.7 | 3.1 |
| lmo1936 (gpsA) | Similar to NAD(P)H-dependent glycerol-3-phosphate dehydrogenase | 2.2 | 4.2 |
| lmo2480 | Similar to acetyltransferase | 2.2 | 2.4 |
| Motility | |||
| lmo0677 | Similar to flagellar biosynthesis protein FliQ | 3.2 | 2.8 |
| Signal transduction | |||
| lmo0287 | Similar to two-component response regulator | 2.5 | 3.5 |
| Transcription or translation | |||
| lmo0866 | Similar to ATP-dependent RNA helicase | 2.4 | 3.0 |
| lmo1067 | Similar to GTP-binding elongation factor | 3.1 | 3.3 |
| lmo1364 (cspL)d | Similar to cold shock protein | 7.1 | 7.0 |
| lmo1722 | Similar to ATP-dependent RNA helicases | 2.4 | 3.9 |
| Amino acid metabolism | |||
| lmo0055 (purA) | Highly similar to adenylosuccinate synthetase | 2.2 | 3.0 |
| Other or hypothetical proteins | |||
| lmo0047 | Unknown | 2.8 | 5.6 |
| lmo0189 | Highly similar to B. subtilis Veg protein | 12.2 | 5.2 |
| lmo0391 | Unknown | 3.8 | 4.1 |
| lmo0392 | Highly similar to B. subtilis YqfA protein | 3.1 | 2.8 |
| lmo0393 | Unknown | 3.7 | 3.8 |
| lmo0592 | Unknown | 3.4 | 2.4 |
| lmo0604 | Similar to B. subtilis YvlA protein | 2.8 | 3.5 |
| lmo0905 | Unknown | 2.5 | 3.0 |
| lmo0954 | Unknown | 2.8 | 5.0 |
| lmo1245 | Unknown | 4.7 | 4.6 |
| lmo1487 | Similar to unknown proteins | 2.3 | 3.0 |
| lmo1670 | Similar to conserved hypothetical proteins | 2.5 | 4.0 |
| lmo1937 (engA) | GTP-binding protein | 2.6 | 5.4 |
| lmo2713 | Secreted protein with one GW repeat | 2.5 | 3.4 |
Only the 30 genes that met the stringent criteria for being upregulated at 4°C in both log- and stationary-phase cells (i.e., a >2-fold change and an adjusted P value of <0.001) are listed here; all genes upregulated at 4°C in either log- or stationary-phase cells are listed in Tables S1 and S2 in the supplemental material.
Protein functions are based on annotations provided by ListiList (http://genolist.pasteur.fr/ListiList/), TIGR (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi), the NCBI (http://www.ncbi.nlm.nih.gov/), and the KEGG Sequence Similarity Database (http://www.genome.jp/kegg/ssdb/).
Change (n-fold) indicates the transcript level ratio between L. monocytogenes 10403S cells grown at 4°C and those grown at 37°C (as determined by microarray analysis); positive values indicate that transcript levels are higher at 4°C than at 37°C (e.g., a value of 2.3 indicates a 2.3-fold-higher transcript level at 4°C than at 37°C).
cspL has also been designated cspLA.
L. monocytogenes genes that show higher transcript levels at 4°C. (i) Genes with higher transcript levels at 4°C in both log- and stationary-phase cells.
Genes with higher transcript levels at 4°C in both log- and stationary-phase (Table 2) could be considered part of the “core” cold growth regulon. The 30 L. monocytogenes genes that were grouped into this category included three operons (Fig. 3) as well as genes encoding a cold shock protein (cspL), two RNA helicases, a two-component response regulator (lmo0287), a flagellar biosynthesis protein (lmo0679), and a number of genes with unknown functions (see Table 2 for a complete listing). A number of these genes encode proteins with confirmed or likely roles in cold growth and cold adaptation. For example, the putative ATP-dependent DEAD-box RNA helicases (encoded by lmo0866 and lmo1722; Table 2) likely act as chaperones facilitating translation at low temperatures, as supported by evidence from studies of Escherichia coli and B. subtilis (42, 91). The GTP-binding elongation factor homolog (encoded by lmo1067), which was also identified here as part of the L. monocytogenes “core” cold growth regulon, may assist in translation elongation at low temperatures; the lmo1067 homolog in B. subtilis was found to be upregulated at 15°C (7), and E. coli bipA (53.2% amino acid [aa] sequence identity to lmo1067) (28) was shown to be required for growth at low temperatures (72). While only one flagellar biosynthesis protein (encoded by lmo0679) showed higher transcript levels in both log- and stationary-phase cells at 4°C with an adjusted P value of <0.001, three more genes in the flagellar biosynthesis operon (consisting of lmo0675 to lmo0689) also showed higher transcript levels in both log- and stationary-phase cells but with an adjusted P value of <0.05 for DE in stationary phase (Fig. 3).
FIG. 3.
Illustration of selected putative L. monocytogenes operons identified by microarray analysis to be upregulated at 4°C in log and stationary phases (A), log phase only (B), and stationary phase only (C). Operons were included even if some genes did not meet the stringent cutoff criteria (i.e., a ≥2-fold difference in transcript levels and an adjusted P value of <0.001). Transcript level ratios are given under each gene. Positive values indicate higher transcript levels at 4°C than at 37°C; while most genes have positive values, some genes have negative values for transcript level ratios, indicating higher transcript levels at 37°C (e.g., a value of −2.1 indicates a 2.1-fold-higher transcript level at 37°C). Transcript level ratios of ≥2 (indicating ≥2-fold differences in transcript levels) are in bold; adjusted P values for significance of differential expression are indicated by ** (P < 0.001), * (P < 0.05), and NS (not significant) (P > 0.05). Known promoters (GenBank accession no. AF0322444 and Y09161) (16a, 81) are marked with small arrows. Rho-independent terminators were predicted for L. monocytogenes as described by de Hoon et al. (19) and are shown as hairpins; the previously determined Rho-dependent terminator downstream of lmaA (81) is shown as a hairpin with a diamond. Genes are not drawn to scale.
Interestingly, seven genes and one operon (consisting of lmo0624 and lmo0625) with higher transcript levels at 4°C in both log- and stationary-phase cells had putative or confirmed functions in lipid metabolism or represented cell membrane proteins (Table 2), consistent with the importance of membrane modifications for cold adaptation (3, 22). One of these genes, lmo0581 (met), encodes a putative S-adenosylmethionine-dependent methyltransferase; an L. monocytogenes deletion in this gene resulted in impaired growth under alkaline conditions and under ethanol stress (77). Another one of these genes, lmo0540, encodes a putative penicillin binding protein; this class of proteins is critical for bacterial cell wall peptidoglycan assembly (35, 100); peptidoglycan polymerization and modification appear to contribute to stress resistance, including osmotic stress, in some bacteria (75).
(ii) Genes with higher transcript levels only in log-phase cells at 4°C.
The 75 genes that were found to have higher transcript levels only in log-phase cells grown at 4°C included at least 10 operons (representing 45 of these genes) (Fig. 3; see also Table S1 in the supplemental material) as well as 30 individual genes. A number of these operons and genes encode proteins and/or pathways with confirmed or likely roles in cold growth and cold adaptation, such as compatible solute transporters, an RNA helicase, and amino acid synthesis pathways providing precursors for low-melting-point, branched-chain fatty acids. For example, both the opuCABCD operon (Fig. 3), which encodes a carnitine transporter, and gbuC, which encodes the binding protein of glycine betaine transporter II (see Table S1 in the supplemental material), had higher transcript levels in log-phase cells at 4°C. These transporters are known to facilitate the uptake of compatible solutes important for L. monocytogenes growth at low temperatures (2). lmo1450 (see Table S1 in the supplemental material) encodes a DEAD-box family RNA helicase that may act as an RNA chaperone at low temperatures (42, 91). Interestingly, the protein encoded by lmaA was previously found to be secreted in cultures growing at 4 and 20°C but not at 37°C, and it has been hypothesized that lmaA may encode a protein transport system or surface structures that may contribute to growth at low temperatures (81). Most genes in a large operon directly downstream of lmaA (consisting of lmo0117 to lmo0129) also showed higher transcript levels in log-phase cells at 4°C (Fig. 3); the genes in this operon encode a number of proteins with similarity to A118 phage proteins.
