Abstract
Bifidobacteria are one of the main microbial inhabitants of the human colon. Usually administered in fermented dairy products as beneficial microorganisms, they have to overcome the acidic pH found in the stomach during the gastrointestinal transit to be able to colonize the lower parts of the intestine. The mechanisms underlying acid response and adaptation in Bifidobacterium longum biotype longum NCIMB 8809 and its acid-pH-resistant mutant B. longum biotype longum 8809dpH were studied. Comparison of protein maps, and protein identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis, allowed us to identify nine different proteins whose production largely changed in the mutant strain. Furthermore, the production of 47 proteins was modulated by pH in one or both strains. These included general stress response chaperones and proteins involved in transcription and translation as well as in carbohydrate and nitrogen metabolism, among others. Significant differences in the levels of metabolic end products and in the redox status of the cells were also detected between the wild-type strain and its acid-pH-resistant mutant in response to, or as a result of, adaptation to acid. Remarkably, the results of this work indicated that adaptation and response to low pH in B. longum biotype longum involve changes in the glycolytic flux and in the ability to regulate the internal pH. These changes were accompanied by a higher content of ammonium in the cytoplasm, likely coming from amino acid deamination, and a decrease of the bile salt hydrolase activity.
Bifidobacteria are anaerobic, gram-positive, nonmotile, and nonsporulating irregular or branched fermentative rod-shaped bacteria in which the acid susceptibility is dependent on the strain (31). As with other intestinal bacteria, they have been shown to provide protection against gastrointestinal disorders, but it has also been proposed that they exercise other health-promoting effects, like the inhibition of pathogenic microorganisms, antimutagenic and anticarcinogenic activities, prevention of diarrhea, immune modulation, and reduction of serum cholesterol levels (42, 48). Because of these properties, several species belonging to the genus Bifidobacterium are considered probiotics, and they are largely applied in dairy products (28).
Acid tolerance in bifidobacteria is of particular importance, as this property is closely related to their use in human nutrition. In general, it can be considered that bifidobacteria have a weak acid tolerance with the exception of Bifidobacterium animalis (32), which can survive acidic pH better than the other species and which is usually detected as the sole viable bifidobacterium species in fermented milks (21). In addition, it has been shown that the effect of pH in the stomach and duodenum promotes changes in the microbiota, excluding the growth of some species like bifidobacteria or Escherichia coli, while Lactobacillus or some yeasts are favored (35). Bifidobacteria are consumed in fermented milks, where they are added at the beginning of the fermentation process together with the starter culture which acidifies milk by lactose fermentation, reaching a pH usually lower than 4.6 (46). After ingestion, and in order to get to their ecological niche, i.e., the large intestine, bacteria must survive passage through the low-pH environment of the stomach. Then, bifidobacteria have to cope with acid stress from their storage, through their distribution and to their delivery, which usually results in their loss of viability and therefore in a reduction of the potential probiotic effects (7, 35).
It is known that exposure of bacteria to stress factors (heat, bile salts, or acid pH) can provide protection against further hostile environmental conditions (5, 17, 34). For instance, the acid tolerance response has been evidenced in Bifidobacterium longum and B. animalis (32, 41) and in other gram-positive bacteria such as Listeria monocytogenes (13), Enterococcus faecalis (16), or Lactococcus lactis (37). Mechanisms underlying acid tolerance used by gram-positive bacteria include not only proton pumping via the F1Fo-ATPase but also changes in the cell membrane and regulatory mechanisms, alterations in different metabolic pathways, and amino acid decarboxylation (12).
With the purpose of understanding how bifidobacteria can adapt to acid stress, we used as a model microorganism B. longum biotype longum NCIMB 8809, a potential probiotic strain (36), and B. longum biotype longum 8809dpH, a mutant derived thereof, which we isolated as acid pH resistant. The physiological properties and proteomes of the two strains were compared.
MATERIALS AND METHODS
Strains and culture conditions.
The bacterial strains used in this study were B. longum biotype longum NCIMB 8809 (wild-type [WT] strain) (NCIMB, Aberdeen, Scotland, United Kingdom), which was isolated from nursling stools, and its acid-pH-resistant mutant B. longum biotype longum 8809dpH. Strains were routinely grown in MRS (BD Diagnostic Systems, Sparks, MD, unless indicated otherwise) supplemented with 0.05% l-cysteine (Sigma, St. Louis, MO) (MRSC), at 37°C and under anaerobic conditions (Bactron anaerobic/environmental chamber; Sheldon Manufacturing Inc., Cornelius, OR) in an atmosphere of 5% CO2-5% H2-90% N2. For the isolation of acid-resistant mutants, cells from an overnight culture of B. longum biotype longum NCIMB 8809, previously subcultured in standard conditions, were washed in phosphate-buffered saline and used to inoculate (1%) fresh MRSC (Scharlau, Barcelona, Spain) adjusted at different pH values (5.0, 4.0, 2.0, and 1.25) with 1 N HCl. These cultures were incubated at 37°C for 16 h, and then aliquots were plated on MRSC at neutral pH and incubated at 37°C for 3 to 4 days to recover possible acid-resistant strains. The acquisition of a stable acid resistance phenotype in the mutants was determined by testing their ability to survive in simulated gastric conditions (3 g/liter pepsin, adjusted at pH 2.0 with HCl; Sigma) after daily cultivation in MRSC for 2 weeks as previously described (10).
For the proteomic and physiological experiments, one colony isolated on MRSC agar was grown overnight. Aliquots of 2.5 ml of those precultures were washed twice in MRSC adjusted to pH 7.0 or pH 4.8, used to inoculate 250 ml of fresh MRSC adjusted to pH 7.0 or pH 4.8, and incubated anaerobically until mid-exponential phase (optical density at 600 nm [OD600], 0.5).
Molecular identification of Bifidobacterium mutant strain.
Chromosomal DNA from Bifidobacterium isolates was extracted according to the method of Wilson et al. (51), which was slightly modified by including an incubation step with lysozyme (50 mg/ml) at 37°C for 30 min prior to the extraction procedure. Then, DNA of B. longum was isolated using a GenElute bacterial genomic DNA kit (Sigma) and used as a template for PCR amplifications. Primers (Bif164, 5′-CATCCGGCATTACCACCC, and Bif662, 5′-CCACCGTTACACCGGGAA) specific for the 16S rRNA gene sequence (22, 29) were used to amplify a partial sequence of the 16S gene (19). For the amplification of the gene coding for O-acetylhomoserine (thiol)-lyase and its 500-bp upstream region, the forward primer 5′-CTCCAATCAATCAGACGGATGTCAC (CysDF1) and the reverse primer 5′-CCCAATATGGAGAGGCATTATGAACG (CysDR2) were used. For the amplification of the 3.8-kb sequence containing the gene of the methionine synthase and the upstream region, we used the forward primer 5′-CGATGAGCGTCAACCTGCTGGACG (MetEF2) and the reverse primer 5′-GCTCGGGTTTATGTAAGGAAATCAGC (MetER). PCR fragments were purified with the GenElute PCR cleanup kit (Sigma) and sequenced in the Servicio de Secuenciación de ADN (SSAD, CIB, Cantoblanco, Madrid, Spain).
Sensitivity of B. longum biotype longum NCIMB 8809 and its acid-pH-resistant mutant B. longum biotype longum 8809dpH to artificial gastrointestinal conditions.
