Abstract
Plasmid pGNB1 was isolated from bacteria residing in the activated sludge compartment of a wastewater treatment plant by using a transformation-based approach. This 60-kb plasmid confers resistance to the triphenylmethane dye crystal violet and enables its host bacterium to decolorize crystal violet. Partial sequencing of pGNB1 revealed that its backbone is very similar to that of previously sequenced IncP-1β plasmids. The two accessory regions of the plasmid, one located downstream of the replication initiation gene trfA and the other located between the conjugative transfer modules Tra and Trb, were completely sequenced. Accessory region L1 contains a transposon related to Tn5501 and a gene encoding a Cupin 2 conserved barrel protein with an unknown function. The triphenylmethane reductase gene tmr and a truncated dihydrolipoamide dehydrogenase gene that is flanked by IS1071 and another putative insertion element were identified in accessory region L2. Subcloning of the pGNB1 tmr gene demonstrated that this gene is responsible for the observed crystal violet resistance phenotype and mediates decolorization of the triphenylmethane dyes crystal violet, malachite green, and basic fuchsin. Plasmid pGNB1 and the associated phenotype are transferable to the α-proteobacterium Sinorhizobium meliloti and the γ-proteobacterium Escherichia coli. This is the first report of a promiscuous IncP-1β plasmid isolated from the bacterial community from a wastewater treatment plant that harbors a triphenylmethane reductase gene. The pGNB1-encoded enzyme activity is discussed with respect to bioremediation of sewage polluted with triphenylmethane dyes.
Bacteria living in wastewater habitats have to adapt rapidly to changing conditions depending on the pollutant composition of the sewage. The horizontally mobile gene pool of bacteria has been recognized to be very important for adaptive responses to selective pressures caused by diverse chemical compounds (6, 11, 43, 45). In particular, mobile broad-host-range plasmids significantly contribute to the dissemination of beneficial genetic traits among the population of a certain species and even beyond species boundaries. In recent years several promiscuous broad-host-range plasmids belonging to the IncP-1 group and carrying different accessory modules conferring resistance and degradation capabilities were identified in different hosts (7, 35). Among these, plasmids encoding resistance to nearly all clinically relevant antimicrobial drug classes were isolated from bacterial populations residing in wastewater treatment plants (15, 33, 34, 39, 41, 42). Other IncP-1 plasmids harbor genes for the degradation of different pollutants, such as the xenobiotics atrazine, 2-chlorobenzoate, 3-chlorobenzoate, 2,5-dichlorobenzoate, 2,4-dichlorophenoxyacetic acid, p-toluenesulfonate, and haloacetates (18, 20, 37, 46-48). Many more catabolic gene regions were identified on plasmids belonging to other Inc groups (7, 8, 28). Catabolic gene clusters on plasmids have frequently been found to be associated with mobile genetic elements, e.g., insertion sequences (IS elements) and transposons, which suggests that these elements play an important role in the distribution of catabolic capacities (27, 38, 40, 45).
Triphenylmethane dyes, such as malachite green, crystal violet, and basic fuchsin, are widely used in the textile, pharmaceutical, food, and cosmetic industries, in paper printing, and in photochemical processes (29). Considerable amounts of these toxic and mutagenic dyes are discharged into wastewater treatment facilities and thus impose a selective pressure on the microbial flora residing in wastewater habitats. In recent years several microorganisms capable of dye decolorization were identified. Among these, an Aeromonas hydrophila strain possessing a broad-spectrum dye decolorization activity was described (30). Likewise, different Mycobacterium species are able to tolerate high concentrations of triphenylmethane dyes (19). Jang et al. (17) described a biochemical analysis of a triphenylmethane reductase purified from Citrobacter sp. strain KCTC. This enzyme catalyzes the NADH-dependent reduction and thus decolorization of certain triphenylmethane dyes.
Here we report the isolation of a conjugative broad-host-range IncP-1β plasmid conferring resistance to and decolorization of triphenylmethane dyes from the bacterial community of a municipal wastewater treatment plant. This plasmid was characterized at the genomic and functional levels.
MATERIALS AND METHODS
Isolation of plasmid pGNB1 from an activated sludge bacterial community.
Activated sludge samples were taken from an activated sludge basin of the Bielefeld-Heepen wastewater treatment plant in Germany in October 2004. Plasmids were isolated from activated sludge bacteria as described previously (2). Briefly, activated sludge bacteria were cultivated on Luria broth (LB) agar containing crystal violet (10 μg ml−1) for 40 h at 30°C. Growing bacteria were collected and washed with 50 ml of an 0.85% NaCl solution, and total plasmid DNA was prepared using a Nucleobond PC100 kit and AX100 columns according to the protocol supplied by the manufacturer (Macherey-Nagel, Düren, Germany). Plasmid DNA preparations were subsequently used to transform CaCl2-competent Escherichia coli KAM3 (25) cells. Transformants were selected on LB agar supplemented with crystal violet (10 μg ml−1).