A number of operons with higher transcript levels in log-phase cells at 4°C have previously been shown to be downregulated by the pleiotropic transcriptional repressor CodY in L. monocytogenes (8); these CodY-repressed operons include the operon consisting of lmo0675 to lmo0689 (encoding flagellar biosynthesis proteins), the operon consisting of lmo0984 to lmo0988 (encoding proteins with unknown functions), and the ilvD-ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD-ilvA operon. Interestingly, the proteins encoded by ilvD-ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD-ilvA are important for the biosynthesis of amino acids, including isoleucine, which is an important precursor for the synthesis of anteiso-branched-chain fatty acids essential for ensuring membrane fluidity in L. monocytogenes cells grown at low temperatures (3, 102). In addition, the CodY-repressed genes flaA, which encodes a flagellin protein, and lmo2824, which encodes a d-3-phosphoglycerate dehydrogenase, were also found to have higher transcript levels in log-phase cells at 4°C.
(iii) Genes with higher transcript levels only in stationary-phase cells at 4°C.
The 140 genes that showed higher transcript levels only in stationary-phase cells at 4°C included at least 24 operons (representing 58 of these genes) (Fig. 3; see also Table S2 in the supplemental material). A number of these operons and genes encode proteins and/or pathways with confirmed or likely roles in cold growth and cold adaptation, including a putative DNA gyrase protein, transcriptional regulators, ribosomal proteins, phosphotransferase systems (PTS), a peptide transporter, a two-component sensor histidine kinase, and a protein-folding enzyme. Specifically, the parE-parC operon, which showed higher transcript levels in stationary-phase cells at 4°C (with an adjusted P value of <0.05 for parC; Fig. 3), encodes DNA gyrase-like proteins that may have a role in relieving negative supercoiling at low temperatures (30, 46). Ribosomal operons with higher transcript levels (Fig. 3) include rplK-rplA (encoding ribosomal proteins L11 and L1), rplU-lmo1541-rpmA (encoding the ribosomal proteins L21 and L27 and a protein with unknown function), and infC-rpmT-rplT (encoding translation initiation factor IF-3, ribosomal protein L35, and ribosomal protein L20). In addition, six individual genes encoding ribosomal proteins were also found to be upregulated at 4°C, including lmo1480, lmo1787, lmo2548, lmo2620, lmo2627, and lmo2856 (see Table S2 in the supplemental material).
Operons encoding PTS-associated proteins (Fig. 3) that showed higher transcript levels at 4°C include a cellobiose-specific PTS operon (consisting of lmo2683 to lmo2685), a mannitol-specific PTS operon (consisting of lmo2799 to lmo2795), and a galactitol-specific PTS operon (consisting of lmo2668 to lmo2665), indicating that the expression of these particular PTS may be important for carbon assimilation during stationary-phase growth at low temperatures. In addition, two operons located immediately adjacent to the galactitol-specific PTS operon (consisting of lmo2668 to lmo2665), including the operon consisting of lmo2664 and lmo2663 (encoding sorbitol dehydrogenase and polyol dehydrogenase, respectively) and that consisting of lmo2661, lmo2660, and lmo2659 (encoding ribulose-5-phosphate-3-epimerase, transketolase, and ribulose-phosphate-3-epimerase, respectively), also showed higher transcript levels at 4°C (Fig. 3). Ribulose-5-phosphate-3-epimerase, transketolase, and ribulose-phosphate-3-epimerase are part of the phosphogluconate pathway, which is involved in the production of NADPH for fatty acid synthesis as well as in the conversion of pentoses and hexoses for nucleotide synthesis and glycolysis (86). The aroF-aroB-lmo1926-hisC-tyrA-aroE amino acid biosynthesis operon (Fig. 3) also showed higher transcript levels in stationary-phase cells at 4°C, consistent with two-dimensional protein gel electrophoresis data that showed the induction of B. subtilis AroF after 3 h of exposure to low temperatures (33).
Individual genes with higher transcript levels in stationary-phase cells at 4°C also included lmo2196 (oppA), which encodes an oligopeptide binding protein previously shown to be required for L. monocytogenes growth at 5°C (13), as well as lisK, which encodes a two-component regulatory system histidine kinase that is part of a response system that affects membrane composition in L. monocytogenes (see Table S2 in the supplemental material) (18). Two genes encoding prolyl isomerases also showed higher transcript levels at 4°C, including tig, which has been reported to be important for protein folding at low temperatures in E. coli (52), and lmo2376. While lmo2376 showed only 1.99-fold-higher transcript levels at 4°C in L. monocytogenes (adjusted P < 0.05) (see Table S2 in the supplemental material), ppiB, the B. subtilis homolog of lmo2376, has been found to encode a protein that shows increased synthesis at low temperatures (33) and that is involved in protein folding (29), supporting that lmo2376 plays a role in cold growth.
Finally, some genes encoding regulatory proteins were also found to show higher transcript levels in stationary-phase cells grown at 4°C. Specifically, the four genes (i.e., rsbRSTU) representing the σA-dependent 5′ portion of the eight-gene L. monocytogenes sigB operon show higher transcript levels at 4°C than at 37°C (Fig. 3). While the codV-clpQ-clpY-codY operon, which includes the gene encoding the transcriptional repressor CodY, showed higher transcript levels in stationary-phase cells at 4°C (Fig. 3), two previously identified CodY-repressed operons (i.e., those consisting of lmo2664 and lmo2663 and lmo2661 to lmo2659; see above) also showed higher transcript levels in stationary-phase cells at 4°C than in cells at 37°C.
L. monocytogenes genes that show lower transcript levels at 4°C. (i) Genes with lower transcript levels at 4°C in both log- and stationary-phase cells.
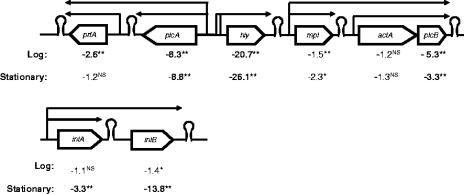
A total of 10 genes, including virulence genes and genes encoding heat shock proteins, showed ≥2.0-fold-lower transcript levels (adjusted P < 0.001) at 4°C in both log- and stationary-phase cells (Table 3). Interestingly, three of the six virulence genes in the major L. monocytogenes virulence gene island (i.e., hly, plcA, and plcB) showed lower transcript levels in log- and stationary-phase cells grown at 4°C (Table 3; Fig. 4), consistent with the fact that the transcription of these virulence genes is activated by PrfA (12, 59), which is preferentially translated at 37°C (45). In addition to groES, which showed significantly lower transcript levels, with an adjusted P value of <0.001, in both log- and stationary-phase cells grown at 4°C, groEL also showed lower transcript levels under both conditions (even though only at an adjusted P value of <0.05 in stationary-phase cells) (Table 3), indicating that the groES-groEL operon, which encodes class I heat shock proteins, is downregulated at 4°C independent of growth phase.
TABLE 3.