The ability of B. longum to survive in in vitro conditions simulating the passage through the stomach and intestine was evaluated. For this purpose, cell suspensions of each strain (108 to 109 cells/ml) were incubated in sterile saline solution (0.5% [wt/vol] NaCl), containing 3 g/liter pepsin (Sigma) and adjusted at pH 2.0 with HCl, at 37°C for 120 min. Aliquots were withdrawn at different times (0, 90, and 120 min) to determine plate counts on MRSC agar. After exposure to simulated gastric conditions, cells were collected by centrifugation (8,000 × g, 10 min) and washed in sterile saline solution. Then, cell pellets were suspended in saline solution containing pancreatin (1 g/liter; Sigma) and bile salts (0.5% [wt/vol] oxgall; Sigma) and incubated at 37°C for 120 min. Aliquots were also withdrawn at different incubation times (0, 90, and 120 min) to determine plate counts.
A more detailed study of the ability of the WT and acid-resistant strains to survive in MRSC adjusted at a range of acid pH values (2.5, 3.5, 4.0, and 5.0) with HCl was also carried out. Cell suspensions were incubated at 37°C, and samples were withdrawn at different times (0 and 90 min) to determine the plate counts and viability with the LIVE/DEAD BacLight Bacterial Viability stain kit (Invitrogen Co., Carlsbad, CA), according to the manufacturer's instructions. Data acquisition was performed using an EPICS-XL-MCL flow cytometer (Coulter Corporation, Miami, FL).
The different abilities of the WT and mutant strains to grow in the presence of different concentrations of bile salts (0.5, 1.0, 2.0, and 3.0% [wt/vol]) were also determined (10). The assayed concentrations of bile included those present in physiological conditions (from 0.3 to 2.0%) (34). Briefly, cells from overnight cultures were harvested by centrifugation (6,000 × g, 4°C, 10 min), washed with phosphate-buffered saline, and diluted in fresh medium supplemented with the respective concentrations of bile salts to reach a final OD655 of 0.1. Bacterial growth was monitored by measuring the OD655 in a Microplate Reader Model 550 (Bio-Rad, Hercules, CA). Growth curves were analyzed with a modified Gompertz model (52).
2D electrophoresis, statistical analysis, and protein identification.
Cell extracts were obtained and two-dimensional (2D) electrophoresis was basically performed as described previously (44). Briefly, 350 μg of protein from bifidobacterial extracts was loaded into strips with a pH range of 4 to 7 and focused for 60,000 V·h, and the second dimension was carried out in a 12.5% polyacrylamide-sodium dodecyl sulfate gel. This pH range covers 71.5% of the theoretical proteins of B. longum NCC2705 (45). Proteic spots were visualized with Bio-Safe Coomassie blue staining. Spot detection and volume quantitation were carried out with ImageMaster 2D Elite (version 3.10; Amersham Biosciences). At least three independent analyses for each growth condition were performed. An effect of acidic pH on the expression of proteins was considered if the mean normalized spot volume varied at least 1.5-fold and was confirmed by analysis of variance at the significance level of P < 0.05 using pH as a factor with two categories, pH 7.0 and pH 4.8. Proteins were identified using the MOWSE (Molecular Weight Search) score (39).
Estimation of the redox balance, the intracellular pH, and the bile salt hydrolase (BSH) activity.
The fluorescence properties of buffered cell suspensions (OD600 of 0.6) in 50 mM Tris-HCl buffer, pH 7.0, were monitored in an Eclipse fluorescence spectrophotometer (Varian, Inc., Palo Alto, CA). The intensity values corresponding to NADH were calculated from the 413-nm emission at λex = 316 nm, whereas for flavin adenine dinucleotide (FAD) the intensity values were calculated from the 436-nm emission at λex = 380 nm, according to the work of Ammor et al. (1). The redox ratio was deduced from the NADH- and FAD-related signals using the equation redoxratio = FADintensity/(FADintensity + NADHintensity) (25).
Measurements of intracellular pH values of buffered cell suspensions, grown at pH 7.0 or 4.8, at two different external pHs (7.0 and 4.8) were recorded for the WT and the acid-resistant mutant as previously described (43), and the quantitative determination of the BSH activity was performed according to the method of Noriega et al. (33).
Intracellular amino acid and NH3 determination.
For the extraction of amino acids and ammonia, 2 ml of B. longum cultures at an OD600 of 0.5 was harvested by centrifugation, washed twice, resuspended in the same volume of 50 mM Tris-HCl adjusted to pH 7, and boiled for 15 min. Cell debris was discarded by centrifugation (13,000 × g, 15 min, 4°C). The supernatants were evaporated in a vacuum concentrator (Eppendorf AG, Hamburg, Germany), resuspended in 20 μl of 0.1 N HCl, and derivatized with dabsyl chloride (Sigma) in the presence of 0.15 mM sodium bicarbonate for 15 min at 70°C, as previously described (26). Ten microliters of the derivatized samples was separated in a Novapack C18 column (Waters Corporation, Milford, CA) at a constant temperature of 50°C, and the amino acid and NH3 derivatives were detected with a UV detector at a wavelength of 436 nm.
High-pressure liquid chromatography and gas chromatography analysis.
For glucose and acetic, lactic, and formic acid determination, studies were performed with buffered cell suspensions. Ten milliliters of cultures at an OD600 of 0.6 was collected by centrifugation (10,000 × g for 15 min). The bacterial pellet was washed twice with 50 mM Tris-HCl buffer, pH 7.0, and resuspended in 10 ml of 50 mM Tris-HCl buffer, pH 5.6, containing 25 mM glucose. The suspension was incubated with constant mild stirring for 4 h at 37°C. Cells were removed from the suspension by centrifugation, and the supernatant was analyzed by high-pressure liquid chromatography according to the method of Sánchez et al. (44) to quantify acid levels. Results presented are the means of at least three separate experiments. The ethanol in supernatants of resting cells was determined by dynamic headspace extraction and gas chromatography-mass spectrometry analysis as previously described by Ruas-Madiedo et al. (40).
Nucleotide sequence accession numbers.
The genes coding for MetE and CysD, and their upstream regions, were sequenced in both strains, NCIMB 8809 and 8809dpH, and the sequences were submitted under accession numbers EF453722 and EF453723.
RESULTS
Isolation and characterization of an acid-resistant mutant in B. longum biotype longum.
In order to isolate acid-resistant mutants from B. longum biotype longum NCIMB 8809, cell suspensions of this strain were incubated at different pH values (1.25, 2.0, 4.0, and 5.0), and then plated on MRSC. Small colonies could be obtained only from cultures at pH 4.0 and 5.0, those of pH 4.0 being selected for further characterization and confirmation of their genus and species identity by specific PCR (22, 29). The 16S rRNA gene of one of these isolates, named B. longum biotype longum 8809dpH, was partially sequenced, showing 100% identity with its counterpart.
B. longum biotype longum 8809dpH showed significantly higher survival than the WT in simulated gastrointestinal conditions after 90 and 120 min of incubation. While the colony counts of the WT decreased more than 5 units after 90 min of incubation (from 8.85 ± 0.17 to 3.83 ± 0.28 log CFU/ml) and were undetectable (<3 log CFU/ml) after 120 min, the mutant did not show a significant decrease in survival after 120 min (from 8.98 ± 0.10 to 8.78 ± 0.30 log CFU/ml; values are expressed as means ± standard deviations of at least triplicate experiments). The stability of the acid resistance phenotype of the mutant was also confirmed by the absence of significant variations (P < 0.05) in its ability to survive in gastric juices after daily cultivation for 2 weeks in nonacidified MRSC (data not shown).