Bacterial strains and growth conditions.
E. coli strains KAM3 (25), DH5α (14), XL1-Blue (4), and CSH52 (24) containing plasmid pGNB1 were grown in LB supplemented with crystal violet (10 μg ml−1) at 37°C.
Standard DNA techniques.
Plasmid pGNB1 is a self-transmissible plasmid containing the complete genetic information for conjugal plasmid transfer and thus could be transferred to other E. coli strains and Sinorhizobium meliloti by conjugation. The plasmid contents of E. coli and S. meliloti strains harboring plasmid pGNB1 were visualized in an Eckhardt gel as described by Hynes et al. (16). Plasmid pGNB1 was isolated from E. coli strains with a Nucleobond PC100 kit (Macherey-Nagel, Düren, Germany), whereas recombinant plasmids containing subcloned pGNB1 restriction fragments were isolated by using a QIAprep Spin miniprep kit (QIAGEN, Hilden, Germany) according to the protocol supplied by the manufacturer. Restriction enzyme digestion, agarose gel electrophoresis, DNA cloning, and transformation of E. coli were carried out as described by Sambrook et al. (32).
Restriction fragment library construction, sequencing, and sequence analysis of the pGNB1 accessory regions.
Purified pGNB1 plasmid DNA was restricted with restriction endonucleases BamHI, EcoRI, PstI, SalI, SphI, and NotI. The restriction fragments obtained were separately subcloned into the sequencing vectors pUC18 (50), pBluescript-II KS (Stratagene, La Jolla, CA), and pZErO-2 (Invitrogen). Sequencing of recombinant plasmids was done by IIT Biotech GmbH (Bielefeld, Germany) as previously described (39). Computer-assisted assembly and sequence quality control were carried out with the CONSED/AUTOFINISH software tool (12, 13). For annotation of the finished sequence, the annotation tool GenDB (version 2.2) was used (23).
Decolorization of triphenylmethane dyes mediated by plasmid pGNB1.
To test for decolorization of triphenylmethane dyes mediated by plasmid pGNB1, 10-μl portions of E. coli strains KAM3 and DH5α with and without plasmid pGNB1 grown to the logarithmic phase (optical density at 580 nm, about 0.5) were applied to LB agar plates containing 15 μg ml−1 crystal violet, malachite green, or basic fuchsin and grown overnight at 37°C. Likewise, 10-μl portions of S. meliloti(pGNB1) and wild-type S. meliloti were applied to TY agar plates (1) containing 15 μg ml−1 crystal violet, malachite green, or basic fuchsin and grown overnight at 30°C. Decolorization of the dyes was visible as a clear halo surrounding a colony. MICs of crystal violet were determined by growing strains in LB in the presence of 5, 10, 15, 20, 30, 50, 65, 70, 80, 90, 100, 110, and 120 μg ml−1 crystal violet.
Antimicrobial susceptibility testing.
Disk diffusion tests to determine antimicrobial susceptibility were carried out using the general experimental conditions recommended in the guidelines of the Clinical and Laboratory Standards Institute (5) and by the manufacturer of the disks (Oxoid GmbH, Wesel, Germany). The following disks purchased from Oxoid were used: amikacin (AK30), ampicillin (AMP25), azithromycin (AZM15), aztreonam (ATM30), bacitracin (B10), cefaclor (CEC30), cefepim (FEP30), cefotaxim (CTX30), cefpirom (CPO30), ceftazidim (CAZ30), ceftibuten (CFT30), cefuroxim (CXM30), chloramphenicol (C10), ciprofloxacin (CIP5), clarithromycin (CLR15), clindamycin (DA10), colistin sulfate (CT10), erythromycin (E30), gentamicin (CN10), kanamycin (K30), lincomycin (MY15), neomycin (N30), norfloxacin (NOR5), novobiocin (NV30), piperacillin (PRL30), polymyxin B (PB300), rifampin (RD30), spectinomycin (SH10), streptomycin (S10, S25), sulfonamide (S3 300), tetracycline (TE10), tobramycin (TOB10), and vancomycin (VA5). Roxithromycin (ROX15) and tylosin (TY30) disks were purchased from MAST Diagnostica (Reinfeld, Germany).
Conjugative transfer of plasmid pGNB1.