Genes identified by microarray analysis to be downregulated at 4°C in both log- and stationary-phase cellsa
| Protein category and gene | Protein functionb | Change (n-fold) in cells grown toc:
|
|
|---|---|---|---|
| Log phase | Stationary phase | ||
| Virulence factors | |||
| lmo0201 (plcA) | Phosphatidylinositol-specific phospholipase C | −8.3 | −8.8 |
| lmo0202 (hly) | Listeriolysin O precursor | −20.7 | −26.1 |
| lmo0205 (plcB) | Phospholipase C | −5.3 | −3.3 |
| Heat shock proteins | |||
| lmo2069 (groES) | Class I heat shock protein (chaperonin) GroES | −5.8 | −4.4 |
| lmo2068 (groEL) | Class I heat shock protein (chaperonin) GroEL | −5.7 | −3.1d |
| Metabolism | |||
| lmo0210 (ldh) | Similar to l-lactate dehydrogenase | −4.0 | −3.8 |
| Transcription or translation | |||
| lmo0822 | Similar to transcriptional regulators | −2.2 | −2.4 |
| Other or hypothetical proteins | |||
| lmo0104 | Unknown | −2.6 | −3.7 |
| lmo0355 | Similar to flavocytochrome c fumarate reductase chain A | −9.2 | −3.9 |
| lmo1219 | Unknown | −2.5 | −4.8 |
| lmo2453 | Similar to lipolytic enzyme | −2.1 | −5.5 |
While only 10 genes met the stringent criteria for being downregulated at 4°C in both log- and stationary-phase cells (i.e., a ≥2-fold change and an adjusted P value of <0.001), 11 genes are listed here. groEL was included (even though groEL differential expression in stationary-phase cells did not meet the criterion of an adjusted P value of <0.001) as this gene is in the same operon as groES (which met the criteria of a ≥2-fold change and an adjusted P value of <0.001); all genes downregulated at 4°C in either log- or stationary-phase cells are listed in Tables S3 and S4 in the supplemental material.
Protein functions are based on annotations provided by ListiList (http://genolist.pasteur.fr/ListiList/), TIGR (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi), the NCBI (http://www.ncbi.nlm.nih.gov/), and the KEGG Sequence Similarity Database (http://www.genome.jp/kegg/ssdb/).
Change (n-fold) indicates the transcript ratio between L. monocytogenes 10403S cells grown at 4°C and those grown at 37°C (as determined by microarray analysis); negative values indicate transcript levels that are lower at 4°C than at 37°C (e.g., a value of −8.6 indicates an 8.6-fold-lower transcript level at 4°C than at 37°C).
For lmo2068 (groEL), differential transcription in stationary-phase cells was significant at an adjusted P value of <0.05 but not at an adjusted P value of <0.001.
FIG. 4.
Illustration of selected L. monocytogenes virulence genes identified by microarray analysis to be downregulated at 4°C. Transcript level ratios are given under each gene. Negative values indicate lower transcript levels at 4°C than at 37°C (e.g., a value of −3.3 indicates a 3.3-fold-lower transcript level at 4°C). Transcript level ratios of ≥2 (indicating ≥2-fold differences in transcript levels) are in bold; adjusted P values for significance of differential expression are indicated by ** (P < 0.001), * (P < 0.05), and NS (not significant) (P > 0.05). Previously reported promoters and terminators (79) are indicated by small arrows and hairpins, respectively. Genes are not drawn to scale.
(ii) Genes with lower transcript levels only in log-phase cells grown at 4°C.
The 64 genes that were found to have lower transcript levels only in log-phase cells at 4°C included a total of seven operons (representing 20 of these genes) (Fig. 3; see also Table S1 in the supplemental material) as well as 44 individual genes, including prfA and an operon encoding a mannose-specific PTS (consisting of lmo0096 to lmo0098) (see Table S3 in the supplemental material). Interestingly, the hrcA-grpE-dnaK operon showed 3.4- to 4.3-fold-lower transcript levels in log-phase cells grown at 4°C (see Table S3 in the supplemental material). This finding was surprising as the HrcA-repressed groES-groEL operon also showed lower transcript levels, but this finding may be a consequence of posttranscriptional regulation of HrcA activity (89). cspB, which encodes a putative cold shock protein, shows 36.1-fold-lower transcript levels at 4°C than at 37°C (see Table S3 in the supplemental material), consistent with observations that the cspB homologs in E. coli and B. subtilis do not seem to be critical for growth at low temperatures (32, 99).
(iii) Genes with lower transcript levels only in stationary-phase cells at 4°C.
The 92 genes found to have lower transcript levels only in stationary-phase cells at 4°C included at least 16 operons (representing 29 of these genes) (see Table S4 in the supplemental material). While approximately 30 of these genes encode proteins with unknown or hypothetical functions (see Table S4 in the supplemental material), three genes encoding internalins also showed lower transcript levels at 4°C, including the inlAB operon (Fig. 4) and inlC. Interestingly, relA, which encodes a (p)ppGpp synthetase involved in the stringent response, was also found to have lower transcript levels in stationary-phase cells grown at 4°C. Furthermore, cspD, which encodes a putative cold shock protein similar to E. coli CspC (59.0% aa identity) (11) and B. subtilis CspD (81.5% aa identity) (57), showed lower transcript levels at 4°C than at 37°C (adjusted P < 0.05). clpB, which encodes an ATP-binding endoprotease, also showed considerably lower transcript levels in stationary-phase cells at 4°C (15.7-fold-lower transcript levels at 4°C, representing one of the genes with the largest differences in transcript levels between 4 and 37°C in stationary phase). Interestingly, ltrC, encoding the low-temperature-requirement C protein (101), and sigL, encoding the alternative sigma factor σL, which appears to be important for cold growth in B. subtilis (95), also showed lower transcript levels at 4°C (6.9- and 2.6-fold-lower transcript levels, respectively, during stationary-phase growth at 4°C than during growth at 37°C) (see Table S4 in the supplemental material).
GSEA of genes in different role categories.
GSEA identified three role categories that are overrepresented among the genes that showed higher transcript levels in log-phase cells grown at 4°C, including (i) “cellular processes: chemotaxis and motility” (q = 0.001), (ii) “amino acid biosynthesis: pyruvate family” (q = 0.008), and (iii) “purines, pyrimidines, nucleosides, and nucleotides: pyrimidine ribonucleotide biosynthesis” (q = 0.025). Chemotaxis and motility-related genes include the operon consisting of lmo0675 to lmo0689, which encodes flagellar biosynthesis proteins, while genes involved in pyruvate family amino acid biosynthesis include the ilvD-ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD-ilvA operon. Two role categories (i.e., “protein synthesis: tRNA aminoacylation” [q = 0.002] and “protein fate: protein folding and stabilization” [q = 0.019]) were overrepresented among the genes with lower transcript levels in log-phase cells grown at 4°C. Genes in the “tRNA aminoacylation” subrole category that showed significantly lower transcript levels in log-phase cells at 4°C encode a number of tRNA synthetases (phenyalanyl-, glycyl-, alanyl-, and asparaginyl-tRNA synthetases), while the “protein folding and stabilization” category includes several genes encoding class I heat shock proteins (i.e., grpE, dnaK, dnaJ, groES, and groEL).
GSEA of stationary-phase microarray data identified four and two gene sets, respectively, that were enriched among genes with higher and lower transcript levels at 4°C. The role categories overrepresented among genes with higher transcript levels at 4°C include (i) “protein synthesis: ribosomal proteins—synthesis and modification” (q = 0.002), (ii) “cell envelope: biosynthesis of murein sacculus and peptidoglycan” (q = 0.001), (iii) “cellular processes: chemotaxis and motility” (q = 0.025), and (iv) energy metabolism: ATP-proton motive force interconversion (q = 0.041). Nine of the 21 genes in the ATP-proton motive force gene set are organized into two operons (one consisting of atpD-atpC and the other consisting of atpI-atpB-atpE-atpF-atpH-atpA-atpG), which encode various chains of the H+-transporting ATP synthase; 8 of these 9 genes were found to have significantly higher transcript levels at 4°C (adjusted P value of <0.05; 1.6- to 2.4-fold change). The role categories overrepresented among genes with lower transcript levels in stationary phase at 4°C include (i) “energy metabolism: amino acids and amines” (q = 0.002) and (ii) cellular processes: pathogenesis” (q = 0.006). The latter subrole category contains many of the internalin genes (i.e., inlA, inlB, and inlC) but none of the genes in the prfA virulence gene island, as many of these genes were categorized into separate subrole categories.