The viabilities of WT and mutant strains after exposure to a wide range of pH values in MRSC as well as their abilities to grow in the presence of bile are shown in Table 1. The numbers of both culturable (plate counts in MRSC) and viable (LIVE/DEAD kit) cells of the mutant strain were significantly higher (P < 0.05) than those of the WT after exposure to a pH range from 2.5 to 5.0. The mutant also showed a significantly (P < 0.05) better ability than the WT to grow in the presence of all tested bile concentrations (0.5 to 3%).
TABLE 1.
Viability of B. longum biotype longum NCIMB 8809 and B. longum biotype longum 8809dpH after exposure to different acid pH values in MRSC and effect of bile concentration on growth ratesa
| pH | % Viability (SYTO9)b
|
Culturable bacteria (log CFU/ml)c
|
Bile (%) | Relative growth rate (% μ)d
|
|||
|---|---|---|---|---|---|---|---|
| NCIMB 8809 | 8809dpH | NCIMB 8809 | 8809dpH | NCIMB 8809 | 8809dpH | ||
| 6.4 | 94.87 ± 6.62 | 97.39 ± 1.15 | 8.08 ± 0.66 | 8.78 ± 0.54 | 0.0 | 100.00 | 100.00 |
| 5.0 | 68.16 ± 4.30 | 96.84e ± 0.86 | 2.67 ± 1.20 | 8.60e ± 0.90 | 0.5 | 45.07 ± 8.20 | 93.00e ± 5.00 |
| 4.0 | 32.89 ± 1.74 | 94.06e ± 1.27 | 1.52 ± 0.70 | 8.64e ± 0.50 | 1.0 | 23.88 ± 7.56 | 81.20e ± 3.45 |
| 3.5 | 1.79 ± 0.50 | 77.58e ± 1.80 | 0.00 ± 0.50 | 6.39e ± 0.33 | 2.0 | 3.94 ± 1.05 | 72.27e ± 5.00 |
| 2.5 | 0.00 ± 0.10 | 44.05e ± 1.70 | 0.00 ± 0.00 | 5.56e ± 0.65 | 3.0 | 0.70 ± 1.00 | 65.64e ± 4.56 |
Values, except for percent bile, are expressed as means ± standard deviations of at least three independent experiments.
Viability was determined by flow cytometry with the LIVE/DEAD kit after 90 min of exposure to acidified MRSC.
CFU were counted on MRSC after 90 min of exposure to acidified MRSC.
Data are expressed as the percentages of the growth rate (μ; h−1) obtained in the absence of bile, which was given a value of 100%.
Significant difference between WT and mutant strains (P < 0.05).
Constitutive differences between WT and acid-pH-resistant mutant at neutral pH.
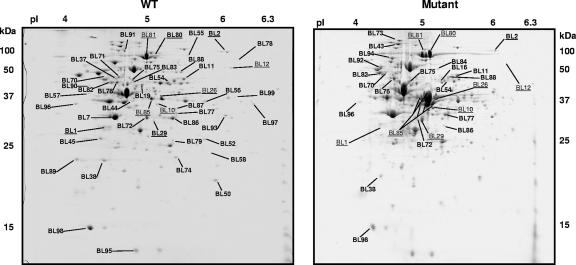
The cytoplasmic protein pools were analyzed by means of 2D electrophoresis, and both WT and mutant strains were compared after growth at pH 7.0 (Fig. 1). Under these conditions, the production of nine proteins was clearly affected in the mutant, five and four of them being significantly over- and underproduced, respectively, compared with the WT (Table 2).
FIG. 1.
Representative 2D gels containing cytosolic extracts from mid-exponential-phase cells of B. longum biotype longum NCIMB 8809 (left) and B. longum biotype longum 8809dpH (right) grown in MRSC initially adjusted to pH 7. Spots identified by peptide mass fingerprinting are labeled, and the corresponding identifications are listed in Tables 2 and 3. Spots that showed variations between the WT and the mutant are underlined, whereas those that modified their quantities at pH 4.8 are in bold.
TABLE 2.
Proteins whose production was affected by the acquisition of acid tolerance in the strain B. longum biotype longum 8809dpH
| COGa | Spot no.b | Putative functionc | Gene | Accession no. | Molecular massd | pIe | Cov.f | MOWSEg | Prod.h |
|---|---|---|---|---|---|---|---|---|---|
| Carbohydrate transport and metabolism | BL81 | Xylulose-5-phosphate/fructose- 6-phosphate phosphoketolase | xfp | BL0959 | 92.5 | 5.0 | 57 | 3.59E + 09 | Down |
| BL26 | Glyceraldehyde-3-phosphate dehydrogenase C | gap | BL1363 | 37.7 | 5.2 | 18 | 2.55E + 07 | Up | |
| BL29 | UDP-glucose 4-epimerase | galE1 | BL1644 | 37.3 | 5.1 | 34 | 1.02E + 05 | Up | |
| Energy production and conversion | BL2 | Aldehyde-alcohol dehydrogenase 2 | adh2 | BL1575 | 98.9 | 5.9 | 16 | 4.92E + 09 | Down |
| Amino acid transport | BL80 | Methionine synthase | metE | BL0798 | 85.4 | 5.2 | 47 | 4.32E + 25 | Up |
| and metabolism | BL10 | Cystathionine gamma-synthase | metB | BL1155 | 41.9 | 5.3 | 77 | 1.08E + 11 | Up |
| BL12 | Probable ATP binding protein of ABC transporter for peptides | dppD | BL1390 | 73.2 | 5.9 | 64 | 1.12E + 15 | Down | |
| BL85 | O-Acetylhomoserine (thiol)-lyase | cysD | BL0933 | 47.6 | 5.1 | 48 | 1.77E + 07 | Up | |
| Cell envelope biogenesis, outer membrane | BL1 | BSH | bsh | BL0796 | 35.1 | 4.7 | 60 | 4.59E + 11 | Down |
COG, cluster of orthologous genes.
Spot numbers refer to the proteins labeled in Fig. 1.
Putative functions were established according to the cluster of orthologous genes for B. longum NCC2705.
Theoretical molecular mass expressed in kDa.
Theoretical isoelectric point expressed in pH.
Percentage of sequence coverage.
Molecular weight search score.
Prod., production. Up-regulation (up) or down-regulation (down) of matching proteins between B. longum biotype longum NCIMB 8809 and B. longum biotype longum 8809dpH.
Interestingly, the main differences between strains were identified as the methionine synthase (MetE, spot BL80); the O-acetylhomoserine (thiol)-lyase (CysD, spot BL85); and, to a lesser extent, the cystathionine gamma-synthase (MetB, spot BL10). To determine if the changes in production of MetE and CysD were related to mutations of their structural gene, the genes coding for these proteins and their upstream regions were sequenced in both strains. No changes were detected between B. longum biotype longum NCIMB 8809 and the mutant DNA sequences (accession numbers EF453722 and EF453723), suggesting a pleiotropic effect of a mutation located somewhere else.
In addition, the mutant showed changes in the isoform distribution of two important proteins of the carbohydrate catabolism, i.e., xylulose-5-phosphate/fructose-6-phosphate phosphoketolase (Xfp) and glyceraldehyde-3-phosphate dehydrogenase C (Gap). Two other enzymes involved in carbohydrate metabolism were affected: UDP-glucose 4-epimerase (GalE1) was overproduced, and the aldehyde-alcohol dehydrogenase 2 (Adh2) was underproduced. Surprisingly, BSH was underproduced in the acid-pH-resistant mutant.