Conjugative transfer tests with plasmid pGNB1 were done as described previously (39). E. coli CSH52 (a γ-proteobacterium) and S. meliloti (fdxN::tet; an α-proteobacterium) (21) were used as recipients in mating experiments. E. coli CSH52 transconjugants were selected on LB containing streptomycin (600 μg ml−1) and crystal violet (15 μg ml−1), whereas S. meliloti transconjugants were selected on TY medium containing tetracycline (10 μg ml−1) and crystal violet (15 μg ml−1). Transfer frequencies were calculated per recipient.
Amplification of 16S rRNA gene and internal tmr gene fragments.
Crude total DNA preparations from activated sludge isolates were obtained as described by Weidner et al. (49). Amplification of the 16S rRNA gene was performed as described previously (49) by using primers 1385r (5′-CGGTGTGTRCAAGGCCC-3′, where R is A or G) and 27f (5′-GAGTTTGATCCTGGCTCA-3′). The numbers in the primer designations refer to the binding positions in the E. coli 16S rRNA gene. Primers specific for the pGNB1 tmr gene were designed by using the Primer 3 program. The primer combination consisting of tmr-L1 (5′-GTCAAATCATTGCCATCGTG-3′) and tmr-R1 (5′-TCGAATGAGACAGGCTGATG-3′) was used to amplify tmr-specific amplicons from total DNA preparations from activated sludge bacteria by PCR. PCRs were done under standard conditions using the BIO-Taq DNA polymerase (BIOLINE). The amplicons obtained were verified by sequencing.
Nucleotide sequence accession numbers.
The annotated nucleotide sequences of pGNB1 accessory gene regions L1 and L2 have been deposited in the GenBank database under accession numbers EF628291 and EF628292, respectively.
RESULTS AND DISCUSSION
Isolation of plasmid pGNB1 from an activated sludge bacterial community from a wastewater treatment plant.
Bacteria in an activated sludge sample from the Bielefeld-Heepen wastewater treatment plant (Bielefeld, Germany) were selected on LB agar containing the triphenylmethane dye crystal violet (final concentration, 10 μg ml−1). Total plasmid DNA was isolated from growing colonies and subsequently used to transform E. coli KAM3 carrying a deletion in the multidrug efflux system genes acrAB (25). E. coli KAM3 transformants were selected on LB agar containing 10 μg ml−1 crystal violet. Three of 12 transformants that were able to grow on this medium carried an approximately 60-kb plasmid as determined by Eckhardt gel analysis. Isolation of the plasmids and restriction with endonucleases BamHI, HindIII, and PstI revealed that the three plasmids had identical restriction patterns. One of the plasmids conferring crystal violet resistance was designated pGNB1 and chosen for further analysis. Interestingly, colonies of E. coli KAM3 carrying pGNB1 were able to decolorize crystal violet. Since E. coli KAM3 also harbors an F′ plasmid, it was necessary to separate pGNB1 from F′, which was achieved by transformation of E. coli DH5α with plasmid DNA isolated from E. coli KAM3(pGNB1). E. coli DH5α transformants carrying pGNB1 were selected on LB agar supplemented with crystal violet (10 μg ml−1), and their plasmid content was checked using Eckhardt gels.
Partial sequencing of plasmid pGNB1 revealed that it is an IncP-1β plasmid.
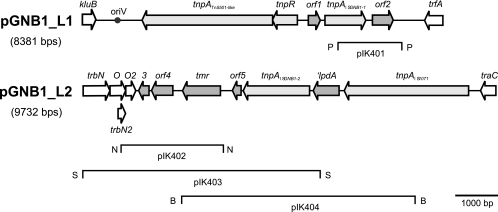
To determine the replicon type of pGNB1, a restriction fragment library of the plasmid was constructed. Restriction fragments generated by the enzymes EcoRI, PstI, SalI, and SphI were cloned into appropriate sequencing vectors. Terminal end sequencing of 17 different recombinant plasmids revealed that pGNB1 is an IncP-1β plasmid closely related to the IncP-1β plasmids pTP6 (36), R751 (44), pB8 (34), pB10 (33), pADP-1 (20), pJP4 (47), and pUO1 (37). The IncP-1β-specific genes kfrC, kleA, klcC, klcB, kluB, trfA, trbB, trbN, trbO, traC, traG, and traI could be identified on the subcloned restriction fragments. Since the backbone regions of IncP-1β plasmids are highly conserved and very well known, we decided to limit our sequencing approach to the accessory regions of the plasmid. Analysis of sequencing data in the restriction fragment library showed that pGNB1 contains two accessory regions, one located downstream of the replication initiation gene trfA1 and the other between the tra and trb conjugative transfer modules. The restriction fragment clones representing accessory regions were completely sequenced by using a primer walking strategy. Gaps between restriction fragments were covered by walking reads on pGNB1 plasmid DNA. This approach led to the establishment of the complete nucleotide sequences of the two pGNB1 accessory regions in Phred40 quality. Accessory region L1 is flanked by the kluB gene (putative postsegregational killing gene) and the trfA1 backbone gene, whereas trbNO and traC flank accessory region L2 (Fig. 1).