GSEA of σB-, CodY-, and PrfA-dependent genes.
GSEA found that genes in the CodY regulon were significantly enriched among genes with higher transcript levels in log-phase cells (q = 0.011) and stationary-phase cells (q < 0.001) at 4°C, suggesting the derepression of CodY at 4°C. Genes in the PrfA regulon were also significantly enriched among genes with lower transcript levels in log-phase cells (q = 0.006) and stationary-phase cells (q = 0.007) at 4°C, consistent with the enhanced expression of PrfA at 37°C (45, 58). Genes in the σB regulon were significantly enriched among genes with higher transcript levels in log-phase cells at 4°C (q = 0.02) but were also significantly enriched among genes with lower transcript levels in stationary-phase cells at 4°C (q < 0.001), suggesting that growth phase may be more important than temperature in regulating transcription of the σB regulon.
qRT-PCR confirmation of selected L. monocytogenes genes that have higher transcript levels at 4°C.
For four genes that were found by microarray analysis to show higher transcript levels at 4°C than at 37°C in both log- and stationary-phase cells, differential transcription was also evaluated by qRT-PCR. Genes selected for qRT-PCR confirmation included lmo0287 (encoding a two-component response regulator) and lmo1364 (cspL, encoding cold shock protein L) as well as lmo0866 and lmo1722, both of which encode ATP-dependent DEAD-box RNA helicases; all four of these genes encode proteins with plausible roles in cold growth and cold adaptation (42, 44, 66, 91). The transcript levels of these genes were normalized to the geometric means for the housekeeping genes rpoB and gap; the factor “temperature” (4°C or 37°C) did not have a significant effect on either rpoB or gap transcript levels (P > 0.1; GLM). The analysis of normalized transcript levels for the four target genes (lmo0287, lmo0866, cspL, and lmo1722) showed that normalized transcript levels for all four genes were significantly (P < 0.0005; GLM) affected by temperature. For all four target genes tested, transcript levels were higher at 4°C than at 37°C in both log- and stationary-phase cells (Fig. 5), confirming the microarray data, which also showed higher transcript levels for these four genes at 4°C in both log- and stationary-phase cells (Table 2).
FIG. 5.
Transcript levels for putative cold stress genes as determined by qRT-PCR. Transcript levels for the housekeeping genes rpoB and gap and for four putative cold stress genes were determined by qRT-PCR for L. monocytogenes 10403S cells grown in BHI broth to log or stationary phase at 4°C or 37°C. Transcript levels for the putative cold stress genes lmo0287, lmo0866, cspL, and lmo1722 were normalized to the geometric means for rpoB and gap. While rpoB and gap transcript levels were significantly affected (P < 0.0001 and P < 0.01, respectively) by growth phase, with lower absolute transcript levels (per 10 μg RNA) observed in stationary-phase cells, consistent with previous qRT-PCR data (16a), normalization of the transcript levels of the four target genes to the geometric means for rpoB and gap transcript levels was performed to adjust transcript levels to account for physiological differences between bacterial cells in different growth phases. The fact that temperature did not significantly (P > 0.1) affect rpoB and gap transcript levels supports that analyses of target gene transcript levels normalized to rpoB and gap transcript levels provide robust data for evaluating the effect of temperature on target gene transcript levels. Data represent the averages for three biological replicates; error bars indicate standard deviations. For a given gene, different letters above the bars indicate significantly different transcript levels.
DISCUSSION
Overall, our data show that (i) a large number of L. monocytogenes genes are differentially expressed at 4 and 37°C, (ii) L. monocytogenes genes with higher transcript levels at 4°C include a number of genes and operons with previously reported or plausible roles in cold adaptation, and (iii) L. monocytogenes genes with lower transcript levels at 4°C include a number of virulence and virulence-associated genes as well as some heat shock genes.
(i) A large number of L. monocytogenes genes are differentially expressed at 4 and 37°C.
Among the 2,857 L. monocytogenes ORFs included in our microarray, a total of 245 genes (8.6% of all ORFs in the array) showed higher transcript levels (≥2.0-fold change; adjusted P value < 0.001) in stationary- or log-phase cells grown at 4°C than in cells grown at 37°C. A previous study using selective capture of transcribed sequences identified 24 genes to be more highly transcribed in L. monocytogenes 10403S cells grown at 10°C than in cells grown at 37°C (62). While some genes (i.e., flaA and aroA) identified in this previous study (62) as upregulated at 10°C also showed higher transcript levels at 4°C in our study, other genes identified in this previous study (62) as upregulated at 10°C were not identified as cold induced here, most likely due to differences in experimental conditions (e.g., use of different growth temperatures) and/or methods used to measure differential transcription. A previous study by Bayles et al. (5), which used two-dimensional gel electrophoresis to identify cold shock proteins in L. monocytogenes 10403S cells exposed to 5°C for up to 120 min and cold acclimation proteins in L. monocytogenes cells exposed to 5°C for up to 5 h, found 12 cold shock proteins and 4 cold acclimation proteins, which were not further identified. Two-dimensional protein gel electrophoresis studies of other psychrotolerant microorganisms (e.g., Pseudomonas fragi and Arthrobacter globiformis) found 20 P. fragi proteins that were present at higher levels in cells grown at 4°C for 270 min than in cells grown at 30°C (38) and 9 A. globiformis proteins that were induced at 4°C after cold shock for 240 min (9). While fairly small numbers of cold-induced proteins are usually identified using traditional two-dimensional gel electrophoresis approaches, including proteins in L. monocytogenes (5, 9, 38, 47), whole-genome microarray analyses can clearly provide more-comprehensive analysis of the cold shock regulon. Similar to our studies, microarray-based transcriptome analyses of other bacteria revealed large numbers of genes that were upregulated during cold shock and cold growth (16, 25). For example, transcriptome analysis of B. subtilis (16) identified 279 of 4,107 ORFs to be upregulated (≥2-fold change; adjusted P value ≤ 0.01) at 15°C compared to those at 37°C. In our study, a total of 166 L. monocytogenes genes (i.e., 5.8% of all ORFs in the array) showed lower transcript levels at 4°C (in either stationary-phase or log-phase cells). By comparison, microarray analyses of B. subtilis found 301 genes (7.3% of ORFs) to be downregulated in cells grown at 15°C (compared to those in cells grown at 37°C) (16).
Transcriptome analysis of bacteria exposed to other stress conditions, e.g., heat shock (1, 41), also revealed stress response regulons of considerable size. For example, in B. subtilis, 124 of approximately 4,100 genes present in a microarray (i.e., 3.0% of genes) were induced during heat shock (i.e., at 3, 10, and 20 min after exposure of cells to 48°C) (41). In Staphylococcus aureus, 98 of approximately 3,300 ORFs included in the Affymetrix S. aureus GeneChip (i.e., 3.0% of genes) were upregulated after 30 min of heat shock at 42°C (1).
(ii) L. monocytogenes genes upregulated at 4°C include a number of genes and operons with previously reported or plausible roles in cold adaptation.
