Abstract
The human intestine harbors both lactate-producing and lactate-utilizing bacteria. Lactate is normally present at <3 mmol liter−1 in stool samples from healthy adults, but concentrations up to 100 mmol liter−1 have been reported in gut disorders such as ulcerative colitis. The effect of different initial pH values (5.2, 5.9, and 6.4) upon lactate metabolism was studied with fecal inocula from healthy volunteers, in incubations performed with the addition of dl-lactate, a mixture of polysaccharides (mainly starch), or both. Propionate and butyrate formation occurred at pH 6.4; both were curtailed at pH 5.2, while propionate but not butyrate formation was inhibited at pH 5.9. With the polysaccharide mix, lactate accumulation occurred only at pH 5.2, but lactate production, estimated using l-[U-13C]lactate, occurred at all three pH values. Lactate was completely utilized within 24 h at pH 5.9 and 6.4 but not at pH 5.2. At pH 5.9, more butyrate than propionate was formed from l-[U-13C]lactate in the presence of polysaccharides, but propionate, formed mostly by the acrylate pathway, was the predominant product with lactate alone. Fluorescent in situ hybridization demonstrated that populations of Bifidobacterium spp., major lactate producers, increased approximately 10-fold in incubations with polysaccharides. Populations of Eubacterium hallii, a lactate-utilizing butyrate-producing bacterium, increased 100-fold at pH 5.9 and 6.4. These experiments suggest that lactate is rapidly converted to acetate, butyrate, and propionate by the human intestinal microbiota at pH values as low as 5.9, but at pH 5.2 reduced utilization occurs while production is maintained, resulting in lactate accumulation.
The metabolic activities of gut bacteria play an important role in human health and disease (22). The end products of bacterial fermentation are diet dependent, with acetate, propionate, and butyrate being major fermentation products that can provide up to 10% of the energy requirements of humans (35, 40). Butyrate, in particular, is the preferred energy source for colonocytes (20, 46) and is considered to protect against colorectal disease (3, 39, 43, 53). Lactate is also produced by many species of gut bacteria but does not normally accumulate in the colon. Furthermore, in fecal samples from healthy donors, lactate either is not detected or is present at low concentrations (<3 mmol liter−1) (15), due to further metabolism within the colon. Lactate does accumulate in certain gut disorders, however, e.g., ulcerative colitis, where concentrations up to 100 mmol liter−1 lactate have been reported elsewhere (29, 51). Colonic and fecal lactate concentration depends on the balance between bacterial production and host absorption plus microbial utilization. In farm animals it is well established that an imbalance can lead to lactic acidosis (1, 50).
Low-mol%-G+C gram-positive bacteria and the gram-negative Bacteroidetes are normally the most dominant bacterial groups present in the human colonic microbiota (16, 17, 25, 27, 32). Many of these bacteria are capable of producing lactate as one of several fermentation products. Certain bacterial groups produce lactate as a major product, however, including high-mol%-G+C-content gram-positive Bifidobacterium spp. and lactic acid bacteria such as Lactobacillus and Enterococcus spp. (10). The abundance of bifidobacteria is usually approximately 2 to 4% of total colonic bacteria but with much interindividual variation (15, 32). Acetate, butyrate, and propionate are potential end products of lactate metabolism in the human colon (7, 13, 42). Propionate-producing bacteria, which include Veillonella sp. and Megasphaera elsdenii, have been shown to utilize lactate, converting it mainly to propionate, in the gut of farm animals (9, 24), and related species are assumed to have a similar role in the human large intestine (26, 49). Recently, however, additional lactate-utilizing bacteria have also been identified and isolated from human feces that produce butyrate from lactate (13); these include Eubacterium hallii and Anaerostipes caccae, which form part of the low-mol%-G+C gram-positive bacterial population (23, 28).