FIG. 1.
Genetic map of pGNB1 accessory regions L1 and L2. Coding regions are indicated by arrows showing the direction of transcription. kluB (putative postsegregational killing gene), trfA (replication initiation gene), trbN and trbO (mating pair formation genes), and traC (conjugative transfer gene) are indicated by open arrows and represent plasmid backbone genes. Accessory region L1 contains a transposon related to Tn5501, a new insertion sequence element (ISGNB1-1), and a gene, orf2, encoding a conserved protein (Cupin 2 conserved barrel protein) having an unknown function. Recombinant plasmid pIK401 contains a PstI (P) restriction fragment with orf2. The origin of vegetative replication (oriV) is indicated by a filled circle. Accessory region L2 contains the triphenylmethane reductase gene (tmr) and a truncated dihydrolipoamide dehydrogenase gene (′lpdA) which is flanked by IS1071 and ISGNB1-2. Subcloning of a 2.4-kb NotI (N) restriction fragment, a 5.8-kb SphI (S) restriction fragment, and a 5.4-BamHI (B) restriction fragment resulted in recombinant plasmids pIK402, pIK403, and pIK404, respectively. These plasmids were tested to determine whether they conferred resistance to antimicrobial drugs and triphenylmethane dyes.
Accessory region L1 on plasmid pGNB1 contains a transposon related to Tn5501.
A transposon bordered by 38-bp inverted repeats and related to Tn5501 was inserted between the pGNB1 kluAB operon (putative postsegregational killing genes) and the replication initiation gene trfA. The insertion site of Tn5501, marked by 5-bp direct repeats (TTCTC), is located 289 bp downstream of trfA between TrfA-binding sites 9 and 10 (numbering of Smalla et al. [36]). A 4,230-bp segment of the transposon consisting of the transposition module genes tnpA (transposase) and tnpR (resolvase), orf1, and the 5′ end of a parE-like gene is 99.5% identical to the Tn5501-like transposon previously identified on IncP-1β plasmid pB8 isolated from an unknown bacterium from an activated sludge community (34). The pGNB1 orf1-parE′ module encoding a putative addiction system was truncated by insertion of a new 1,047-bp insertion sequence, designated ISGNB1-1, which is flanked by 15-bp inverted repeat motifs. The encoded transposase of ISGNB1-1 belongs to the Transposase_11 family (accession no. pfam01609) and is 73% identical to TnpA of Burkholderia vietnamiensis G4 IS4 (accession no. EAM29391). Adjacent to ISGNB1-1 the orf2 gene encoding a conserved protein which contains double-stranded β-helix domains was identified. The orf2 gene product is identical to the Cupin 2 conserved barrel protein of plasmid 2 residing in Ralstonia metallidurans CH34 (accession no. YP_582148).
The remaining part of the sequenced fragment (717 bp) contains the replication initiation gene trfA and is 100% identical to Burkholderia cepacia AMMD IncP-1β plasmid 1 (accession no. CP000443).
Second accessory region on plasmid pGNB1, L2, contains a triphenylmethane reductase gene flanked by an insertion sequence element.
To establish the complete nucleotide sequence of pGNB1 accessory region L2, restriction fragments covering this region (Fig. 1) were subcloned and subsequently sequenced. Gaps were closed by using a primer walking strategy with pGNB1 plasmid DNA. The left part of the sequenced fragment contains the IncP-1-specific gene trbN encoding a lytic transglycosylase involved in mating pair formation and the 5′ part of trbO. A 832-bp region spanning trbN-trbO′ is 99.9% identical to the corresponding segments of IncP-1β plasmids pJP4, pTSA, pUO1, pB8, pB10, and pTP6, indicating that these plasmids originated from a common ancestor. A 368-bp region containing ′trbN-trbO′ is reiterated downstream of the truncated first copy of trbO′. The accessory module inserted next to trbO′ encodes a predicted triphenylmethane reductase that is 99.7% identical to the corresponding enzyme previously identified in Citrobacter sp. strain KCTC 18061P (17). Triphenylmethane reductase was shown to catalyze the NADH-dependent reduction of triphenylmethane dyes, such as crystal violet, malachite green, and basic fuchsin. The dinucleotide-binding motif G-X2-G-X2-G conserved in NAD(P)H-dependent enzymes is also present in the N terminus of pGNB1 triphenylmethane reductase. The gene downstream of pGNB1 tmr encodes a conserved hypothetical protein (Orf4) that is identical to a corresponding protein of plasmid pLM80 identified in the gram-positive bacterium Listeria monocytogenes (26). Interestingly, the pLM80 gene for the hypothetical protein is also clustered with a tmr-like gene. There is no indication that the pGNB1 tmr-orf4 module is part of a mobile genetic element. Thus, it is not known how the gene region was incorporated into the plasmid.