Bacteria that transition to low temperatures have to overcome a number of well-recognized problems, including decreased membrane fluidity, which reduces nutrient uptake capabilities, increased superhelical coiling of DNA, which may negatively affect a bacterium's ability to replicate or transcribe DNA, secondary structures in RNA, which affect translation, reduced enzyme activities, inefficient or slow protein folding, and reduced ability of ribosomes to function properly at low temperatures (33). A considerable number of the L. monocytogenes genes and operons we identified to be upregulated in cells grown at 4°C appear to facilitate bacterial responses to these challenges, as discussed in more detail below.
L. monocytogenes genes that are upregulated at 4°C and encode proteins likely to facilitate replication at low temperatures include parE and parC, which encode subunits of DNA gyrase. DNA gyrases likely play a role in relieving negative supercoiling at low temperatures, supported by observations that DNA gyrases are important for cellular processes in B. subtilis at low temperatures (30) and that gyrase-encoding genes are cold inducible in E. coli (46). Three genes encoding RNA helicases were also found to be upregulated at 4°C in L. monocytogenes, consistent with the observation that some DEAD-box RNA helicases can act as RNA chaperones that resolve secondary structures in mRNA that are formed at low temperatures (42, 91). Similar to our findings, the transcription of genes encoding RNA helicases has also been found to be induced in other bacteria exposed to low temperatures, including E. coli, where rhlE (csdA), encoding a putative ATP-dependent RNA helicase, showed higher transcript levels in cells exposed to 16°C (76), and Yersinia pestis, where four genes (rhlE, srmB1, dbpA, and deaD), encoding RNA helicases, showed higher transcript levels at 10°C (36). The importance of helicases for cold growth is also supported by the observation that an E. coli strain with a site-directed mutation in the DEAD motif of the helicase CsdA (91) showed reduced growth at low temperatures. In addition to RNA helicases, some cold-inducible cold shock proteins also facilitate translation by binding to single-stranded mRNA (presumably preventing mRNA that has been unwound by RNA helicases from refolding) until the ribosome initiates translation at low temperatures (33, 42). For example, in B. subtilis, RNA helicases such as CshA and CshB have been found to work together with cold shock proteins (e.g., CspB) to help keep mRNA single stranded so that the ribosome can initiate translation (42). Interestingly, we also found that a gene encoding an L. monocytogenes cold shock protein (cspL) is induced in cells grown at 4°C, consistent with the fact that B. subtilis cspC, which shares 90% aa sequence identity with cspL (57), has also been found to have higher transcript levels at 15°C (32), which may at least be partially due to increased cspC mRNA stability in cells grown at low temperatures (50). In addition to the upregulation of genes facilitating translation at 4°C, we also found that tig, which encodes a putative prolyl isomerase important for protein folding (33), shows higher transcript levels at 4°C, consistent with the observation that an E. coli tig mutant is cold sensitive (52). Thus, genes upregulated in L. monocytogenes cells grown at 4°C include genes encoding proteins important for replication as well as both translation and proper folding of translated proteins.
We also found that a number of genes encoding translation initiation factors and ribosomal proteins showed higher transcript levels in stationary-phase L. monocytogenes cells grown at 4°C. Previous two-dimensional protein gel electrophoresis studies have also found that some ribosomal proteins are induced at low temperatures in B. subtilis (i.e., ribosomal proteins S6 and L7/L12) (31) and in E. coli (i.e., initiation factor IF-2) (47). Translation initiation factors may aid tRNA binding to the ribosomal subunit in E. coli (47), and ribosomal proteins may be important for the correct assembly of rRNA at low temperatures (31). Genome-wide transcriptional profiling of B. subtilis also found that genes encoding ribosomal proteins and translation initiation factors were upregulated in bacteria exposed to cold shock at 15 and 18°C (7, 49), further supporting the importance of ribosomal proteins and translation initiation factors in bacterial cold adaptation.
L. monocytogenes has two major compatible solute transport systems, i.e., carnitine and glycine betaine transporters, that are known to be critical for adaptation at low temperatures (2). In our study, gbuC, encoding a glycine betaine binding protein, and the opuC operon, encoding the carnitine transport system in L. monocytogenes, were induced at 4°C during log-phase growth, consistent with previous evidence supporting a critical role for these osmolyte transporter systems in growth at low temperatures (2). Similarly, transcription of oppA, which encodes an oligopeptide binding protein transporter that is required for L. monocytogenes growth at low temperatures (13), was also upregulated at 4°C in stationary-phase cells. The upregulation of similar oligopeptide and osmolyte transporter genes (e.g., the opuCABCD operon, oppC) has previously been observed in B. subtilis exposed to low temperatures (16, 49), indicating a conserved role for these transporters in cold growth across different gram-positive genera.
Genes with higher transcript levels in L. monocytogenes exposed to low temperatures also included a number of genes with apparent involvement in modifications of membrane fatty acid composition, which may facilitate adjustments of membrane fluidity to allow for growth at low temperatures. Specifically, upregulation of the amino acid biosynthesis operon ilvD-ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD-ilvA may be important at low temperatures as the amino acid isoleucine is a critical precursor for the synthesis of anteiso-branched-chain fatty acids (102), which have a lower melting temperature than iso-branched-chain fatty acids and thus allow L. monocytogenes to maintain membrane fluidity at low temperatures (3, 22, 69, 80, 102). Genes in this operon were also induced in B. subtilis grown at 15°C (33, 49). A critical role for this pathway in adaptation to growth at low temperatures is further supported by findings that anteiso-C15:0 fatty acids are more abundant in the cell membrane of L. monocytogenes cells grown at low temperatures than in that of L. monocytogenes cells grown at 37°C (3). Interestingly, L. monocytogenes lmo0287, which was upregulated at 4°C, encodes a two-component response regulator with 83.5% aa sequence identity to B. subtilis yccF (57). While this two-component response regulator appears to be essential in L. monocytogenes (51, 96) and in B. subtilis (23), as supported by the inability to generate null mutants in the respective gene in either of these two species, a null mutation in the nonessential yccF ortholog in Streptococcus pneumoniae was found to affect the transcription of fatty acid biosynthesis genes and transport-associated genes that alter membrane composition (66), suggesting that this regulator contributes to alterations of cell membrane composition that may facilitate cold growth.
Additional noteworthy genes with higher transcript levels in L. monocytogenes at 4°C include a flagellum-associated operon (consisting of lmo0675 to lmo0689), which showed higher transcript levels in log phase at 4°C, as well as other flagellum-associated genes that showed higher transcript levels in stationary-phase cells at 4°C. This is consistent with previous studies that found the transcription of flaA and the operon consisting of lmo0675 to lmo0689 to be temperature regulated (20, 70). The observation that chemotaxis and motility genes were found to be enriched among genes with higher transcript levels at 4°C likely represents MogR-mediated repression of flagellar gene transcription at elevated temperatures (34) and is consistent with the observation that L. monocytogenes motility is generally observed only in cells grown at temperatures below 37°C (20, 34). Interestingly, while transcriptional analysis of E. coli also revealed that flagellum-associated genes show higher transcript levels in cells exposed to 15°C (73), in B. subtilis, chemotaxis and motility genes appeared to be repressed at low temperatures (16).
