Abstract
Wood-grown cultures of Daldinia concentrica oxidized a permethylated β-14C-labeled synthetic lignin to 14CO2 and also cleaved a permethylated α-13C-labeled synthetic lignin to give Cα-Cβ cleavage products that were detected by 13C nuclear magnetic resonance spectrometry. Therefore, this ascomycete resembles white-rot basidiomycetes in attacking the recalcitrant nonphenolic structures that predominate in lignin.
The degradation of lignocellulose by ascomycetes, a process generally called soft rot, is an important route for carbon cycling in plant litter and soils (2). Little is known about the decay mechanisms of these fungi, in contrast to those of the better-studied lignocellulolytic basidiomycetes that degrade wood. Nevertheless, it is clear that some soft-rot fungi can degrade lignin, because they erode the secondary cell wall and decrease the content of acid-insoluble material (Klason lignin) in angiosperm wood (13, 21). Some ascomycetes have also been shown to mineralize radiolabeled synthetic lignins (known as dehydrogenative polymerizates [DHPs]), but the low extents of 14CO2 production reported—invariably less than 10% of the lignin—indicate that the ligninolytic capabilities of ascomycetes are more limited than those of white-rot basidiomycetes (11, 17).
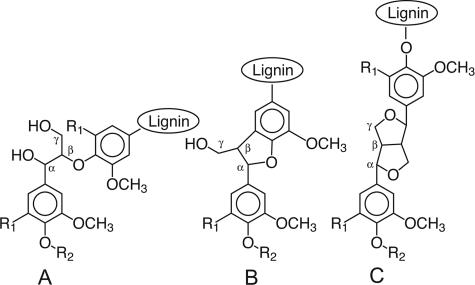
One possibility is that ascomycetes attack only the phenolic units in lignin, which comprise roughly 10% of the polymer and are chemically more labile than the ether-linked, nonphenolic units that make up the remainder (Fig. 1). Phenolic lignin structures are oxidized and directly cleaved by a variety of mild biological oxidants such as manganic chelates and phenol oxidases (7, 20). By contrast, the cleavage of nonphenolic lignin structures requires the action of stronger oxidants, including oxyradicals produced by the action of peroxidases or laccases on various redox mediators. The production of these more vigorous ligninolytic systems is typical of white-rot basidiomycetes (5, 12) and has not yet been convincingly shown in soft-rot ascomycetes. However, the production of phenol oxidases that could act directly on phenolic lignin is widespread among ascomycetes (11, 14, 19).
FIG. 1.
Principal structures of natural lignins and DHPs. (A) Arylglycerol-β-aryl ether structures. (B) Phenylcoumaran structures. (C) Resinol structures. R1 is H for guaiacyl structures and OCH3 for syringyl structures. R2 is H for phenolic structures, CH3 for permethylated phenolic structures, and lignin for nonphenolic structures.
Mineralization of DHPs.
To address this question, we compared the extents to which Daldinia concentrica mineralized phenolic and nonphenolic DHPs. D. concentrica is a xylariaceous ascomycete that produces phenol oxidase activity (14) and degrades angiosperm wood extensively (13). Because chemical analyses of wood degraded by this fungus have shown that it degrades syringyl lignin units faster than guaiacyl ones (Fig. 1) (13), we performed these experiments with a syringyl/guaiacyl DHP (i.e., a polymeric model of angiosperm lignin) that was 14C labeled at Cβ on the side chains of its syringyl units. The radiolabeled DHP, prepared as described earlier (4, 9), had a specific activity of 0.01 mCi/mmol of syringyl subunits and a syringyl/guaiacyl ratio of approximately 4:1. A portion of this phenolic syringyl/guaiacyl DHP was then permethylated with diazomethane as described earlier (8) to obtain a nonphenolic syringyl/guaiacyl DHP, i.e., a polymer in which all of the phenolic hydroxyl groups had been blocked as methyl ethers. In addition, a phenolic guaiacyl DHP (i.e., a polymeric model of gymnosperm lignin), prepared previously with 14C at Cβ of its side chains (0.01 mCi/mmol of guaiacyl subunits) (4), was included in the experiment for comparison.