Environmental conditions are likely to have an impact on lactate metabolism and may influence the relative proportions of lactate producers and utilizers present. Studies with a continuous-flow fermentor have shown that pH alters the end products of fermentation and the abundance of bacterial populations (52). The pH of the proximal colon has been reported to be mildly acidic in healthy subjects consuming nondigestible carbohydrates (8, 37) that are actively fermented, while stool samples from patients with active ulcerative colitis often have lower pH (44, 51). Although many lactate-producing bacteria are considered to be tolerant of acidic pH (45), less is known about the sensitivity of lactate utilizers. A differential response to pH between producers and utilizers would explain why lactate may accumulate in the colon in certain disease states.
The aim of this work, therefore, was to investigate both lactate formation from mixed dietary polysaccharides by fecal microbes at different pHs and the capacity for lactate removal by lactate-utilizing bacteria. To this end, lactate fluxes were quantified by use of stable isotopes and 16S rRNA-targeted fluorescent in situ hybridization (FISH) probes were employed to track the populations of potentially relevant groups of gut bacteria.
MATERIALS AND METHODS
Collection and preparation of fecal samples.
Fecal samples were provided by four adult volunteers (two male and two female), aged 31 to 61 years and all consuming a Western-style diet. The volunteers (referred to as donors A, B, C, and D) did not take any antibiotics or drugs known to influence fecal microbiota for the last 6 months prior to the start of the studies. For the batch culture incubations a fecal slurry (20%) was prepared within 2 h of collection in anaerobic phosphate-buffered saline (0.14 M NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.8 mM KH2PO4, and 0.05% cysteine HCl; pH 7.4) and 0.1 ml slurry was added to 9.0 ml medium under anaerobic conditions to give a final concentration of approximately 0.2%.
Batch culture incubations.
Batch culture incubations were performed using an anaerobic medium based on the work of Macfarlane et al. (36) as modified by Walker et al. (52). The carbohydrate sources present in the mixed substrate medium were potato starch (0.14%, wt/vol) in addition to xylan, pectin, amylopectin, and arabinogalactan each at 0.015% (wt/vol). Three different substrate conditions were studied: a mixture of carbohydrates (total of 0.2%, as described above), a mixture of carbohydrates plus dl-lactate (11 to 14 mmol liter−1), and dl-lactate alone (11 to 14 mmol liter−1). The total peptide concentrations (comprising equal amounts of casein hydrolysate and peptone water) were 0.2%. Bile salts were added at 0.005%, and the medium was buffered by the addition of 0.32% NaHCO3 and reduced by the addition of 0.05% cysteine HCl. Short-chain fatty acids (SCFA) were also added to the medium to give initial concentrations of approximately 35 mmol liter−1 acetate; 9 mmol liter−1 propionate; 5 mmol liter−1 butyrate; and 1 mmol liter−1 each of valerate, isovalerate, and isobutyrate. For the lactate metabolism studies a solution of l-[U-13C]lactate was added initially to the medium to give approximately 8 molar % excess (MPE). The fermentor medium was boiled for 10 to 15 min to drive off oxygen, prior to addition of the reducing agent cysteine-HCl, and then dispensed into Hungate tubes under a stream of CO2 (41). The medium was heat sterilized at 121°C (15 min). After the medium was cooled, heat-labile vitamins were added and the medium was inoculated with the fecal slurry under CO2 and incubated at 37°C. Tubes were inoculated in duplicate, and samples were taken from the same tube at 0, 3, 6, and 24 h to measure SCFA and lactate concentrations and 13C enrichments. This experiment was performed in media that had been adjusted to three (mean ± standard deviation, 5.2 ± 0.2, 5.9 ± 0.2, and 6.4 ± 0.2) different initial pH values.
Enumeration of bacteria in the fecal and batch culture samples by FISH analysis.