Different response mechanisms are induced at low pH in the WT and mutant strains.
When the WT strain was grown at pH 4.8 and its growth was compared to that at neutral pH, 41 spots were detected to be differentially produced, 18 of which were overproduced and 23 underproduced. When the mutant strain was cultured at pH 4.8, it showed variations in the production levels of 20 spots, which were smaller in number than the WT strain, 17 and 3 of them being over- and underproduced, respectively (Fig. 1; Table 3). In a general view, up-regulated spots are predominant in the mutant, whereas down-regulated ones are more abundant in the WT strain. On the other hand, enzymes involved in carbohydrate metabolism, energy production and conversion, and amino acid metabolism groups are generally overproduced, and down-regulated spots are spread among the other functional groups.
TABLE 3.
Spot identification by mass spectrometry against the theoretical peptide database of B. longum NCC2705 for the strains B. longum biotype longum NCIMB 8809 and B. longum biotype longum 8809dpH in MRSC adjusted initially to pH 4.8 or 7a
| COGb | Spot no.c | Putative functiond | Gene | Accession no. | Cov.e | MOWSEf | WT ratiog | Mutant ratioh |
|---|---|---|---|---|---|---|---|---|
| Carbohydrate transport and | BL70 | Probable alpha-1,4-glucosidase | aglA | BL0529 | 50 | 3.49E + 13 | 11.90 | 1.60 |
| metabolism | BL71 | Phosphoglucomutase | pgm | BL1630 | 60 | 8.17E + 12 | 1.67 | —i |
| BL16 | Pyruvate kinase | pyk | BL0988 | 55 | 2.64E + 10 | — | 2.18 | |
| BL7 | Transketolase | tkt | BL0716 | 25 | 1.08E + 10 | 2.09 | — | |
| BL29 | UDP-glucose 4-epimerase | galE1 | BL1644 | 34 | 1.02E + 05 | 2.13 | — | |
| BL72 | Transaldolase | tal | BL0715 | 65 | 1.00E + 08 | −29.19 | 5.56 | |
| BL73 | Beta-galactosidase | lacZ | BL0978 | 39 | 2.14 + E16 | — | 2.14 | |
| BL55 | Sugar kinase | BL1341 | 30 | 1.52E + 07 | −14.94 | — | ||
| BL74 | Ribulose-phosphate 3-epimerase | rpe | BL0753 | 36 | 1.06E + 03 | −1.68 | — | |
| Energy production and | BL75 | ATP synthase alpha chain | atpA | BL0359 | 65 | 6.70E + 16 | 1.68 | 5.09 |
| conversion | BL76 | ATP synthase beta chain | atpD | BL0357 | 78 | 3.83E + 17 | 1.75 | 3.72 |
| BL77 | Lactate dehydrogenase | ldh2 | BL1308 | 77 | 3.44E + 10 | 3.50 | 1.93 | |
| BL2 | Aldehyde-alcohol dehydrogenase 2 | adh2 | BL1575 | 16 | 4.92E + 09 | −17.42 | −3.61 | |
| BL78 | Phosphoenolpyruvate carboxylase | ppc | BL0604 | 45 | 4.85E + 16 | −1.83 | — | |
| BL79 | Succinyl coenzyme A synthetase alpha chain | sucD | BL0733 | 50 | 3.74E + 10 | −4.29 | — | |
| Amino acid transport and | BL80 | Methionine synthase | metE | BL0798 | 19 | 1.45E + 15 | 1.87 | — |
| metabolism | BL37 | Xaa-Pro aminopeptidase I | pepP | BL1350 | 67 | 5.05E + 16 | 1.79 | — |
| BL82 | Glutamine synthetase 1 | glnA1 | BL1076 | 58 | 3.04E + 12 | 1.74 | 4.30 | |
| BL83 | Acetylornithine aminotransferase | argD | BL1061 | 71 | 2.97E + 09 | 2.73 | — | |
| BL44 | Probable BCAA aminotransferase | ilvE | BL0852 | 13 | 2.09E + 07 | 1.51 | — | |
| BL84 | Dihydroxy acid dehydratase | ilvD | BL1788 | 48 | 1.14E + 11 | — | 2.02 | |
| BL57 | Peptidase M20 homolog | BL1072 | 48 | 1.28E + 10 | −2.79 | — | ||
| BL86 | Ketol-acid reductoisomerase | ilvC2 | BL0531 | 50 | 2.96E + 09 | 20.41 | 7.17 | |
| Nucleotide transport and | BL87 | GMP reductase | BL1500 | 72 | 4.17E + 10 | 2.50 | — | |
| metabolism | BL11 | Bifunctional purine protein PurH | purH | BL0735 | 19 | 3.89E + 13 | −1.54 | −1.68 |
| BL88 | IMP dehydrogenase | guaB | BL1722 | 76 | 1.38E + 17 | −2.03 | 2.18 | |
| BL89 | Probable MTA/SAH nucleosidase | pfs | BL0318 | 74 | 4.69E + 06 | −285.4 | — | |
| Coenzyme metabolism | BL90 | Probable bifunctional folate synthesis | sulD | BL1685 | 20 | 2.48E + 04 | −7.53 | — |
| Translation, ribosomal structure, and biogenesis | BL91 | Polyribonucleotide nucleotidyltransferase | pnpA | BL1546 | 58 | 5.08E + 20 | 4.21 | — |
| BL92 | 30S ribosomal protein S1 | rpsA | BL0992 | 74 | 2.05E + 13 | — | 2.09 | |
| BL19 | Glutamyl-tRNA synthetase | gltX | BL0469 | 66 | 1.27E + 18 | 1.61 | — | |
| BL93 | Tryptophanyl-tRNA synthetase | trpS | BL0600 | 23 | 3.86E + 03 | 1.76 | — | |
| BL94 | Elongation factor G | fusA | BL1098 | 65 | 1.39E + 19 | — | 2.29 | |
| BL38 | Probable 50S ribosomal protein L25 | rplY | BL0853 | 10 | 3.23E + 07 | −1.82 | 1.53 | |
| BL50 | Ribosome recycling factor | frr | BL1506 | 15 | 1.51E + 04 | −2.59 | — | |
| BL95 | 30S ribosomal protein S6 | rpsF | BL0416 | 82 | 3.70E + 04 | −8.80 | — | |
| RNA processing and modification | BL58 | Hypothetical RNA methyltransferase | BL1394 | 19 | 3.29E + 03 | −5.92 | — | |
| Transcription | BL56 | N utilization substance homolog | nusA | BL1615 | 57 | 4.42E + 08 | −1.82 | — |
| BL96 | Probable transcription antitermination protein | nusG | BL1288 | 41 | 2.97E + 07 | −2.15 | 2.49 | |
| Posttranslational modification, | BL97 | Chaperone protein | dnaJ | BL0719 | 56 | 1.11E + 10 | −3.01 | — |
| protein turnover, and chaperones | BL98 | GroES | groES | BL1558 | 68 | 4.18E + 02 | −2.29 | 2.02 |
| Cell envelope biogenesis, outer membrane | BL1 | BSH | bsh | BL0796 | 60 | 4.59E + 11 | −12.88 | — |
| DNA replication, | BL99 | RecA protein | recA | BL1415 | 14 | 6.29E + 02 | −2.97 | — |
| recombination, and repair | BL54 | Putative ATPase involved in DNA repair | BL0016 | 53 | 3.65E + 07 | 1.73 | 5.87 | |
| BL43 | DNA-directed RNA polymerase beta chain | rpoB | BL1205 | 47 | 7.89E + 22 | — | −1.87 | |
| Signal transduction mechanisms | BL45 | Response regulator of two-component system | BL0005 | 56 | 5.21E + 05 | −3.88 | — | |
| General function prediction only | BL52 | Probable ATP binding protein of ABC transporter | BL0870 | 71 | 3.11E + 08 | −3.88 | — |
Only spots which were over- or underproduced by a factor of greater than 1.5 are included.