Our microarray analyses revealed that codY showed higher transcript levels in stationary-phase cells grown at 4°C than in stationary-phase cells grown at 37°C. codY encodes a pleiotropic transcriptional regulator, which appears to actively repress transcription during log phase at 37°C and appears to have reduced repressing activity as L. monocytogenes enters stationary phase at 37°C (8). CodY represses genes involved in amino acid metabolism, nitrogen assimilation, and sugar uptake, and derepression of the CodY regulon has been found to be important for virulence in L. monocytogenes (8, 67). Interestingly, we found that a number of CodY-repressed genes (8, 67) showed higher transcript levels in log-phase cells grown at 4°C than in log-phase cells grown at 37°C, including a flagellar biosynthesis operon and an amino acid metabolism-associated operon (ilvD-ilvB-ilvN-ilvC-leuA-leuB-leuC-leuD-ilvA), which was found to be CodY dependent in B. subtilis (67). In stationary-phase cells, most of these CodY-repressed genes and operons were no longer differentially expressed at 4°C and 37°C, even though two other operons that have been shown to be repressed by CodY at 37°C (8) showed higher transcript levels in stationary-phase cells at 4°C than in stationary-phase cells at 37°C, despite the fact that codY transcription itself was upregulated. This may be explained by the fact that CodY needs corepressors to repress transcription (85); therefore, CodY-dependent genes may not be repressed even if codY transcription is enhanced. Overall, our data indicate that codY transcription as well as transcription of a number of genes in the CodY regulon may be affected by growth temperature, consistent with the proposed role for CodY in virulence (8), which would suggest temperature-dependent regulation of CodY to appropriately express virulence genes at the host body temperature. Our data indicating that L. monocytogenes CodY appears to contribute to the regulation of genes important for growth at low temperatures are consistent with the observation that B. subtilis CodY regulates a large number of genes that encode proteins important for adaptation to poor growth conditions (67). L. monocytogenes CodY thus appears to contribute to both virulence and stress response (e.g., cold stress), similar to σB (53). While both the CodY and PrfA regulons showed consistent differential expression patterns in log- and stationary-phase cells (with genes in the CodY regulon more likely to show higher transcript levels at 4°C and genes in the PrfA regulon more likely to show lower transcript levels at 4°C), differential transcription of the σB regulon appeared to be affected more by growth phase than by temperature. Genes in the σB regulon were enriched among genes with higher transcript levels in log-phase cells at 4°C and among genes with lower transcript levels in stationary-phase cells at 4°C, suggesting induction of the σB regulon in log-phase cells at 4°C (compared to that in log-phase cells at 37°C), consistent with observations by Becker et al. (6). While genes in the σB regulon seemed to show lower transcript levels in stationary-phase cells at 4°C, the 5′ portion of the sigB operon, which encodes four regulators of σB (RsbS), showed higher transcript levels in stationary-phase cells grown at 4°C, indicating that higher transcript levels for rsb genes does not necessarily result in higher σB activity, consistent with extensive posttranslational regulation of Rsb and σB activities (17). While L. monocytogenes sigB null mutants did not show a cold growth defect in rich media (16a), sigB null mutants showed reduced growth at refrigeration temperatures in minimal media and in meats (6, 68). In B. subtilis, a sigB null mutant showed reduced growth at 15°C (14). A number of B. subtilis genes induced during growth at low temperatures appear to be part of the σB regulon (16, 71, 92). While σB thus appears to contribute to the regulation of gene expression during growth at low temperatures, further studies are needed to more clearly define specific roles for σB and its regulon in L. monocytogenes cold adaptation, including their roles in different growth phases and under different growth conditions.
(iii) L. monocytogenes genes downregulated at 4°C include a number of virulence and virulence-associated genes as well as some heat shock genes.
Stress response genes with lower transcript levels in L. monocytogenes cells grown at 4°C included the groES-groEL operon, consistent with the fact that these genes encode well-documented class I heat shock proteins (24, 37, 40). Interestingly, some L. monocytogenes genes annotated as genes encoding cold shock proteins were also found to be downregulated in cells grown at 4°C, including cspD as well as cspB, a homolog of E. coli cspC (11) and B. subtilis cspD (57). These findings are consistent with observations that E. coli cspC is not cold inducible and is expressed at 37°C (98) and with data suggesting that B. subtilis cspD has a more important role at 37°C (32). Some of the conserved features of cold shock proteins thus may be important for functions required under conditions other than cold growth.
Consistent with the well-documented, temperature-dependent transcription of PrfA-dependent L. monocytogenes virulence genes (45, 58), we also found that many of the PrfA-dependent genes in the prfA virulence gene island, as well as PrfA-dependent genes in other locations (e.g., bsh), showed higher transcript levels at 37°C than at 4°C, generally regardless of growth phase. On the other hand, inlA, inlB, and inlC, which are all coregulated by σB and PrfA (60, 63, 64), showed higher transcript levels at 37°C than at 4°C in stationary-phase cells but not in log-phase cells. Overall, these findings suggest different patterns of temperature-dependent regulation for σB- and PrfA-coregulated internalins, compared to genes regulated only by PrfA, consistent with previous observations of temperature-dependent regulation of PrfA- and σB-dependent genes (64). Finally, relA, which encodes a (p)ppGpp synthetase reported to be involved in the regulation of L. monocytogenes virulence (90), also showed higher transcript levels at 37°C than at 4°C in stationary-phase cells, thus identifying another protein with regulatory function, in addition to PrfA, σB, and CodY, that may be involved in temperature-dependent regulation of L. monocytogenes virulence genes.
Supplementary Material
Acknowledgments
Y. C. Chan and S. Raengpradub were both supported by a USDA National Needs Fellowship grant (to K.J.B.). This work was also supported in part by the National Institutes of Health (award no. RO1-AI052151-01A1 [to K.J.B.]) and the USDA National Research Initiative (project no. 2005-35201-15330 [to K.J.B.]).
We thank Teresa Bergholz for help with enrichment analyses.
Footnotes
Published ahead of print on 24 August 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anderson, K. L., C. Roberts, T. Disz, V. Vonstein, K. Hwang, R. Overbeek, P. D. Olson, S. J. Projan, and P. M. Dunman. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 188:6739-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelidis, A. S., and G. M. Smith. 2003. Role of the glycine betaine and carnitine transporters in adaptation of Listeria monocytogenes to chill stress in defined medium. Appl. Environ. Microbiol. 69:7492-7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annous, B. A., L. A. Becker, D. O. Bayles, D. P. Labeda, and B. J. Wilkinson. 1997. Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63:3887-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett, T., T. O. Suzek, D. B. Troup, S. E. Wilhite, W. C. Ngau, P. Ledoux, D. Rudnev, A. E. Lash, W. Fujibuchi, and R. Edgar. 2005. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 33:D562-D566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayles, D. O., B. A. Annous, and B. J. Wilkinson. 1996. Cold stress proteins induced in Listeria monocytogenes in response to temperature downshock and growth at low temperatures. Appl. Environ. Microbiol. 62:1116-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, L. A., S. N. Evans, R. W. Hutkins, and A. K. Benson. 2000. Role of σB in adaptation of Listeria monocytogenes to growth at low temperature. J. Bacteriol. 182:7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckering, C. L., L. Steil, M. H. Weber, U. Völker, and M. A. Marahiel. 2002. Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis. J. Bacteriol. 184:6395-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett, H. J., D. M. Pearce, S. Glenn, C. M. Taylor, M. Kuhn, A. L. Sonenshein, P. W. Andrew, and I. S. Roberts. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63:1453-1467. [DOI] [PubMed] [Google Scholar]
- 9.Berger, F., N. Morellet, F. Menu, and P. Potier. 1996. Cold shock and cold acclimation proteins in the psychrotrophic bacterium Arthrobacter globiformis SI55. J. Bacteriol. 178:2999-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 11.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 12.Bockmann, R., C. Dickneite, W. Goebel, and J. Bohne. 2000. PrfA mediates specific binding of RNA polymerase of Listeria monocytogenes to PrfA-dependent virulence gene promoters resulting in a transcriptionally active complex. Mol. Microbiol. 36:487-497. [DOI] [PubMed] [Google Scholar]
- 13.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brigulla, M., T. Hoffmann, A. Krisp, A. Völker, E. Bremer, and U. Völker. 2003. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broad Institute of Harvard and MIT. 2006. Listeria monocytogenes Sequencing Project. http://www.broad.mit.edu/annotation/genome/listeria_group/MultiHome.html.