Samples were taken from the feces (0.5 g) and the batch culture incubations (1 ml) after 24 h for FISH analysis. Fecal samples were diluted with phosphate buffer, and both diluted feces and incubation samples were fixed by being mixed 1:3 in 4% (wt/vol) paraformaldehyde at 4°C for 16 h and stored at −20°C. FISH analysis was performed as described by Harmsen et al. (23). Diluted cell suspensions were applied to gelatin-coated slides. A total of 10 μl of a 50-ng μl−1 concentration of the oligonucleotide probes plus 100 μl of hybridization buffer was added, and the slides were hybridized overnight. To prevent fading of fluorescence, 50 μl of Vectashield (Vector Laboratories, Burlingame, CA) was applied to each slide. Cells were counted automatically using image analysis software (12) with a Leica DMRXA epifluorescence microscope, except when the number of cells was less than 10 per field of view, in which case the cells were counted manually. For each sample 25 microscopic fields were counted. The samples were all assessed with the following probes: Bac303 (38), Erec482 (19), Bif164 (31), Prop853 (52), Ehal1469 (23), and Acac194 (28). Total bacterial numbers were estimated by using the universal probe Eub338 (2).
Determination of concentrations and 13C enrichments in SCFA and lactate.
Duplicate derivatized samples were routinely prepared for estimation of concentrations of SCFA and lactate by capillary gas chromatography (GC) (47). l-Lactate concentrations were analyzed enzymatically in those tubes containing the medium with the mixture of carbohydrates and incubated at pH 5.2. d-Lactate was then determined by the difference between total dl-lactate, determined by GC, and l-lactate concentrations. Lactate and SCFA enrichments and lactate concentrations, estimated by isotope dilution, were measured by GC-mass spectrometry analysis of the tert-butyldimethylsilyl derivatives. Procedures were performed as described previously (5, 14). The mass spectrometer was operated under electron impact ionization conditions. For acetate, the ions M+, M + 1, and M + 2 at mass/charge ratios (m/z) of 117, 118, and 119, respectively, were monitored; for butyrate, M+, M + 1, M + 2, and M + 4 (i.e., m/z 145, 146, 147, and 149, respectively) were determined, the last to quantify butyrate formation from two [1,2-13C]acetate molecules; for propionate, M+, M + 1, M + 2, and M + 3 (i.e., m/z 131, 132, 133, and 134, respectively) were measured, while for lactate, M+, M + 1, and M + 3 ion fragments were analyzed (m/z 261, 262, and 264, respectively). For the concentration determinations, appropriate corrections were applied for the enrichments in the sample.
Kinetic modeling of lactate metabolism.
Let C and E denote the lactate concentration (mmol liter−1) and enrichment (MPE), respectively. Let i denote the interval between any two times t0 and t1, and let Fl(i) denote the flow of labeled and unlabeled lactate during i. E(i) denotes the average enrichment during i. As the system was not in steady state, inflows (subscript “in”) and outflows (subscript “out”) to the lactate pool were calculated separately; therefore, at any time point inflows and outflows may differ.
Lactate formation (Flin) and utilization (Flout) are obtained from the changes in labeled and total (labeled plus unlabeled) lactate: E(t1) C(t1) = E(t0) C(t0) − E(i) Flout(i) and C(t1) = C(t0) − Flout(i) + Flin(i).
Occasionally, Flin and Flout were slightly negative, and here they were set equal to zero. Lactate utilization at 24 h was estimated assuming no lactate formation from 6 to 24 h.
Statistical analysis.
SCFA data were averaged over duplicate tubes. Data were analyzed for each time point separately. The SCFA data and bacterial log counts were analyzed as two-way analysis of variance (ANOVA) with volunteer as random effect and substrate, pH, and their interaction as treatment effects. In order to compare treatment means, post-hoc t tests, based on the least significant difference, were performed.