COG, cluster of orthologous genes.
Spot numbers refer to the proteins labeled in Fig. 1.
Putative functions were assigned according to the cluster of orthologous genes for B. longum NCC2705.
Percentage of sequence coverage.
Molecular weight search score.
Ratio of WT strain value for growth at pH 4.8 to WT strain value for growth at pH 7 for each spot.
Ratio of mutant strain value for growth at pH 4.8 to mutant strain value for growth at pH 7 for each spot.
—, not significant.
Influence of low-pH adaptation on energy recycling under pH challenge.
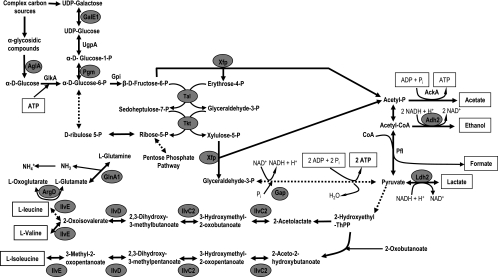
The production of several enzymes involved in different steps of glycolysis was found to be modulated by acid pH in both strains (Fig. 2). Alpha-1,4-glucosidase was overproduced in both strains. Furthermore, the WT strain also showed an increase in the production of phosphoglucomutase and UDP-glucose 4-epimerase. All these enzymes are related to the utilization of complex carbohydrate sources, and they fuel the bifid shunt. Interestingly, transaldolase was highly underproduced after growth at acidic pH in the WT (ratio, −29.19) whereas the mutant increased its amount by a factor of 5.56. Theoretically, this should favor the formation of glyceraldehyde 3-phosphate from fructose 6-phosphate faster in the mutant than in the WT strain. In accordance, a decrease of the glucose consumption was observed for the mutant strain when grown at pH 4.8, together with a moderate increase of the total carbon balance of the bifid shunt (Table 4). However, in the WT strain no noticeable changes in these two features were detected.
FIG. 2.
Schematic representation of the carbon catabolic pathway, the bifid shunt, and the BCAA metabolism. Enzymes whose production was modified after adaptation or exposure to acid pH in B. longum are marked with a gray circle.
TABLE 4.
Glucose consumed; ethanol and lactic, formic, and acetic acid production; acetic/lactic acid ratio; and carbon balance of buffered resting cells of B. longum biotype longum NCIMB 8809 and B. longum biotype longum 8809dpH grown in MRSC adjusted to pH 7.0 or pH 4.8a
| Strain and condition | Glucose consumed (mM) | Organic acid formation (mM)
|
Ethanol formation (mM) | Acetic/lactic ratio | Carbon balance | ||
|---|---|---|---|---|---|---|---|
| Lactic acid | Formic acid | Acetic acid | |||||
| NCIMB 8809 | |||||||
| pH 7 | 6.235 ± 0.281 | 3.936 ± 0.270 | 0.000 ± 0.000 | 9.190 ± 0.692 | 0.204 ± 0.037 | 2.334 ± 0.041 | 0.811 ± 0.026 |
| pH 4.8 | 6.461 ± 0.527 | 4.168 ± 0.235 | 0.459 ± 0.022 | 9.646 ± 0.519 | 0.205 ± 0.056 | 2.314 ± 0.022 | 0.838 ± 0.042 |
| 8809dpH | |||||||
| pH 7 | 6.470 ± 0.337 | 3.190 ± 0.251 | 0.214 ± 0.010 | 7.808 ± 0.499 | 0.233 ± 0.012 | 2.459 ± 0.246 | 0.659 ± 0.012 |
| pH 4.8 | 3.621 ± 1.096 | 2.694 ± 0.391 | 0.503 ± 0.073 | 5.424 ± 0.493 | 0.240 ± 0.064 | 2.025 ± 0.106 | 0.792 ± 0.048 |
Values are expressed as means ± standard deviations of at least three independent experiments.
Impact of low pH on the BSH of B. longum biotype longum.
BSH was constitutively down-regulated in the mutant, as well as in the WT, as a response to acidic pH (Tables 2 and 3). Furthermore, cytoplasmic BSH activity followed the same pattern, correlating with the enzyme levels. Indeed, at pH 7.0 the BSH activity is constitutively diminished in cell extracts of the mutant (2.73 ± 0.41 U/mg protein) compared with the WT (10.55 ± 2.79 U/mg protein) and decreases in the WT at pH 4.8 (2.75 ± 1.72 U/mg protein) but not in the mutant (3.11 ± 1.04 U/mg protein).
Impact of low pH on the physiology of B. longum biotype longum.
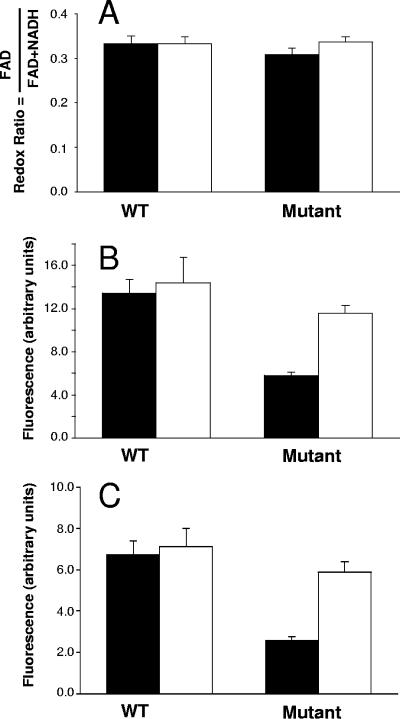
Proteomic analysis showed several changes in the levels of enzymes of pyruvate catabolism (Tables 2 and 3). The production of aldehyde-alcohol dehydrogenase 2 was diminished after growth at pH 4.8 by a factor of 17.42 in B. longum biotype longum NCIMB 8809 and by a factor of 3.61 in the mutant, while lactate dehydrogenase (Ldh2) in the same conditions was up-regulated in both strains. No changes in the production of pyruvate formate-lyase (Pfl) or acetate kinase (AckA) were detected. In bifidobacteria, pyruvate is mainly transformed into lactic and acetic acids, giving a theoretical final acetic/lactic ratio of 3:2. Pyruvate can also theoretically be transformed to formate by pyruvate formate-lyase. Our proteomic data indicate that the exposure to low pH in B. longum (WT and mutant) predominantly directed the reduction of pyruvate to lactate by Ldh2. These changes may have some consequences for the various end products generated from pyruvate and may thus also affect the regeneration of NAD+ and the NADH/NAD+ ratio. Therefore, we measured the different end products, lactic, formic, and acetic acids as well as ethanol (Table 4), and the fluctuations of intracellular pools of NADH and FAD+ (Fig. 3), whose fluorescence could be monitored under in vivo conditions (1). We did not detect a strong modification of the acetic/lactic acid ratio and of the ethanol production. However, we observed an increase in formate production in both strains after exposure to low pH. The WT strain did not produce any detectable formic acid at pH 7.0, whereas the mutant produced 0.214 ± 0.01 mM. At low pH the two strains produced similar amounts of formate (Table 4). As shown in Fig. 3, the growth of the WT and the mutant at pH 4.8 did not influence the global redox ratio (FAD/[FAD + NADH]), although in the case of the mutant strain the quantities of both FAD+ and NADH increased significantly. This was correlated with the preservation of the relationship between acetic and lactic acid production (Table 4).