The module upstream of tmr on pGNB1 encodes a truncated dihydrolipoamide dehydrogenase (LpdA) possessing an intact pyridine nucleotide-disulfide oxidoreductase dimerization domain (accession no. pfam02852). Compared to other dihydrolipoamide dehydrogenase genes, only the 3′ part (591 bp) of lpdA is retained on pGNB1. Interestingly, the truncated pGNB1 lpdA gene has a GTG start codon that is preceded by a convincing ribosome-binding site (AGAGGAG). Accordingly, the lpdA gene product might be produced. LpdA of pGNB1 is identical to the corresponding part of dihydrolipoamide dehydrogenase encoded in the genome of the nitroaromatic compound degrader Acidovorax sp. strain JS42 (accession no. ZP_01383531). Complete lpdA genes were previously identified on plasmids pADP-1 and pAA1 of the atrazine-degrading bacteria Pseudomonas sp. strain ADP (20) and Arthrobacter aurescens, respectively (31). Truncation of lpdA on pGNB1 was caused by insertion of IS1071 four bases upstream of lpdA. Transcription of the IS1071 tnpA gene is oriented towards lpdA, so that read-through transcription of lpdA starting from the tnpAIS1071 promoter seems to be possible. IS1071 elements nearly identical (99.9%) to the element on pGNB1 were previously identified on R906 of Bordetella bronchiseptica (accession no. DQ471307), pJP4 of Ralstonia eutropha (47), pAA1 of A. aurescens (31), pUO1 of Delftia acidovorans (37), and pADP-1 of Pseudomonas sp. strain ADP (20). IS1071 was frequently found to be associated with degradative genes (9). On the other side lpdA of pGNB1 is flanked by a putative IS element (ISGNB1-2) closely related to ISPps1 (IS91 family) of Herbaspirillum huttiensis. The tnpA gene of this insertion sequence encodes a transposase belonging to the Transposase_32 family (accession no. pfam04986) that is 97% identical to TnpA of H. huttiensis (10).
The remaining part of the sequenced fragment (495 bp) containing the 3′ end of the conjugative transfer gene traC is identical to the corresponding regions of IncP-1β plasmids pTP6, R751, pB8, pB10, pADP-1, pJP4, and pUO1, again supporting the close relationship of these plasmids.
In summary, plasmid pGNB1 accessory region L2 contains a module encoding a triphenylmethane dye degradation gene and another element consisting of two insertion sequences which flank a truncated dihydrolipoamide dehydrogenase gene.
pGNB1 tmr gene confers resistance to triphenylmethane dyes.
To further characterize the pGNB1 triphenylmethane reductase gene, the corresponding region was subcloned on a 5.6-kb SphI restriction fragment, on a 2.4-kb NotI fragment, and on a 5.4-kb BamHI fragment (Fig. 1). The resulting constructs, pIK402, pIK403, and pIK404, respectively, conferred resistance to 15 μg ml−1 crystal violet and 15 μg ml−1 malachite green to the host bacterium E. coli DH5α, whereas DH5α strains carrying the empty cloning vectors were not able to grow in the presence of these dyes. E. coli DH5α carrying either pIK402, pIK403, or pIK404 decolorized the dyes crystal violet, malachite green, and basic fuchsin in the vicinity of a colony, whereas the corresponding control strains carrying empty cloning vectors did not. The described decolorization phenotype was also observed for E. coli KAM3(pGNB1) and E. coli DH5α(pGNB1). The plasmid-free strains KAM3 and DH5α are sensitive to crystal violet and malachite green and could not decolorize these dyes. The MIC of crystal violet for E. coli strain DH5α carrying pIK402 (tmr) is 120 μg ml−1, whereas the corresponding value for DH5α without the plasmid is 2.5 μg ml−1. E. coli DH5α(pGNB1) was able to decolorize LB medium contaminated with up to 110 μg ml−1 crystal violet during overnight growth (15 h), which is a property that could have biotechnological relevance.
To summarize, the pGNB1 tmr gene confers the ability to degrade triphenylmethane dyes to the host bacterium.