In addition, linear relationships between the increase in bacterial counts of Eubacterium hallii and Anaerostipes caccae over 24 h and butyrate production and lactate utilization were investigated for data from pH 5.9 and 6.4 (at pH 5.2 there was little change in the abundances of E. hallii and A. caccae and lactate utilization was small or negligible, and so this pH was excluded from analyses). For butyrate production, data from all three substrate conditions were used, whereas for lactate utilization only the lactate and mixture of polysaccharides plus lactate substrates were used (as utilization was not measured for the polysaccharide mixture on its own). The effect of volunteer on intercept and slope was initially included as a fixed effect but was found not to be significant and was excluded from subsequent analyses. P < 0.05 was regarded as statistically significant. All data were analyzed using Genstat, 8th edition, release 8.1 (VSN International Ltd., Hemel Hempstead, Herts, United Kingdom).
RESULTS
Investigation of the effects of pH and carbohydrate supply on lactate metabolism in batch cultures.
In vitro batch culture incubations were conducted following inoculation with fecal slurries from four different volunteers. For these experiments, parallel incubations were conducted with dietary polysaccharides (mainly potato starch) alone, with lactate alone, or with a combination of the two substrates (see Materials and Methods), and the effect of pH was studied by setting different initial pH values (5.2, 5.9, and 6.4) for the incubations. The pH remained stable in these incubations (difference between initial and final pH was <0.3), except when lactate was the only substrate present at pH 5.2, in which case pH increased (≤0.6 units). l-[U-13C]lactate was added to the incubations containing lactate (with or without mixed polysaccharide) in order to track lactate metabolism.
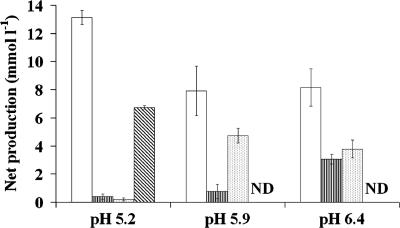
The 24-h incubation at the lowest pH (5.2) resulted in similar metabolite concentrations for samples from all four volunteers when only the polysaccharide mixture was present in the medium (Table 1; Fig. 1). The acetate/lactate ratio was approximately 3:2 in the absence of added lactate, with only small amounts of propionate and butyrate formed. Both d- and l-lactate enantiomers were formed, with l-lactate being dominant. The proportion of total lactate as d-lactate varied between volunteers (28.8, 8.8, 10.8, and 24.5% for volunteers A, B, C, and D, respectively). For the two lower pH values, addition of lactate to the polysaccharide mix resulted in reduced net formation of acetate (P < 0.001). Incubations conducted at pH 5.9 resulted in the formation mainly of butyrate and acetate, while pH 6.4 resulted in increased formation of propionate as well as butyrate relative to pH 5.2 (Table 1; Fig. 1). At the two higher pHs, 5.9 and 6.4, when lactate was added together with the polysaccharide mix, butyrate concentrations at 24 h were further increased by 2.8-fold on average. Net propionate formation also increased with pH but was greater in the presence of lactate alone for the two higher pHs (P < 0.001; Table 1). Although the responses to the type of substrate and pH were similar for the volunteers, there were significant differences between volunteers for net acetate (P < 0.001), butyrate (P < 0.001), and propionate (P = 0.014) concentrations.
TABLE 1.