- 16.Budde, I., L. Steil, C. Scharf, U. Volker, and E. Bremer. 2006. Adaptation of Bacillus subtilis to growth at low temperature: a combined transcriptomic and proteomic appraisal. Microbiology 152:831-853. [DOI] [PubMed] [Google Scholar]
- 16a.Chan, Y. C., K. J. Boor, and M. Wiedmann. 2007. σB-Dependent and -independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth. Appl. Environ. Microbiol. 73:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaturongakul, S., and K. J. Boor. 2004. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotter, P. D., C. M. Guinane, and C. Hill. 2002. The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob. Agents Chemother. 46:2784-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Hoon, M. J., Y. Makita, K. Nakai, and S. Miyano. 2005. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 1:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dons, L., O. F. Rasmussen, and J. E. Olsen. 1992. Cloning and characterization of a gene encoding flagellin of Listeria monocytogenes. Mol. Microbiol. 6:2919-2929. [DOI] [PubMed] [Google Scholar]
- 21.Dussurget, O., D. Cabanes, P. Dehoux, M. Lecuit, C. Buchrieser, P. Glaser, and P. Cossart. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45:1095-1106. [DOI] [PubMed] [Google Scholar]
- 22.Edgcomb, M. R., S. Sirimanne, B. J. Wilkinson, P. Drouin, and R. D. Morse. 2000. Electron paramagnetic resonance studies of the membrane fluidity of the foodborne pathogenic psychrotroph Listeria monocytogenes. Biochim. Biophys. Acta 1463:31-42. [DOI] [PubMed] [Google Scholar]
- 23.Fabret, C., and J. A. Hoch. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 180:6375-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gahan, C. G., J. O'Mahony, and C. Hill. 2001. Characterization of the groESL operon in Listeria monocytogenes: utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect. Immun. 69:3924-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao, H., Z. K. Yang, L. Wu, D. K. Thompson, and J. Zhou. 2006. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J. Bacteriol. 188:4560-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerhardt, P. N., L. Tombras Smith, and G. M. Smith. 2000. Osmotic and chill activation of glycine betaine porter II in Listeria monocytogenes membrane vesicles. J. Bacteriol. 182:2544-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 29.Gothel, S. F., C. Scholz, F. X. Schmid, and M. A. Marahiel. 1998. Cyclophilin and trigger factor from Bacillus subtilis catalyze in vitro protein folding and are necessary for viability under starvation conditions. Biochemistry 37:13392-13399. [DOI] [PubMed] [Google Scholar]
- 30.Grau, R., D. Gardiol, G. C. Glikin, and D. de Mendoza. 1994. DNA supercoiling and thermal regulation of unsaturated fatty acid synthesis in Bacillus subtilis. Mol. Microbiol. 11:933-941. [DOI] [PubMed] [Google Scholar]
- 31.Graumann, P., K. Schroder, R. Schmid, and M. A. Marahiel. 1996. Cold shock stress-induced proteins in Bacillus subtilis. J. Bacteriol. 178:4611-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graumann, P., T. M. Wendrich, M. H. Weber, K. Schroder, and M. A. Marahiel. 1997. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 25:741-756. [DOI] [PubMed] [Google Scholar]
- 33.Graumann, P. L., and M. A. Marahiel. 2000. Cold shock response in Bacillus subtilis, p. 27-40. In M. Inouye and K. Yamanaka (ed.), Cold shock response and adaptation. Horizon Scientific Press, Wymondham, England.
- 34.Grundling, A., L. S. Burrack, H. G. A. Bouwer, and D. E. Higgins. 2004. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl. Acad. Sci. USA 101:12318-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guinane, C. M., P. D. Cotter, R. P. Ross, and C. Hill. 2006. Contribution of penicillin-binding protein homologs to antibiotic resistance, cell morphology, and virulence of Listeria monocytogenes EGDe. Antimicrob. Agents Chemother. 50:2824-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han, Y., D. Zhou, X. Pang, L. Zhang, Y. Song, Z. Tong, J. Bao, E. Dai, J. Wang, Z. Guo, J. Zhai, Z. Du, X. Wang, J. Wang, P. Huang, and R. Yang. 2005. DNA microarray analysis of the heat- and cold-shock stimulons in Yersinia pestis. Microbes Infect. 7:335-348. [DOI] [PubMed] [Google Scholar]
- 37.Hanawa, T., T. Yamamoto, and S. Kamiya. 1995. Listeria monocytogenes can grow in macrophages without the aid of proteins induced by environmental stresses. Infect. Immun. 63:4595-4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hebraud, M., E. Dubois, P. Potier, and J. Labadie. 1994. Effect of growth temperatures on the protein levels in a psychrotrophic bacterium, Pseudomonas fragi. J. Bacteriol. 176:4017-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hebraud, M., and J. Guzzo. 2000. The main cold shock protein of Listeria monocytogenes belongs to the family of ferritin-like proteins. FEMS Microbiol. Lett. 190:29-34. [DOI] [PubMed] [Google Scholar]
- 40.Hecker, M., W. Schumann, and U. Völker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 41.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunger, K., C. L. Beckering, F. Wiegeshoff, P. L. Graumann, and M. A. Marahiel. 2006. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J. Bacteriol. 188:240-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.ILSI Research Foundation and Risk Science Institute Expert Panel on Listeria monocytogenes in Foods. 2005. Achieving continuous improvement in reductions in foodborne listeriosis—a risk-based approach. J. Food Prot. 68:1932-1994. [DOI] [PubMed] [Google Scholar]
- 44.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 45.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 46.Jones, P. G., and M. Inouye. 1994. The cold-shock response—a hot topic. Mol. Microbiol. 11:811-818. [DOI] [PubMed] [Google Scholar]
- 47.Jones, P. G., R. A. VanBogelen, and F. C. Neidhardt. 1987. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 169:2092-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Junttila, J. R., S. I. Niemela, and J. Hirn. 1988. Minimum growth temperatures of Listeria monocytogenes and non-haemolytic Listeria. J. Appl. Bacteriol. 65:321-327. [DOI] [PubMed] [Google Scholar]
- 49.Kaan, T., G. Homuth, U. Mader, J. Bandow, and T. Schweder. 2002. Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiology 148:3441-3455. [DOI] [PubMed] [Google Scholar]
- 50.Kaan, T., B. Jurgen, and T. Schweder. 1999. Regulation of the expression of the cold shock proteins CspB and CspC in Bacillus subtilis. Mol. Gen. Genet. 262:351-354. [DOI] [PubMed] [Google Scholar]
- 51.Kallipolitis, B. H., and H. Ingmer. 2001. Listeria monocytogenes response regulators important for stress tolerance and pathogenesis. FEMS Microbiol. Lett. 204:111-115. [DOI] [PubMed] [Google Scholar]
- 52.Kandror, O., and A. L. Goldberg. 1997. Trigger factor is induced upon cold shock and enhances viability of Escherichia coli at low temperatures. Proc. Natl. Acad. Sci. USA 94:4978-4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2006. Contributions of Listeria monocytogenes σB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 152:1827-1838. [DOI] [PubMed] [Google Scholar]
- 55.Ko, R., L. T. Smith, and G. M. Smith. 1994. Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J. Bacteriol. 176:426-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kreft, J., J.-A. Vasquez-Boland, E. Ng, and W. Goebel. 1999. Virulence gene clusters and putative pathogenicity islands in listeriae, p. 219-232. In J. B. Kaper and J. H. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, DC.