Each DHP was fractionated by gel permeation chromatography (GPC) on Sephadex LH-20 in N,N-dimethylformamide, and the excluded fractions were taken to eliminate low-molecular-mass material (≤∼1 kDa) that might have been susceptible to uptake and intracellular metabolism. This step is essential in microbiological experiments with DHPs because ligninolysis is strictly an extracellular process (9). The excluded fractions were then subjected again to GPC on a 1.8- by 33-cm column of Sephacryl S-100 in N,N-dimethylformamide (15), and fractions eluting within the sieving range of the column were pooled so that the phenolic and nonphenolic DHPs had similar molecular mass distributions. We included this extra purification step in case the size of a DHP might influence its bioavailability. Figure 2 shows the molecular mass distributions of the three purified DHPs as determined by analytical GPC on Sephacryl S-100 in N,N-dimethylformamide-0.1 M LiCl (15).
FIG. 2.
Gel permeation chromatography of the radiolabeled DHPs used in mineralization experiments. The distributions shown, with their weight average molecular masses (Mw) and number average molecular masses (Mn), are as follows: phenolic syringyl/guaiacyl DHP (solid line), Mw = 33 kDa, Mn = 16 kDa; nonphenolic syringyl/guaiacyl DHP (long-dash line), Mw = 25 kDa Mn = 14 kDa; phenolic guaiacyl DHP (short-dash line), Mw = 21 kDa, Mn = 5 kDa. Elution positions and masses of standards are shown for 35-kDa, 15-kDa, and 0.5-kDa polystyrenes; a 1.1-kDa lignin model tetramer (15); and 3,4-dimethoxybenzaldehyde (0.17 kDa). Vo indicates the excluded volume of the column.
The sized DHPs were dissolved in acetone-water (4:1), and 80,000 dpm of each was added to the end grain of replicate sterile aspen wafers (approximately 300 mg each, 3 by 10 by 30 mm, with the large face perpendicular to the grain). Additional sterile water was added to each block to bring the final volume of added water to 300 μl, and the acetone was allowed to evaporate. The wafers were then placed on spacers over potato dextrose agar in 125-ml Erlenmeyer flasks that already contained a lawn of D. concentrica (FP-140074-sp; Center for Forest Mycology, USDA Forest Products Laboratory). This medium was chosen because lignocellulose decay by ascomycetes is generally stimulated when auxiliary sources of carbon and nitrogen are present (21). An agar plug of the fungus was placed atop each wafer, and the flasks were stoppered with gassing manifolds. The cultures were incubated at 26°C and flushed with humidified, sterile air at 2- or 3-day intervals. The headspace gas from each flask was sparged through an alkaline scintillation cocktail to trap 14CO2 evolved from the DHPs, and the amount of radiocarbon in each sample was then determined by scintillation counting (10).
We grew these cultures on wood wafers because weight loss in the substrate provides an independent test of whether the fungus has expressed its entire lignocellulolytic system. The extensive loss that occurred over the 12-week experiment—55% on average—leaves no doubt that this was the case. The mineralization data (Table 1) show that the phenolic syringyl/guaiacyl DHP was degraded more extensively than the phenolic guaiacyl DHP, in agreement with previous analyses of the lignin from wood degraded by D. concentrica (13). Nevertheless, the results establish that this fungus can degrade guaiacyl lignin structures extracellularly, because the proportion of 14C with a molecular mass less than 1 kDa in our guaiacyl DHP was only about 2% (Fig. 2). Most interestingly, the data also show that blockage of the phenolic groups in the syringyl/guaiacyl DHP had no significant effect on its biodegradation. Therefore, contrary to our initial hypothesis, attack on nonphenolic lignin structures is a significant route for ligninolysis by D. concentrica.
TABLE 1.