Net change in concentrations of lactate (L), acetate (A), propionate (P), and butyrate (B) after 24 h of incubation in batch cultures containing a mix of carbohydrates, lactate, or both and inoculated with fecal slurries from four different volunteersa
| Substrate | pH | Net change in concn (mmol liter−1) of acid
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Volunteer A
|
Volunteer B
|
Volunteer C
|
Volunteer D
|
Mean of all volunteers
|
|||||||||||||||||
| L | A | P | B | L | A | P | B | L | A | P | B | L | A | P | B | L | A | P | B | ||
| Mixture | 5.2 | 6.6 | 12.0 | 0.2 | 0.1 | 7.1 | 13.9 | 0.1 | 0.1 | 6.8 | 14.1 | 0.4 | 0.5 | 6.5 | 12.6 | 0.9 | 0.2 | 6.7 | 13.1 | 0.4 | 0.2 |
| 5.9 | 0.0 | 7.7 | 0.4 | 4.8 | 0.0 | 13.0 | 0.3 | 4.0 | 0.0 | 5.5 | 0.2 | 6.2 | 0.0 | 5.4 | 2.3 | 3.9 | 0.0 | 7.9 | 0.8 | 4.7 | |
| 6.4 | 0.0 | 7.8 | 2.3 | 4.5 | 0.0 | 11.9 | 2.9 | 2.7 | 0.0 | 5.6 | 3.0 | 5.2 | 0.0 | 7.3 | 4.0 | 2.7 | 0.0 | 8.2 | 3.1 | 3.8 | |
| Mixture plus | 5.2 | 5.9 | 7.4 | −0.8 | −0.4 | 6.3 | 8.4 | −0.9 | −0.4 | 6.2 | 7.8 | −0.8 | −0.2 | 6.1 | 7.1 | −0.4 | −0.4 | 6.1 | 7.7 | −0.7 | −0.4 |
| lactate | 5.9 | −11.4 | 2.0 | 0.3 | 12.6 | −12.4 | 7.2 | 0.8 | 11.2 | −10.8 | 0.8 | 0.8 | 14.0 | −10.8 | 2.2 | 3.1 | 10.4 | −11.4 | 2.8 | 1.2 | 12.1 |
| 6.4 | −12.9 | 5.9 | 2.9 | 13.2 | −12.8 | 15.6 | 5.7 | 7.8 | −13.1 | 4.7 | 3.9 | 13.5 | −12.8 | 9.9 | 6.9 | 10.2 | −12.9 | 9.0 | 4.9 | 11.2 | |
| Lactate | 5.2 | −3.4 | −4.1 | −0.9 | 1.6 | −0.4 | 1.3 | 0.2 | 0.4 | −7.8 | −2.3 | 0.4 | 5.2 | −1.5 | 1.0 | 0.6 | 0.9 | −3.3 | −1.0 | 0.1 | 2.0 |
| 5.9 | −11.7 | 1.7 | 3.2 | 7.1 | −11.7 | 4.5 | 5.7 | 4.6 | −11.4 | 2.7 | 4.3 | 6.3 | −11.6 | 4.9 | 4.9 | 4.3 | −11.6 | 3.5 | 4.5 | 5.6 | |
| 6.4 | −12.1 | 6.6 | 7.7 | 3.3 | −12.1 | 6.7 | 7.1 | 1.8 | −12.2 | 4.3 | 6.2 | 3.0 | −12.1 | 6.0 | 5.4 | 2.5 | −12.1 | 5.9 | 6.6 | 2.6 | |
| SEDb | 0.81 | 1.35 | 0.60 | 0.71 | |||||||||||||||||
|
P valuec
|
||||
|---|---|---|---|---|
| L | A | P | B | |
| pH | <0.001 | <0.003 | <0.001 | <0.001 |
| Substrate | <0.001 | <0.001 | <0.001 | <0.001 |
| pH × substrate | <0.001 | <0.001 | <0.001 | <0.001 |
Calculated C recoveries ranged from 84 to 119% for all incubations. Data were determined by GC-mass spectrometry for lactate and GC for acetate, propionate, and butyrate. Initial lactate concentrations ranged from 11 to 14 mmol liter−1 in the lactate-alone and mixture-plus-lactate cultures.
SED, standard error of the difference.
Analyzed as two-way ANOVA with random effect for volunteer and fixed effect for substrate, pH, and their interaction.
FIG. 1.
Effect of initial pH on net production of the major SCFA (acetate [open bars], propionate [vertically striped bars], and butyrate [dotted bars]) and lactate (diagonally striped bars) from 24-h batch culture incubations with a mixture of carbohydrates. Error bars indicate standard errors of the means. ND, not detected.
Rates of lactate formation and utilization.