FIG. 3.
Redox ratios (A), NADH-associated fluorescence (B), and FAD-associated fluorescence (C) of B. longum biotype longum NCIMB 8809 and B. longum biotype longum 8809dpH grown at neutral pH (solid bars) or acid pH (white bars). Vertical lines on the bars represent standard deviations.
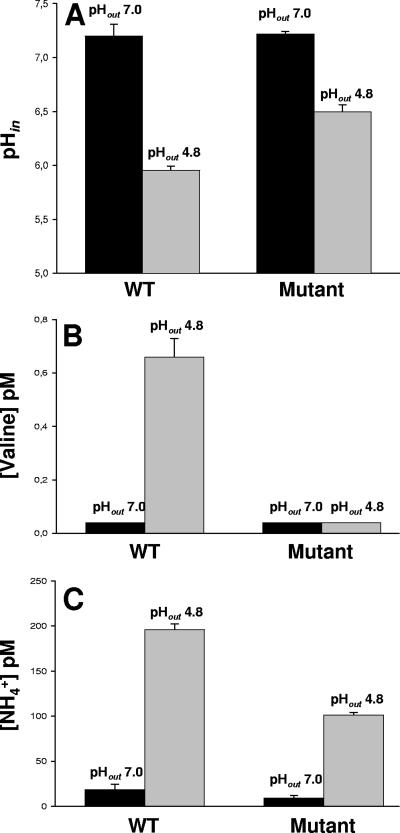
Several enzymes involved in amino acid metabolism are overproduced at low pH in the WT, some are also induced in the mutant, and one is up-regulated by low pH only in the mutant (Table 3). Among those, we observed IlvC2, IlvD, and IlvE, which are involved in the biosynthesis pathway of branched-chain amino acids (BCAA), and the glutamine synthetase GlnA1. As seen in Fig. 2, pyruvate can be driven to the formation of BCAA. Deamination of BCAA has been postulated as a mechanism of maintaining the internal pH of the cells (27). We have thus measured internal pH as well as the BCAA and NH4+ concentrations in cytoplasmic extracts of the WT and the mutant strains grown at neutral and acidic pHs (Fig. 4). A higher concentration of valine in B. longum biotype longum NCIMB 8809, together with a sharp increase of the ammonium content in both strains, was detected after growth at pH 4.8. This increase was less pronounced in the mutant. Moreover, acetylornithine aminotransferase (ArgD; overproduced in the WT) catalyzes a reaction that could serve as a way of recycling the 2-oxoglutarate formed by the BCAA aminotransferase as well as feeding the glutamine synthesis. Remarkably, the overproduction of glutamyl-tRNA synthetase detected in the WT under acidic conditions could also be related to this fact.
FIG. 4.
(A) Intracellular pH of B. longum cells (grown in MRSC at pH 7.0) after suspension in buffer at pH 7.0 or pH 4.8. (B and C) Valine concentration (B) and NH4+ concentration (C) measured in cell extracts of B. longum biotype longum NCIMB 8809 and B. longum biotype longum 8809dpH grown at neutral pH (black bars) or acid pH (grey bars).
Finally, measurements of intracellular pH (pHin) values at two different external pHs (pHout) were recorded for the WT and the mutant strains (Fig. 4). As expected, the higher the pHout, the higher the pHin that was observed. Remarkably, when cells were grown at pH 7.0 and they were put in contact with a pHout of 4.8, the WT was not able to maintain the pHin above 6 whereas the mutant maintained it above 6.5. No significant differences between WT and mutant strains were observed when cells were grown at pH 4.8 (data not shown). Agreeing with this, we found the F1Fo-ATPase subunits AtpA and AtpD in higher amounts under acidic conditions in both strains, although the increase was much more pronounced in the mutant strain (Table 3).
DISCUSSION
Acid resistance has been proposed as a selection marker for potential probiotic bifidobacteria (8, 9, 38). To understand the mechanisms underlying this process, molecular techniques for disrupting genes and controlling gene expression have been extensively used. However, the lack of efficient transformation systems and effective molecular tools for gene inactivation severely limits functional studies in Bifidobacterium species (50). In the present work we have undertaken this challenge by isolating an acid-resistant mutant of a strain of B. longum biotype longum.
To unravel the functions involved in acid response in B. longum and to search for those responsible for the acid-resistant phenotype of the mutant, we chose a proteomic approach. Three proteins involved in the synthesis of the sulfur amino acids methionine and cysteine, MetE, CysD, and MetB, were strongly overexpressed in the mutant, indicating the impact that the mutation(s) produced could have on sulfur metabolism in B. longum biotype longum 8809dpH.
It has been suggested that B. longum cysteine biosynthesis and sulfur assimilation may be accomplished by an atypical pathway involving homologs of cystathionine γ-synthase, cystathionine β-synthetase, and cystathionine γ-lyase, using succinylhomoserine and the H2S or methanethiol produced by other colonic microbiota as substrates (45). However, we have to take into account that the mutant strain was obtained by natural selection, and the large number of proteins identified as being differentially produced between the WT and the mutant point to a mutation(s) in a global regulation system rather than in an individual gene. Therefore, the pleiotropically affected genes might partially mask some of the genes directly involved in acid resistance.
Remarkably, growth rates were higher for the mutant than for the WT in the presence of bile, which pointed to an association between the acquisition of resistance to acid pH and bile salt tolerance in B. longum, as postulated before for other Bifidobacterium strains (34, 41, 43). Related to this, this work shows that BSH was underproduced in the mutant and underproduced in the WT under acidic conditions. This enzyme is present in many bacterial species of the gastrointestinal tract and catalyzes the release of the amino acid moiety from the conjugated bile salt, rendering deconjugated bile acids. Its activity is one of the positive factors for probiotic selection (3), and its function has been extensively studied (6, 11, 23, 24, 47), but its physiological role in probiotic bacteria remains unclear. Deconjugated bile salts are more toxic than the conjugated ones for Bifidobacterium (18, 33), due to their higher detergent activity. Thus, the lower BSH activity detected as a result of the response of the WT and the adaptation of the mutant to low pH in Bifidobacterium could represent a way of counteracting bile toxicity. Interestingly, the BSH-encoding gene (BL0796) is located downstream from metF (BL0797) and metE (BL0798). As mentioned above, MetE is sensitive to low pH and regulated in the opposite way from BSH. We cannot thus yet exclude the possibility that those genes belong to a gene cluster that is pH regulated.