Plasmid pGNB1 does not confer resistance to commonly used antimicrobial compounds.
Annotation of the available pGNB1 nucleotide sequence revealed the presence of genes for which it was not possible to predict any function. These genes are orf2, orf3, orf4, orf5, and lpdA. To analyze whether one of these genes is involved in mediating antibiotic resistance, antimicrobial susceptibility tests using the disk diffusion method were carried out. E. coli DH5α strains containing pGNB1 or recombinant plasmid pIK401, pIK402, pIK403, or pIK404, each of which harbors subcloned restriction fragments of pGNB1 accessory regions L1 and L2 (Fig. 1), were tested to determine whether they had reduced susceptibilities to 35 different antimicrobial compounds (see Materials and Methods). No reduced susceptibilities were detected for any of these strains. Thus, the function of pGNB1 accessory region L1 remains unknown.
Plasmid pGNB1 is self-transmissible and can be transferred to α- and γ-proteobacteria.
Restriction analysis and partial sequencing of plasmid pGNB1 showed that its backbone, including the conjugative transfer (tra) and mating pair formation (trb) modules, is conserved. IncP-1 plasmids generally are self-transmissible and possess a broad host range. To determine the transfer properties of plasmid pGNB1, mating experiments with the recipient bacteria E. coli (a γ-proteobacterium) and S. meliloti (an α-proteobacterium) were carried out. Plasmid pGNB1 transfers at high frequencies (calculated per recipient cell) from E. coli DH5α to the streptomycin-resistant E. coli strain CSH52 (transfer frequency, 2.8 × 10−1 per recipient) and from E. coli DH5α to S. meliloti (transfer frequency, 2.2 × 10−3 per recipient). The transfer frequencies for pGNB1 are comparable to those for other IncP-1β plasmids isolated from wastewater treatment plant bacteria, and hence the classification of pGNB1 as a self-transmissible, broad-host-range plasmid is justified. S. meliloti(pGNB1) was tested for the ability to decolorize triphenylmethane dyes, and it was found that the plasmid-containing strain, in contrast to the plasmid-free parental strain, could reduce crystal violet, malachite green, and basic fuchsin. Thus, the decolorization phenotype was transferred by acquisition of plasmid pGNB1.
Wastewater treatment plant bacteria related to Delftia acidovorans harbor a triphenylmethane reductase gene (tmr).
To determine the species affiliation of wastewater treatment plant isolates able to decolorize crystal violet, crude total DNA was prepared from four randomly selected faint white colonies growing on LB agar plates supplemented with crystal violet (final concentration, 10 μg ml−1). These DNA preparations were used as template DNAs to generate 16S rRNA gene-specific PCR amplicons by using primers 27f and 1385r. Sequencing of the obtained 16S rRNA gene amplicons of the four different colonies revealed that they were 99, 98, 98.5, and 97% identical to the 16S rRNA gene nucleotide sequence of the β-proteobacterium Delftia acidovorans (synonym, Comamonas acidovorans) (22). All four isolates also gave PCR amplicons with primers designed for the pGNB1 triphenylmethane reductase gene tmr. D. acidovorans and Comamonas sp. are known to be natural hosts for IncP-1β plasmids (3, 37, 46).
Concluding remarks.
To our knowledge, this is the first report of an IncP-1 plasmid encoding a gene product that is able to confer resistance to and decolorize triphenylmethane dyes. Since IncP-1 plasmids are very well known for their high transfer activities and their extremely broad host ranges, it is very likely that pGNB1 facilitates dissemination of the encoded catabolic activity among a wide range of bacterial species.
Genomic analysis of plasmid pGNB1 revealed that it is very closely related to the IncP-1β antibiotic resistance plasmid pB8, which was also isolated from a bacterial community from a wastewater treatment plant. It thus appears that nearly identical plasmids can serve as vehicles for both antibiotic resistance determinants and catabolic traits, probably depending on the selective pressure to which the corresponding host bacterium has recently been exposed. Consequently, it is quite conceivable that catabolic plasmids can easily be rearranged by incorporation of antibiotic resistance modules, especially in environments contaminated with antibiotics and other pollutants, which is the case for sewage. Thus, catabolic plasmids should be considered vehicles for the emergence of new antibiotic resistance plasmids. Maintenance of this kind of plasmids in sewage bacteria is selected for by the presence of pollutants, such as triphenylmethane dyes.
Since pollution of water with triphenylmethanes may cause serious ecological damage, the possibility that plasmid pGNB1 might be useful for biotechnological processes aimed towards bioremediation of liquid wastes contaminated with triphenylmethane dyes should be considered.
Footnotes
Published ahead of print on 3 August 2007.