Rates of lactate formation (Flin) and utilization (Flout) were estimated from the incubations with l-[U-13C]lactate (initial enrichment, 7.8 MPE) by examining 0-, 3-, 6-, and 24-h time points (Table 2). Lactate formation could be estimated up to 6 h for all treatments regardless of the rate of lactate utilization. The rate of lactate production at 6 h from the incubations with the polysaccharide mix plus lactate was greater at pH 5.9 than at pH 5.2 or 6.4 (P < 0.001). Lactate production at pH 5.2 was similar (6.6 to 6.8 mmol liter−1) for all four volunteers after 24 h when both polysaccharides and lactate were present (Table 2).
TABLE 2.
Total lactate production (Flin) and utilization (Flout) over time (3, 6, or 24 h) in batch cultures containing lactate, with or without a range of carbohydrates (mixture), and inoculated with fecal slurries from four different volunteers
| Substrate | pH | Time (h) | Lactate (mmol liter−1)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Volunteer A
|
Volunteer B
|
Volunteer C
|
Volunteer D
|
Mean of all volunteers
|
||||||||
| Flin | Flout | Flin | Flout | Flin | Flout | Flin | Flout | Flin | Flout | |||
| Mixture plus lactate | 5.2 | 3 | 0.42 | 0.02 | 0.22 | 0.01 | 0.21 | 0.00 | 0.39 | 0.08 | 0.31 | 0.03 |
| 6 | 2.13 | 0.21 | 0.43 | 0.11 | 1.38 | 0.09 | 1.97 | 0.12 | 1.48 | 0.13 | ||
| 24 | 6.61 | 0.68 | 6.81 | 0.52 | 6.67 | 0.47 | 6.66 | 0.57 | 6.69 | 0.56 | ||
| 5.9 | 3 | 0.89 | 0.24 | 0.24 | 1.25 | 0.77 | 0.51 | 0.90 | 0.60 | 0.70 | 0.65 | |
| 6 | 2.71 | 1.45 | 2.30 | 1.61 | 4.35 | 0.55 | 2.89 | 0.94 | 3.06 | 1.14 | ||
| 24 | NDa | 14.1 | ND | 14.7 | ND | 15.1 | ND | 13.8 | ND | 14.4 | ||
| 6.4 | 3 | 0.70 | 0.28 | 0.15 | 0.25 | 1.12 | 0.31 | 0.67 | 0.25 | 0.66 | 0.27 | |
| 6 | 1.01 | 1.71 | 1.13 | 0.40 | 1.19 | 0.86 | 1.65 | 0.95 | 1.24 | 0.98 | ||
| 24 | ND | 13.9 | ND | 13.8 | ND | 14.3 | ND | 14.5 | ND | 14.1 | ||
| Lactate | 5.2 | 3 | 0.00 | 0.16 | 0.13 | 0.04 | 0.04 | 0.18 | 0.01 | 0.02 | 0.05 | 0.10 |
| 6 | 0.08 | 0.16 | 0.2 | 0.04 | 0.18 | 0.21 | 0.05 | 0.07 | 0.13 | 0.12 | ||
| 24 | 0.08 | 3.42 | 0.2 | 0.57 | 0.18 | 7.98 | 0.05 | 1.57 | 0.13 | 3.39 | ||
| 5.9 | 3 | 0.07 | 0.07 | 0.09 | 0.17 | 0.10 | 0.15 | 0.05 | 0.03 | 0.08 | 0.10 | |
| 6 | 0.18 | 0.08 | 0.14 | 0.17 | 0.12 | 0.15 | 0.20 | 0.03 | 0.16 | 0.11 | ||
| 24 | ND | 11.8 | ND | 11.9 | ND | 11.8 | ND | 11.9 | ND | 11.9 | ||
| 6.4 | 3 | 0.17 | 0.00 | 0.13 | 0.00 | 0.09 | 0.10 | 0.58 | 0.00 | 0.24 | 0.03 | |
| 6 | 0.17 | 0.11 | 0.17 | 0.46 | 0.09 | 0.32 | 0.64 | 0.08 | 0.27 | 0.41 | ||
| 24 | ND | 12.2 | ND | 12.4 | ND | 12.2 | ND | 12.8 | ND | 12.4 | ||
| Time (h) |
P valueb
|
||||
|---|---|---|---|---|---|
| Flin
|
Flout
|
||||
| 3 | 6 | 3 | 6 | 24 | |
| pH | 0.05 | 0.005 | 0.013 | 0.007 | <0.001 |
| Substrate | <0.001 | <0.001 | 0.007 | 0.001 | 0.40 |
| pH × substrate | 0.28 | 0.003 | 0.016 | 0.025 | 0.003 |
| SEDc | 0.153 | 0.348 | 0.132 | 0.234 | 0.963 |
ND, no lactate was detected, and Flin could not be determined.