Two of the main changes in protein production were noted for proteins involved in nucleotide metabolism and in transcription/translation. Noticeably, 5′-methylthioadenosine/S-adenosylhomocysteine (MTA/SAH) nucleosidase displayed the highest down-regulation ratio at pH 4.8 (−285.4) in the WT, although such a change could not be detected in the mutant. MTA/SAH nucleosidase is a dual substrate-specific enzyme that plays a key role in regulating the cellular levels of the metabolites MTA and SAH, products involved in bacterial intercellular communication in Salmonella enterica serovar Typhimurium 14028 and other gram-negative bacteria (4). In relation to transcriptional/translational processes, differences in behavior between the two strains were observed. Whereas as a response to low pH the strain NCIMB 8809 decreased the amount of the ribosomal protein homologs RplY, Frr, and RpsF, the mutant increased the production of FusA, RpsA, and RplY. The production of some ribosomal proteins has been shown to be dependent on several stress factors in gram-positive bacteria, such as osmotic shock, phosphate and amino acid starvation, and ethanol and heat treatment (2, 14, 15). Concerning chaperone proteins, GroES was overproduced in the mutant at low pH whereas the WT showed a diminution in the amount of both GroES and DnaJ. It has been shown that these proteins are up-regulated when the acid tolerance response is achieved, for example, in Propionibacterium freudenreichii, where the overproduction of GroES and GroEL corresponded with a late response to acid pH (20). Those findings agree with our own results as long as chaperone overproduction can be detected only in the late response to acid pH, represented in our model of study by the mutant strain.
On the other hand, the slightly more efficient carbon utilization by B. longum biotype longum 8809dpH under acidic conditions suggests that carbons coming from the carbohydrate metabolism may be more efficiently driven to acid formation. The subsequent ATP formation at the substrate level may be directed to a higher yield in the ATP production of the cell, which could feed the mechanisms of proton extrusion mediated by the F1Fo-ATPase. The maintenance of internal pH in bacteria lacking a respiratory chain, such as Bifidobacterium, is mainly due to this enzyme (30, 43, 49). The overproduction of the two intracellular subunits of the F1Fo-ATPase in the WT and mutant strains under acidic conditions strongly points to the involvement of this enzyme in the acid response of B. longum, which is in good agreement with the measurements of the intracellular pH. In this respect, it is worthwhile to remark that the higher NH4+ concentration detected in both strains at low pH may help to buffer the internal pH (27). Furthermore, the higher capacity of the mutant strain to overproduce AtpA and AtpD, together with the lower levels of ammonium and valine in its cytoplasm under low-pH conditions, with respect to the WT, is likely due to the acquisition of different mechanisms and mutations during the adaptation process.
In conclusion, acid adaptation and response to acid pH involve different mechanisms in B. longum biotype longum. We suggest that the response of this bacterium to acid stress involves a number of changes in the levels of different proteins that are jointly dedicated to reduce the impact of acidic conditions in the cell physiology, mainly by controlling the intracellular pH through different mechanisms. Some differences in how the WT or the mutant strain overcomes the acidic environment were evidenced, which could reflect the preadaptation of the mutant strain to the acidic challenge. As a response to low pH, the decrease of the levels of the enzyme BSH and its activity and the overproduction of enzymes related to the utilization of complex carbohydrates may indicate that a pH challenge triggers a response that might allow adaptation for further environmental stresses encountered in the gastrointestinal tract. This study establishes the basis for understanding the low-pH response and adaptation in the probiotic bacterium B. longum.
Acknowledgments
This work was financed by European Union FEDER funds and the Spanish Plan Nacional de I + D (project AGL2004-06727-C02-01/ALI and AGL2005-05788-C02-01/ALI). B. Sánchez was the recipient of a FPI predoctoral fellowship from the Spanish Ministerio de Ciencia y Tecnología and from the MICA department of the National Institute for Research in Agronomy (INRA).
Matrix-assisted laser desorption ionization-time of flight mass spectrometry experiments were performed on the PAPPS platform of the INRA Center at Jouy en Josas. We thank Patricia Ruas Madiedo (IPLA-CSIC) for her help in several aspects of this work and Ana Hernández Barranco (IPLA-CSIC) for technical assistance.
Footnotes
Published ahead of print on 24 August 2007.
REFERENCES
- 1.Ammor, S., K. Yaakoubi, I. Chevallier, and E. Dufour. 2004. Identification by fluorescence spectroscopy of lactic acid bacteria isolated from a small-scale facility producing traditional dry sausages. J. Microbiol. Methods 59:271-281. [DOI] [PubMed] [Google Scholar]
- 2.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araya, M., L. Morelli, G. Reid, M. E. Sanders, and C. Stanton. 2002. Guidelines for the evaluation of probiotics in foods. FAO/WHO report. Food and Agriculture Organization of the United Nations and World Health Organization, Geneva, Switzerland. http://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf.
- 4.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begley, M., C. G. Gahan, and C. Hill. 2002. Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 68:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begley, M., C. Hill, and C. G. Gahan. 2006. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72:1729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champagne, C. P., N. J. Gardner, and D. Roy. 2005. Challenges in the addition of probiotic cultures to foods. Crit. Rev. Food Sci. Nutr. 45:61-84. [DOI] [PubMed] [Google Scholar]
- 8.Collado, M. C., and Y. Sanz. 2006. Method for direct selection of potentially probiotic Bifidobacterium strains from human feces based on their acid-adaptation ability. J. Microbiol. Methods 66:560-563. [DOI] [PubMed] [Google Scholar]
- 9.Collado, M. C., M. Gueimonde, Y. Sanz, and S. Salminen. 2006. Adhesion properties and competitive pathogen exclusion ability of bifidobacteria with acquired acid resistance. J. Food Prot. 69:1675-1679. [DOI] [PubMed] [Google Scholar]
- 10.Collado, M. C., Y. Moreno, E. Hernández, J. M. Cobo, and M. Hernández. 2005. In vitro viability of Bifidobacterium strains isolated from commercial dairy products exposed to human gastrointestinal conditions. Food Sci. Technol. Int. 11:307-314. [Google Scholar]
- 11.Corzo, G., and S. E. Gilliland. 1999. Bile salt hydrolase activity of three strains of Lactobacillus acidophilus. J. Dairy Sci. 82:472-480. [DOI] [PubMed] [Google Scholar]
- 12.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, M. J., P. J. Coote, and C. P. O'Byrne. 1996. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology 142:2975-2982. [DOI] [PubMed] [Google Scholar]
- 14.Duche, O., F. Tremoulet, A. Namane, and J. Labadie. 2002. A proteomic analysis of the salt stress response of Listeria monocytogenes. FEMS Microbiol. Lett. 215:183-188. [DOI] [PubMed] [Google Scholar]
- 15.Eymann, C., and M. Hecker. 2001. Induction of sigma(B)-dependent general stress genes by amino acid starvation in a spo0H mutant of Bacillus subtilis. FEMS Microbiol. Lett. 199:221-227. [DOI] [PubMed] [Google Scholar]
- 16.Flahaut, S., A. Hartke, J. C. Giard, A. Benachour, P. Boutibonnes, and Y. Auffray. 1996. Relationship between stress response towards bile salts, acid and heat treatment in Enterococcus faecalis. FEMS Microbiol. Lett. 138:49-54. [DOI] [PubMed] [Google Scholar]