Mineralization of β-14C-labeled synthetic lignins by D. concentrica on wood
| Polymer added | Amt mineralized ± SD (%) at:
|
Wt loss in wood ± SD (%) at 12 wk | ||
|---|---|---|---|---|
| 4 wk | 8 wk | 12 wk | ||
| Nonphenolic syringyl/guaiacyl DHP (n = 6) | 6.2 ± 2.7 | 10.4 ± 4.1 | 11.3 ± 4.4 | 56.1 ± 10.1 |
| Phenolic syringyl/guaiacyl DHP (n = 5) | 7.5 ± 2.4 | 12.3 ± 3.9 | 13.3 ± 4.1 | 51.4 ± 10.2 |
| Phenolic guaiacyl DHP (n = 5) | 2.1 ± 0.6 | 4.0 ± 1.1 | 4.6 ± 1.2 | 58.4 ± 6.4 |
Identification of cleavage products in a nonphenolic DHP.
To address the question of how D. concentrica cleaves nonphenolic lignin structures, we supplemented aspen wafers with a nonphenolic DHP enriched with 13C at Cα of its propyl side chain, extracted this DHP from the wood after decay, and then identified chemical changes at Cα by 13C nuclear magnetic resonance (NMR) spectrometry. The [α-13C]DHP was a guaiacyl polymer prepared earlier (4) with a 13C content greater than 99% at Cα. It was first permethylated to block its phenolic structures and then fractionated on Sephadex LH-20 as described above to provide a high-molecular-mass fraction. The 13C NMR spectrum of this fraction was obtained, after which two sterile aspen wafers (dry weights: 334 mg and 350 mg) were each infused with 9 mg of it in acetone-water and then inoculated with D. concentrica as described above. At the same time, five replicate cultures were set up on aspen wafers (average dry weight: 385 ± 22 mg) without [α-13C]DHP so that the contribution of endogenous lignin to the NMR spectra could be assessed. The cultures were covered with aluminum foil and incubated at 26°C and 70% relative humidity for 7 weeks.
At the conclusion of the experiment, the two supplemented wafers had a combined dry weight of 464 mg and showed weight losses of 31% and 33%, compared with a mean value of 46% ± 3% for the five control wafers without [α-13C]DHP. Apparently, the presence of DHP inhibited wood decay somewhat, but it is evident that a complete biodegradative system was nevertheless expressed in supplemented wafers. Two wafers, weighing a total of 461 mg, were chosen from the decayed set lacking [α-13C]DHP, and each pair was then ball milled. The milled wood samples were treated with a crude cellulase mixture (Cellulysin; Calbiochem, San Diego, CA) for 48 h thrice as described elsewhere (16), after which the lignin-enriched fraction was extracted for 24 h thrice with dioxane-water (9:1) to solubilize as much lignin as possible. The two wafers originally supplemented with [α-13C]DHP yielded 97 mg of solubles, whereas the two unsupplemented wafers yielded 82 mg.
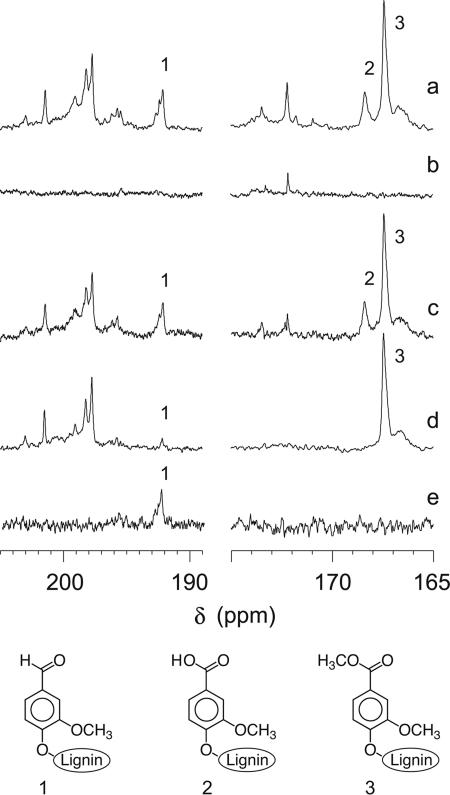
Qualitative 13C NMR analyses of these samples were obtained, and the normalized spectra (Fig. 3) were inspected for signals indicative of ligninolysis. No significant changes were discernible in the alcohol and ether range (70 to 90 ppm), corresponding to Cα in major structures (Fig. 1) of the undegraded DHP, but new structures were apparent in the carbonyl (190 to 205 ppm and 165 to 175 ppm) regions. Figure 3a shows the spectrum in these regions for the [α-13C]DHP that was extracted from degraded wood, and Fig. 3b shows the spectrum in the same regions for the extract from degraded wood that had received no [α-13C]DHP. Figure 3c shows the difference between these two spectra and is thus a spectrum of degraded [α-13C]DHP that has been corrected for the small contribution made by material originating from the wood. To see the changes that D. concentrica caused in the nonphenolic [α-13C]DHP, the difference spectrum in Fig. 3c should be compared with Fig. 3d, which shows the spectrum of the original, undegraded, permethylated polymer.