- 57.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 58.Leimeister-Wächter, M., E. Domann, and T. Chakraborty. 1992. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 174:947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leimeister-Wächter, M., C. Haffner, E. Domann, W. Goebel, and T. Chakraborty. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 87:8336-8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lingnau, A., E. Domann, M. Hudel, M. Bock, T. Nichterlein, J. Wehland, and T. Chakraborty. 1995. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect. Immun. 63:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu, S., D. O. Bayles, T. M. Mason, and B. J. Wilkinson. 2006. A cold-sensitive Listeria monocytogenes mutant has a transposon insertion in a gene encoding a putative membrane protein and shows altered (p)ppGpp levels. Appl. Environ. Microbiol. 72:3955-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu, S., J. E. Graham, L. Bigelow, P. D. Morse II, and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo, Q., M. Rauch, A. K. Marr, S. Muller-Altrock, and W. Goebel. 2004. In vitro transcription of the Listeria monocytogenes virulence genes inlC and mpl reveals overlapping PrfA-dependent and -independent promoters that are differentially activated by GTP. Mol. Microbiol. 52:39-52. [DOI] [PubMed] [Google Scholar]
- 64.McGann, P., R. Ivanek, M. Wiedmann, and K. J. Boor. 2007. Temperature-dependent expression of Listeria monocytogenes internalin and internalin-like genes suggests functional diversity for these proteins among the listeriae. Appl. Environ. Microbiol. 73:2806-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Milohanic, E., P. Glaser, J.-Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 66.Mohedano, M. L., K. Overweg, A. de la Fuente, M. Reuter, S. Altabe, F. Mulholland, D. de Mendoza, P. Lopez, and J. M. Wells. 2005. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 187:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moorhead, S. M., and G. A. Dykes. 2004. Influence of the sigB gene on the cold stress survival and subsequent recovery of two Listeria monocytogenes serotypes. Int. J. Food Microbiol. 91:63-72. [DOI] [PubMed] [Google Scholar]
- 69.Nichols, D. S., K. A. Presser, J. Olley, T. Ross, and T. A. McMeekin. 2002. Variation of branched-chain fatty acids marks the normal physiological range for growth in Listeria monocytogenes. Appl. Environ. Microbiol. 68:2809-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peel, M., W. Donachie, and A. Shaw. 1988. Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and Western blotting. J. Gen. Microbiol. 134:2171-2178. [DOI] [PubMed] [Google Scholar]
- 71.Petersohn, A., J. Bernhardt, U. Gerth, D. Hoper, T. Koburger, U. Volker, and M. Hecker. 1999. Identification of σB-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 181:5718-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pfennig, P. L., and A. M. Flower. 2001. BipA is required for growth of Escherichia coli K12 at low temperature. Mol. Genet. Genomics 266:313-317. [DOI] [PubMed] [Google Scholar]
- 73.Phadtare, S., and M. Inouye. 2004. Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J. Bacteriol. 186:7007-7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phan-Thanh, L., and T. Gormon. 1995. Analysis of heat and cold shock proteins in Listeria by two-dimensional electrophoresis. Electrophoresis 16:444-450. [DOI] [PubMed] [Google Scholar]
- 75.Piuri, M., C. Sanchez-Rivas, and S. M. Ruzal. 2005. Cell wall modifications during osmotic stress in Lactobacillus casei. J. Appl. Microbiol. 98:84-95. [DOI] [PubMed] [Google Scholar]
- 76.Polissi, A., W. De Laurentis, S. Zangrossi, F. Briani, V. Longhi, G. Pesole, and G. Deho. 2003. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154:573-580. [DOI] [PubMed] [Google Scholar]
- 77.Rea, R. B., C. G. Gahan, and C. Hill. 2004. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect. Immun. 72:717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Renzoni, A., P. Cossart, and S. Dramsi. 1999. PrfA, the transcriptional activator of virulence genes, is upregulated during interaction of Listeria monocytogenes with mammalian cells and in eukaryotic cell extracts. Mol. Microbiol. 34:552-561. [DOI] [PubMed] [Google Scholar]
- 79.Roberts, A. J., and M. Wiedmann. 2003. Pathogen, host and environmental factors contributing to the pathogenesis of listeriosis. Cell. Mol. Life Sci. 60:904-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Russell, N. J. 1990. Cold adaptation of microorganisms. Philos. Trans. R. Soc. Lond. B 326:595-611. [DOI] [PubMed] [Google Scholar]
- 81.Schäferkordt, S., and T. Chakraborty. 1997. Identification, cloning, and characterization of the lma operon, whose gene products are unique to Listeria monocytogenes. J. Bacteriol. 179:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schwab, U., B. Bowen, C. Nadon, M. Wiedmann, and K. J. Boor. 2005. The Listeria monocytogenes prfAP2 promoter is regulated by sigma B in a growth phase dependent manner. FEMS Microbiol. Lett. 245:329-336. [DOI] [PubMed] [Google Scholar]
- 83.Smyth, G. K. 2005. LIMMA: linear models for microarray data, p. 397-420. In R. Gentleman, V. Carey, S. Dodoit, R. Irizarry, and W. Huber (ed.), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY.
- 84.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods 31:265-273. [DOI] [PubMed] [Google Scholar]
- 85.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203-207. [DOI] [PubMed] [Google Scholar]
- 86.Sprenger, G. A. 1995. Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch. Microbiol. 164:324-330. [DOI] [PubMed] [Google Scholar]
- 87.Subramanian, A., P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, and J. P. Mesirov. 2005. Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102:15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sue, D., D. Fink, M. Wiedmann, and K. J. Boor. 2004. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150:3843-3855. [DOI] [PubMed] [Google Scholar]
- 89.Susin, M. F., H. R. Perez, R. L. Baldini, and S. L. Gomes. 2004. Functional and structural analysis of HrcA repressor protein from Caulobacter crescentus. J. Bacteriol. 186:6759-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Taylor, C. M., M. Beresford, H. A. Epton, D. C. Sigee, G. Shama, P. W. Andrew, and I. S. Roberts. 2002. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J. Bacteriol. 184:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Turner, A.-M. W., C. F. Love, R. W. Alexander, and P. G. Jones. 2007. Mutational analysis of Escherichia coli DEAD box protein CsdA. J. Bacteriol. 189:2769-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Walker, S. J., and M. F. Stringer. 1987. Growth of Listeria monocytogenes and Aeromonas hydrophila at chill temperatures. J. Appl. Bacteriol. 63:R20. [Google Scholar]
- 94.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wiegeshoff, F., C. L. Beckering, M. Debarbouille, and M. A. Marahiel. 2006. Sigma L is important for cold shock adaptation of Bacillus subtilis. J. Bacteriol. 188:3130-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Williams, T., S. Bauer, D. Beier, and M. Kuhn. 2005. Construction and characterization of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect. Immun. 73:3152-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamanaka, K. 2000. Cold shock response in Escherichia coli, p. 6-26. In M. Inouye and K. Yamanaka (ed.), Cold shock response and adaptation. Horizon Scientific Press, Wymondham, England.
- 99.Yamanaka, K., L. Fang, and M. Inouye. 1998. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Microbiol. 27:247-255. [DOI] [PubMed] [Google Scholar]
- 100.Zawadzka-Skomial, J., Z. Markiewicz, M. Nguyen-Distèche, B. Devreese, J.-M. Frère, and M. Terrak. 2006. Characterization of the bifunctional glycosyltransferase/acyltransferase penicillin-binding protein 4 of Listeria monocytogenes. J. Bacteriol. 188:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng, W., and S. Kathariou. 1994. Transposon-induced mutants of Listeria monocytogenes incapable of growth at low temperature (4°C). FEMS Microbiol. Lett. 121:287-291. [DOI] [PubMed] [Google Scholar]
- 102.Zhu, K., D. O. Bayles, A. Xiong, R. K. Jayaswal, and B. J. Wilkinson. 2005. Precursor and temperature modulation of fatty acid composition and growth of Listeria monocytogenes cold-sensitive mutants with transposon-interrupted branched-chain α-keto acid dehydrogenase. Microbiology 151:615-623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.