REFERENCES
- 1.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 2.Bönemann, G., M. Stiens, A. Pühler, and A. Schlüter. 2006. Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob. Agents Chemother. 50:3075-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boon, N., J. Goris, P. De Vos, W. Verstraete, and E. M. Top. 2001. Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl. Environ. Microbiol. 67:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1Blue—a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing. 15th informational supplement. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Davison, J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73-91. [DOI] [PubMed] [Google Scholar]
- 7.Dennis, J. 2005. The evolution of IncP catabolic plasmids. Curr. Opin. Biotechnol. 16:291-298. [DOI] [PubMed] [Google Scholar]
- 8.Dennis, J. J., and G. J. Zylstra. 2004. Complete sequence and genetic organization of pDTG1, the 83 kilobase naphthalene degradation plasmid from Pseudomonas putida strain NCIB 9816-4. J. Mol. Biol. 341:753-768. [DOI] [PubMed] [Google Scholar]
- 9.Di Gioia, D., M. Peel, F. Fava, and R. C. Wyndham. 1998. Structures of homologous composite transposons carrying cbaABC genes from Europe and North America. Appl. Environ. Microbiol. 64:1940-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton, R. W., and J. S. Karns. 1991. Cloning and comparison of the DNA encoding ammelide aminohydrolase and cyanuric acid amidohydrolase from three s-triazine-degrading bacterial strains. J. Bacteriol. 173:1363-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frost, L. S., R. Leplae, A. O. Summers, and A. Toussaint. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722-732. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, D., C. Desmarais, and P. Green. 2001. Automated finishing with autofinish. Genome Res. 11:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heuer, H., R. Szczepanowski, S. Schneiker, A. Pühler, E. M. Top, and A. Schlüter. 2004. The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1β group without any accessory genes. Microbiology 150:3591-3599. [DOI] [PubMed] [Google Scholar]
- 16.Hynes, M. F., R. Simon, and A. Pühler. 1985. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtC58. Plasmid 13:99-105. [DOI] [PubMed] [Google Scholar]
- 17.Jang, M. S., Y. M. Lee, C. H. Kim, J. H. Lee, D. W. Kang, S. J. Kim, and Y. C. Lee. 2005. Triphenylmethane reductase from Citrobacter sp. strain KCTC 18061P: purification, characterization, gene cloning, and overexpression of a functional protein in Escherichia coli. Appl. Environ. Microbiol. 71:7955-7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jencova, V., H. Strnad, Z. Chodora, P. Ulbrich, W. J. Hickey, and V. Paces. 2004. Chlorocatechol catabolic enzymes from Achromobacter xylosoxidans A8. Int. Biodeterior. Biodegrad. 54:175-181. [Google Scholar]
- 19.Jones, J. J., and J. O. Falkinham III. 2003. Decolorization of malachite green and crystal violet by waterborne pathogenic mycobacteria. Antimicrob. Agents Chemother. 47:2323-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masepohl, B., M. Kutsche, K. U. Riedel, M. Schmehl, W. Klipp, and A. Pühler. 1992. Functional analysis of the cysteine motifs in the ferredoxin-like protein FdxN of Rhizobium meliloti involved in symbiotic nitrogen fixation. Mol. Gen. Genet. 233:33-41. [DOI] [PubMed] [Google Scholar]
- 22.Mayer, J., K. Denger, T. H. Smits, K. Hollemeyer, U. Groth, and A. M. Cook. 2006. N-Acetyltaurine dissimilated via taurine by Delftia acidovorans NAT. Arch. Microbiol. 186:61-67. [DOI] [PubMed] [Google Scholar]
- 23.Meyer, F., A. Goesmann, A. C. McHardy, D. Bartels, T. Bekel, J. Clausen, J. Kalinowski, B. Linke, O. Rupp, R. Giegerich, and A. Pühler. 2003. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Morita, Y., K. Kodama, S. Shiota, T. Mine, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1998. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42:1778-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nojiri, H., M. Shintani, and T. Omori. 2004. Divergence of mobile genetic elements involved in the distribution of xenobiotic-catabolic capacity. Appl. Microbiol. Biotechnol. 64:154-174. [DOI] [PubMed] [Google Scholar]
- 28.Park, H. S., and H. S. Kim. 2000. Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12. J. Bacteriol. 182:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rafii, F., W. Franklin, and C. E. Cerniglia. 1990. Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl. Environ. Microbiol. 56:2146-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren, S., J. Guo, G. Zeng, and G. Sun. 2006. Decolorization of triphenylmethane, azo, and anthraquinone dyes by a newly isolated Aeromonas hydrophila strain. Appl. Microbiol. Biotechnol. 72:1316-1321. [DOI] [PubMed] [Google Scholar]