FIG. 3.
13C NMR results showing (a) a spectrum of the extract from wood decayed by D. concentrica in the presence of permethylated guaiacyl [α-13C]DHP, (b) a spectrum of the extract from decayed wood that received no DHP, (c) the difference between spectra a and b, (d) a spectrum of undegraded, permethylated, guaiacyl [α-13C]DHP, and (e) a DEPT spectrum of the extract from wood decayed in the presence of permethylated guaiacyl [α-13C]DHP. Spectra a and b were normalized with respect to the amount of wood lignin present by equalizing the signal from C-2/C-6 in syringyl aromatic carbons (guaiacyl DHP does not contribute in this region). Spectra a and d were normalized with respect to the amount of DHP present by equalizing the signal from Cα in resinol structures (this carbon was present at a very low abundance in the wood lignin). The samples were dissolved in 400 μl of acetone-d6-D2O (4:1), and the spectra were obtained with a Bruker DPX 250-MHz spectrometer (62.9 MHz for 13C). The qualitative experiments shown in spectra a to d were acquired with a standard power-gated sequence, a 0.5-s delay, and 150,000 scans. Quantitative experiments (not shown) were performed with an inverse-gated sequence, a 15-s delay, a 90° pulse, and 25,000 scans. For spectrum e, the standard Bruker DEPT135 sequence was run for 30,000 scans. In all cases, the acetone signal at 29.8 ppm was used as the internal reference. Structures corresponding to signal assignments 1 to 3 are shown below the spectra. The assignments were confirmed by measuring carbonyl chemical shifts (δ) for 3,4-dimethoxybenzaldehyde, 3,4-dimethoxybenzoic acid, and 3,4-dimethoxybenzoate methyl ester. The new signals at 172 to 174 ppm in spectrum c may represent aliphatic carboxyl groups resulting from Cα-aryl cleavage of the DHP (1).
In [α-13C]DHP recovered from degraded wood, a signal in the region characteristic of benzaldehyde carbonyls was apparent (Fig. 3a and 3c, signal 1) and was confirmed to be from a protonated (i.e., aldehyde as opposed to ketone) carbonyl in a distortionless enhancement with polarization transfer (DEPT) experiment (18) (Fig. 3e). This benzaldehyde signal was absent from the lignin extracted from degraded wood that had received no [α-13C]DHP (Fig. 3b) and therefore must have originated from the DHP. Benzaldehyde residues were also present in the original, undegraded [α-13C]DHP (Fig. 3d), as also found in earlier research on another DHP (3), but they occurred at a much higher level in the degraded sample.
Similarly, the [α-13C]DHP from degraded wood exhibited benzoic acid carboxyl signals (Fig. 3a and c, signal 2) that were detectable neither in the lignin from wood without [α-13C]DHP (Fig. 3b) nor in the original undegraded [α-13C]DHP (Fig. 3d). The undegraded [α-13C]DHP did originally contain benzoic acid residues, in agreement with earlier work (3), but they all appear in our spectra as methyl benzoates (Fig. 3d, signal 3) because the [α-13C]DHP had been permethylated before the experiment. Since these preexisting methyl benzoates were unaffected by fungal decay (Fig. 3a and c) they cannot have been the source of the new benzoic acid residues.