Analyzed as two-way ANOVA with volunteer as random effect and substrate, pH, and their interaction as fixed effects.
SED, standard error of the difference.
Incubations at the lowest pH (5.2) with the mixture of carbohydrates plus lactate resulted in low lactate consumption in samples from all volunteers (<0.7 mmol liter−1; Table 2). With lactate only, however, lactate disposal at 24 h varied more than 10-fold between volunteers (Table 2). Nonetheless, at pH 5.9 and 6.4 all volunteers showed similar rates of utilization of lactate, with complete consumption within 24 h.
Metabolic fate of added dl-lactate.
Proportions of lactate converted to acetate, butyrate, and propionate were estimated from the 13C label appearing in the final products at 24 h (Table 3). In the presence of polysaccharides, lactate was predominantly converted to acetate and butyrate rather than propionate. In the absence of polysaccharides, however, more lactate was converted to propionate, especially at initial pH 6.4 (0.54 to 0.73). The M + 3 propionate enrichment was sevenfold the M + 2 enrichment (2.99 versus 0.40, respectively) and indicates the dominance of the acrylate route as opposed to the randomizing pathway for propionate formation. At pH 6.4 with lactate as the only carbon source, conversion to butyrate was low (<5%).
TABLE 3.
Proportion of lactate converted to acetate, butyrate, and propionate after 24 h of incubation in batch cultures containing lactate, with or without a range of carbohydrates, and inoculated with fecal slurries from four different volunteers
| Substrate | Acetate | Butyrate | Propionate |
|---|---|---|---|
| Mixture plus lactate | |||
| pH 5.9 | 0.61 | 0.34 | 0.05 |
| pH 6.4 | 0.61 | 0.24 | 0.15 |
| Lactate | |||
| pH 5.9 | 0.52 | 0.11 | 0.37 |
| pH 6.4 | 0.31 | 0.03 | 0.66 |
| SEDa | 0.046 | 0.028 | 0.064 |
| P valueb | |||
|---|---|---|---|
| pH | 0.010 | 0.001 | 0.002 |
| Substrate | <0.001 | <0.001 | <0.001 |
| pH × substrate | 0.011 | 0.55 | 0.07 |
SED, standard error of the difference.
Analyzed as two-way ANOVA with volunteer as random effect and substrate, pH, and their interaction as fixed effects.
An increase in lactate enrichment (from 7.8 to 9.7 and 13.3 MPE) was observed in 24-h incubations when lactate was the only substrate at pH 5.2 in the batch culture medium inoculated with fecal slurries from the two volunteers (A and C) who showed higher rates of lactate utilization. Thus, lactate-utilizing bacteria appeared to preferentially convert unlabeled lactate.
Shifts in microbial populations monitored by FISH.