- 31.Sajjaphan, K., N. Shapir, L. P. Wackett, M. Palmer, B. Blackmon, J. Tomkins, and M. J. Sadowsky. 2004. Arthrobacter aurescens TC1 atrazine catabolism genes trzN, atzB, and atzC are linked on a 160-kilobase region and are functional in Escherichia coli. Appl. Environ. Microbiol. 70:4402-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Schlüter, A., H. Heuer, R. Szczepanowski, L. J. Forney, C. M. Thomas, A. Pühler, and E. M. Top. 2003. The 64,508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1β group. Microbiology 149:3139-3153. [DOI] [PubMed] [Google Scholar]
- 34.Schlüter, A., H. Heuer, R. Szczepanowski, S. M. Poler, S. Schneiker, A. Pühler, and E. M. Top. 2005. Plasmid pB8 is closely related to the prototype IncP-1β plasmid R751 but transfers poorly to Escherichia coli and carries a new transposon encoding a small multidrug resistance efflux protein. Plasmid 54:135-148. [DOI] [PubMed] [Google Scholar]
- 35.Schlüter, A., R. Szczepanowski, A. Pühler, and E. M. Top. 2007. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 31:449-477. [DOI] [PubMed] [Google Scholar]
- 36.Smalla, K., A. S. Haines, K. Jones, E. Krögerrecklenfort, H. Heuer, M. Schloter, and C. M. Thomas. 2006. Increased abundance of IncP-1β plasmids and mercury resistance genes in mercury-polluted river sediments: first discovery of IncP-1β plasmids with a complex mer transposon as the sole accessory element. Appl. Environ. Microbiol. 72:7253-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sota, M., H. Kawasaki, and M. Tsuda. 2003. Structure of haloacetate-catabolic IncP-1β plasmid pUO1 and genetic mobility of its residing haloacetate-catabolic transposon. J. Bacteriol. 185:6741-6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Springael, D., and E. M. Top. 2004. Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from ecological studies. Trends Microbiol. 12:53-58. [DOI] [PubMed] [Google Scholar]
- 39.Szczepanowski, R., I. Krahn, N. Bohn, A. Pühler, and A. Schlüter. 2007. Novel macrolide resistance module carried by the IncP-1β resistance plasmid pRSB111, isolated from a wastewater treatment plant. Antimicrob. Agents Chemother. 51:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan, H. M. 1999. Bacterial catabolic transposons. Appl. Microbiol. Biotechnol. 51:1-12. [DOI] [PubMed] [Google Scholar]
- 41.Tauch, A., A. Schlüter, N. Bischoff, A. Goesmann, F. Meyer, and A. Pühler. 2003. The 79,370-bp conjugative plasmid pB4 consists of an IncP-1β backbone loaded with a chromate resistance transposon, the strA-strB streptomycin resistance gene pair, the oxacillinase gene bla(NPS-1), and a tripartite antibiotic efflux system of the resistance-nodulation-division family. Mol. Genet. Genomics 268:570-584. [DOI] [PubMed] [Google Scholar]
- 42.Tennstedt, T., R. Szczepanowski, I. Krahn, A. Pühler, and A. Schlüter. 2005. Sequence of the 68,869 bp IncP-1α plasmid pTB11 from a waste-water treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements. Plasmid 53:218-238. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, C. M., and K. M. Nielsen. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711-721. [DOI] [PubMed] [Google Scholar]
- 44.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 45.Top, E. M., and D. Springael. 2003. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14:262-269. [DOI] [PubMed] [Google Scholar]
- 46.Tralau, T., A. M. Cook, and J. Ruff. 2001. Map of the IncP1β plasmid pTSA encoding the widespread genes (tsa) for p-toluenesulfonate degradation in Comamonas testosteroni T-2. Appl. Environ. Microbiol. 67:1508-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trefault, N., R. De la Iglesia, A. M. Molina, M. Manzano, T. Ledger, D. Perez-Pantoja, M. A. Sanchez, M. Stuardo, and B. Gonzalez. 2004. Genetic organization of the catabolic plasmid pJP4 from Ralstonia eutropha JMP134(pJP4) reveals mechanisms of adaptation to chloroaromatic pollutants and evolution of specialized chloroaromatic degradation pathways. Environ. Microbiol. 6:655-668. [DOI] [PubMed] [Google Scholar]
- 48.Vedler, E., M. Vahter, and A. Heinaru. 2004. The completely sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J. Bacteriol. 186:7161-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weidner, S., W. Arnold, and A. Pühler. 1996. Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl. Environ. Microbiol. 62:766-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]