- 17.Flahaut, S., J. Frere, P. Boutibonnes, and Y. Auffray. 1997. Relationship between the thermotolerance and the increase of DnaK and GroEL synthesis in Enterococcus faecalis ATCC19433. J. Basic Microbiol. 37:251-258. [DOI] [PubMed] [Google Scholar]
- 18.Grill, J. P., S. Perrin, and F. Schneider. 2000. Bile salt toxicity to some bifidobacteria strains: role of conjugated bile salt hydrolase and pH. Can. J. Microbiol. 46:878-884. [DOI] [PubMed] [Google Scholar]
- 19.Gueimonde, M., S. Delgado, B. Mayo, P. Ruas-Madiedo, A. Margolles, and C. G. de los Reyes-Gavilán. 2004. Viability and diversity of probiotic Lactobacillus and Bifidobacterium populations included in commercial fermented milks. Food Res. Int. 37:839-850. [Google Scholar]
- 20.Jan, G., P. Leverrier, V. Pichereau, and P. Boyaval. 2001. Changes in protein synthesis and morphology during acid adaptation of Propionibacterium freudenreichii. Appl. Environ. Microbiol. 67:2029-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayamanne, V. S., and M. R. Adams. 2006. Determination of survival, identity and stress resistance of probiotic bifidobacteria in bio-yoghurts. Lett. Appl. Microbiol. 42:189-194. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann, P., A. Pfefferkorn, M. Teuber, and L. Meile. 1997. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl. Environ. Microbiol. 63:1268-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, G. B., C. M. Miyamoto, E. A. Meighen, and B. H. Lee. 2004. Cloning and characterization of the bile salt hydrolase genes (bsh) from Bifidobacterium bifidum strains. Appl. Environ. Microbiol. 70:5603-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, G. B., M. Brochet, and B. H. Lee. 2005. Cloning and characterization of a bile salt hydrolase (bsh) from Bifidobacterium adolescentis. Biotechnol. Lett. 27:817-822. [DOI] [PubMed] [Google Scholar]
- 25.Kirkpatrick, N. D., C. Zou, M. A. Brewer, W. R. Brands, R. A. Drezek, and U. Utzinger. 2005. Endogenous fluorescence spectroscopy of cell suspensions for chemopreventive drug monitoring. Photochem. Photobiol. 81:125-134. [DOI] [PubMed] [Google Scholar]
- 26.Krause, I., A. Bockhardt, H. Neckermann, T. Henle, and H. Klostermeyer. 1995. Simultaneous determination of amino-acids and biogenic-amines by reversed-phase high-performance liquid-chromatography of the dabsyl derivatives. J. Chromatogr. A 715:67-79. [Google Scholar]
- 27.Len, A. C., D. W. Harty, and N. A. Jacques. 2004. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiology 150:1353-1366. [DOI] [PubMed] [Google Scholar]
- 28.Masco, L., G. Huys, E. de Brandt, R. Temmerman, and J. Swings. 2005. Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int. J. Food Microbiol. 102:221-230. [DOI] [PubMed] [Google Scholar]
- 29.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Kado, T. Takada, K. Matsumoto, and R. Tanaka. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto, M., H. Ohishi, and Y. Benno. 2004. H+-ATPase activity in Bifidobacterium with special reference to acid tolerance. Int. J. Food Microbiol. 93:109-113. [DOI] [PubMed] [Google Scholar]
- 31.Mättö, J., E. Malinen, M. L. Suihko, M. Alander, A. Palva, and M. Saarela. 2004. Genetic heterogeneity and functional properties of intestinal bifidobacteria. J. Appl. Microbiol. 97:459-470. [DOI] [PubMed] [Google Scholar]
- 32.Maus, J. E., and S. C. Ingham. 2003. Employment of stressful conditions during culture production to enhance subsequent cold- and acid-tolerance of bifidobacteria. J. Appl. Microbiol. 95:146-154. [DOI] [PubMed] [Google Scholar]
- 33.Noriega, L., I. Cuevas, A. Margolles, and C. G. de los Reyes-Gavilán. 2006. Deconjugation and bile salts hydrolase activity by Bifidobacterium strains with acquired resistance to bile. Int. Dairy J. 16:850-855. [Google Scholar]
- 34.Noriega, L., M. Gueimonde, B. Sánchez, A. Margolles, and C. G. de los Reyes-Gavilán. 2004. Effect of the adaptation to high bile salts concentrations on glycosidic activity, survival at low pH and cross-resistance to bile salts in Bifidobacterium. Int. J. Food Microbiol. 94:79-86. [DOI] [PubMed] [Google Scholar]
- 35.O'May, G. A., N. Reynolds, A. R. Smith, A. Kennedy, and G. T. Macfarlane. 2005. Effect of pH and antibiotics on microbial overgrowth in the stomachs and duodena of patients undergoing percutaneous endoscopic gastrostomy feeding. J. Clin. Microbiol. 43:3059-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Riordan, K., and G. F. Fitzgerald. 1998. Evaluation of bifidobacteria for the production of antimicrobial compounds and assessment of performance in cottage cheese at refrigeration temperature. J. Appl. Microbiol. 85:103-114. [DOI] [PubMed] [Google Scholar]
- 37.O'Sullivan, E., and S. Condon. 1999. Relationship between acid tolerance, cytoplasmic pH, and ATP and H+-ATPase levels in chemostat cultures of Lactococcus lactis. Appl. Environ. Microbiol. 65:2287-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouwehand, A. C., S. Salminen, and E. Isolauri. 2002. Probiotics: an overview of beneficial effects. Antonie Leeuwenhoek 82:279-289. [PubMed] [Google Scholar]
- 39.Pappin, D. J., P. Hojrup, and A. J. Bleasby. 1993. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 3:327-332. [DOI] [PubMed] [Google Scholar]
- 40.Ruas-Madiedo, P., A. Hernández-Barranco, A. Margolles, and C. G. de los Reyes-Gavilán. 2005. A bile salt-resistant derivative of Bifidobacterium animalis has an altered fermentation pattern when grown on glucose and maltose. Appl. Environ. Microbiol. 71:6564-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saarela, M., M. Rantala, K. Hallamaa, L. Nohynek, I. Virkajärvi, and J. Mättö. 2004. Stationary-phase acid and heat treatments for improvement of the viability of probiotic lactobacilli and bifidobacteria. J. Appl. Microbiol. 96:1205-1214. [DOI] [PubMed] [Google Scholar]
- 42.Salminen, S., and M. Gueimonde. 2004. Human studies on probiotics: what is scientifically proven? J. Food Sci. 69:M137-M140. [Google Scholar]
- 43.Sánchez, B., C. G. de los Reyes-Gavilán, and A. Margolles. 2006. The F1F0-ATPase of Bifidobacterium animalis is involved in bile tolerance. Environ. Microbiol. 8:1825-1833. [DOI] [PubMed] [Google Scholar]
- 44.Sánchez, B., M. C. Champomier-Vergès, P. Anglade, F. Baraige, C. G. de los Reyes-Gavilán, A. Margolles, and M. Zagorec. 2005. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187:5799-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, P. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamime, A. Y. 2002. Fermented milks: a historical food with modern applications—a review. Eur. J. Clin. Nutr. 56:S2-S15. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka, H., H. Hashiba, J. Kok, and I. Mierau. 2000. Bile salt hydrolase of Bifidobacterium longum—biochemical and genetic characterization. Appl. Environ. Microbiol. 66:2502-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tannock, G. W. 2002. The bifidobacterial and lactobacillus microflora of humans. Clin. Rev. Allergy Immunol. 22:231-253. [DOI] [PubMed] [Google Scholar]
- 49.Ventura, M., C. Canchaya, D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Bifidobacterium lactis DSM 10140: identification of the atp (atpBEFHAGDC) operon and analysis of its genetic structure, characteristics, and phylogeny. Appl. Environ. Microbiol. 70:3110-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ventura, M., D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Leeuwenhoek 86:205-223. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, K. H., R. B. Blitchington, and R. C. Greene. 1990. Amplification of bacterial-16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwietering, M. H., I. Jongenburger, F. M. Rombouts, and K. Vantriet. 1990. Modeling of the bacterial-growth curve. Appl. Environ. Microbiol. 56:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]