There are two possible sources for new benzaldehydes and benzoic acids in a biodegraded lignin. First, if the polymer originally contained primary benzyl alcohols, these might simply become oxidized at Cα. Such reactions would not be ligninolytic, but we can rule them out as an explanation for our data because a two-dimensional 1H-13C heteronuclear spin quantum correlation (HSQC) NMR spectrum of the original permethylated [α-13C]DHP showed that it lacked the 1Hα-13Cα cross-peak at a δ of 4.6, 65 ppm, that is expected for primary benzyl alcohols (data not shown; for typical NMR chemical shifts of lignin-related structures see the U.S. Dairy Forage Research Center NMR Database of Lignin and Cell Wall Model Compounds at http://ars.usda.gov/Services/docs.htm?docid=10491). This result leaves the second explanation, which is that the new benzaldehydes and benzoic acids arose via biodegradative Cα-Cβ cleavage of propyl side chains in the [α-13C]DHP. A quantitative 13C NMR spectrum (data not shown) showed that new benzoic acid residues were roughly 10-fold more abundant than new benzaldehyde residues in the degraded [α-13C]DHP and that approximately 1% of the residual polymer had been cleaved between Cα and Cβ.
Conclusion.
Our results show that D. concentrica degrades the recalcitrant nonphenolic structures in lignin and that Cα-Cβ cleavage of the lignin propyl side chain is one of the processes responsible. The ligninolytic agents that D. concentrica produces remain unidentified because no method has yet been found to elicit their production on any medium except wood. However, it is reasonable to predict that they resemble some of the extracellular one-electron oxidants produced by white-rot basidiomycetes, because the latter fungi and their isolated ligninolytic systems also cleave nonphenolic lignin structures between Cα and Cβ (1, 4). The relatively low ability of ascomycetes to depolymerize guaiacyl lignin structures has led researchers to suggest that they produce weaker ligninolytic oxidants (13), but this inference is contradicted by the observation that guaiacyl lignin structures are actually slightly easier to oxidize than syringyl structures (6). We consider it more likely that the ascomycetes produce a relatively small quantity of ligninolytic oxidants and consequently fail to overcome the tendency that guaiacyl lignin fragments have to repolymerize during one-electron oxidation (4).
Acknowledgments
We thank Carl Houtman for calculating the DHP molecular mass distributions, Hoon Kim for milling the wood samples, and John Ralph for advice on sample preparation for NMR analysis.
This study was supported by U.S. Department of Energy grants DE-FG02-94ER20140 and DE-AI02-07ER64480 to K.E.H.
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Chua, M. G. S., C.-L. Chen, H.-M. Chang, and T. K. Kirk. 1982. 13C NMR spectroscopic study of spruce lignin degraded by Phanerochaete chrysosporium. 1. New structures. Holzforschung 36:165-172. [Google Scholar]
- 2.Dix, N. J., and J. Webster. 1995. Fungal ecology. Chapman and Hall, London, United Kingdom.
- 3.Gagnaire, D., and D. Robert. 1977. A polymer model of lignin (DHP) 13C selectively labeled at benzylic positions—synthesis and NMR study. Makromol. Chem. 178:1477-1495. [Google Scholar]
- 4.Hammel, K. E., K. A. Jensen, M. D. Mozuch, L. L. Landucci, M. Tien, and E. A. Pease. 1993. Ligninolysis by a purified lignin peroxidase. J. Biol. Chem. 268:12274-12281. [PubMed] [Google Scholar]
- 5.Hammel, K. E., A. N. Kapich, K. A. Jensen, and Z. C. Ryan. 2002. Reactive oxygen species as agents of wood decay by fungi. Enzyme Microb. Technol. 30:445-453. [Google Scholar]
- 6.Hammel, K. E., P. J. Tardone, M. A. Moen, and L. A. Price. 1989. Biomimetic oxidation of nonphenolic lignin models by Mn(III). New observations on the oxidizability of guaiacyl and syringyl substructures. Arch. Biochem. Biophys. 270:404-409. [DOI] [PubMed] [Google Scholar]
- 7.Kawai, S., T. Umezawa, and T. Higuchi. 1988. Degradation mechanisms of phenolic β-1 lignin substructure model compounds by laccase of Coriolus versicolor. Arch. Biochem. Biophys. 262:99-110. [DOI] [PubMed] [Google Scholar]
- 8.Kirk, T. K., and E. Adler. 1970. Methoxyl-deficient structural elements in lignin of sweetgum decayed by a brown-rot fungus. Acta Chem. Scand. 24:3379-3390. [Google Scholar]
- 9.Kirk, T. K., and G. Brunow. 1988. Synthetic 14C-labeled lignins. Methods Enzymol. 161:65-73. [Google Scholar]
- 10.Kirk, T. K., W. J. Connors, R. D. Bleam, W. F. Hackett, and J. G. Zeikus. 1975. Preparation and microbial decomposition of synthetic [14C]lignins. Proc. Natl. Acad. Sci. USA 72:2515-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liers, C., R. Ullrich, K. T. Steffen, A. Hatakka, and M. Hofrichter. 2006. Mineralization of 14C-labelled synthetic lignin and extracellular enzyme activities of the wood-colonizing ascomycetes Xylaria hypoxylon and Xylaria polymorpha. Appl. Microbiol. Biotechnol. 69:573-579. [DOI] [PubMed] [Google Scholar]
- 12.Martínez, A. T. 2002. Molecular biology and structure-function of lignin-degrading heme peroxidases. Enzyme Microb. Technol. 30:425-444. [Google Scholar]
- 13.Nilsson, T., G. Daniel, T. K. Kirk, and J. R. Obst. 1989. Chemistry and microscopy of wood decay by some higher ascomycetes. Holzforschung 43:11-18. [Google Scholar]
- 14.Nsolomo, V. R., K. Venn, and H. Solheim. 2000. The ability of some fungi to cause decay in the East African camphor tree, Ocotea usambarensis. Mycol. Res. 104:1473-1479. [Google Scholar]
- 15.Ortiz-Bermúdez, P., E. Srebotnik, and K. E. Hammel. 2003. Chlorination and cleavage of lignin structures by fungal chloroperoxidases. Appl. Environ. Microbiol. 69:5015-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ralph, J., T. Akiyama, H. Kim, F. C. Lu, P. F. Schatz, J. M. Marita, S. A. Ralph, M. S. S. Reddy, F. Chen, and R. A. Dixon. 2006. Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J. Biol. Chem. 281:8843-8853. [DOI] [PubMed] [Google Scholar]
- 17.Regalado, V., A. Rodríguez, F. Perestelo, A. Carnicero, G. de la Fuente, and M. A. Falcón. 1997. Lignin degradation and modification by the soil-inhabiting fungus Fusarium proliferatum. Appl. Environ. Microbiol. 63:3716-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders, J. K. M., and B. K. Hunter. 1987. Modern NMR spectroscopy—a guide for chemists. Oxford University Press, Oxford, United Kingdom.
- 19.Torres y Torres, J. L., and J. P. N. Rosazza. 2001. Microbial transformations of p-coumaric acid by Bacillus megaterium and Curvularia lunata. J. Nat. Prod. 64:1408-1414. [DOI] [PubMed] [Google Scholar]
- 20.Tuor, U., H. Wariishi, H. E. Schoemaker, and M. H. Gold. 1992. Oxidation of phenolic arylglycerol β-aryl ether lignin model compounds by manganese peroxidase from Phanerochaete chrysosporium. Oxidative cleavage of an α-carbonyl model compound. Biochemistry 31:4986-4995. [DOI] [PubMed] [Google Scholar]
- 21.Worrall, J. J., S. E. Anagnost, and R. A. Zabel. 1997. Comparison of wood decay among diverse lignicolous fungi. Mycologia 89:199-219. [Google Scholar]