FISH probes were used to follow changes in bacterial populations over the 24 h of incubation. Probes targeted to clostridial clusters IX and XIVa, Bacteroides spp., and Bifidobacterium spp. accounted for 77 to 87% of the bacteria present in the fecal inocula from the four volunteers. Total numbers of bacteria at 24 h, estimated with the Eub338 probe, increased when polysaccharides were present in the medium (P < 0.001), especially at pH 6.4 (P < 0.001; Table 4), while there was no increase at pH 5.2 when lactate was the only added carbon source. Bacteria belonging to clostridial clusters XIVa and XIVb, detected with the Erec482 probe, increased along with increasing initial pH, and these were greater at pH 6.4 and 5.9 than at 5.2 (P < 0.001; Table 4). On the other hand, Bacteroides numbers tended to decrease during incubation, except at pH 6.4, although final values were variable. Bacteria detected with the Prop853 probe (clostridial cluster IX) also declined at pH 5.9 and 5.2, while, in contrast, Bifidobacterium spp. increased sixfold or more in all incubations with added polysaccharides. At the lowest pH with polysaccharides, Bifidobacterium spp. became the dominant group, accounting in many cases for >50% of the total bacteria present following 24 h of incubation. Lactobacilli were detected in incubations at pH 5.2 and in the presence of the polysaccharide mix alone. Numbers were low (<107/g) and represented less than 0.3% of total bacteria in incubations with fecal slurries from three volunteers, but for volunteer C the abundance was greater (5.8%).
TABLE 4.
Counts (log10) of total bacteria per ml (using the universal probe Eub338) and several bacterial groups (using the probes Bac303, Erec482, Bif164, Prop853, Ehal1469, and Acac194) at 0 (initial) and 24 h of incubation in batch cultures containing a mixture of carbohydrates, lactate, or both and inoculated with fecal slurries from four different volunteers
| Substrate and condition | Count (log10) of total bacteria per ml
|
||||||
|---|---|---|---|---|---|---|---|
| Eub | Bac | Erec | Bif | Prop | Ehal | Acac | |
| Initial counta | 7.93 | 7.29 | 7.43 | 6.74 | 7.19 | 4.88 | 4.18 |
| SDb | 0.095 | 0.111 | 0.091 | 0.243 | 0.207 | 0.464 | 0.429 |
| Substrate | |||||||
| Mixture | |||||||
| pH 5.2 | 8.24 | 6.60 | 7.27 | 7.96 | 6.58 | 5.53 | 5.06 |
| pH 5.9 | 8.29 | 7.09 | 7.50 | 7.83 | 7.02 | 6.05 | 5.21 |
| pH 6.4 | 8.62 | 7.58 | 7.76 | 7.76 | 7.59 | 6.79 | 5.39 |
| Mixture plus lactate | |||||||
| pH 5.2 | 8.26 | 6.84 | 7.02 | 7.97 | 6.49 | 5.42 | 4.88 |
| pH 5.9 | 8.44 | 7.16 | 7.70 | 7.94 | 7.17 | 7.23 | 5.34 |
| pH 6.4 | 8.69 | 7.26 | 7.99 | 7.93 | 7.52 | 7.25 | 5.29 |
| Lactate | |||||||
| pH 5.2 | 7.93 | 6.88 | 7.00 | 6.83 | 6.82 | 5.95 | 4.69 |
| pH 5.9 | 8.02 | 6.92 | 7.62 | 6.47 | 6.83 | 6.06 | 5.22 |
| pH 6.4 | 8.30 | 7.18 | 7.48 | 6.67 | 7.02 | 6.73 | 5.48 |
| SEDc | 0.098 | 0.257 | 0.144 | 0.169 | 0.220 | 0.197 | 0.141 |
|
P valued
|
|||||||
|---|---|---|---|---|---|---|---|
| Eub | Bac | Erec | Bif | Prop | Ehal | Acac | |
| pH | <0.001 | 0.003 | <0.001 | 0.20 | <0.001 | <0.001 | <0.001 |
| Substrate | <0.001 | 0.76 | 0.006 | <0.001 | 0.32 | <0.001 | 0.56 |
| pH × substrate | 0.92 | 0.38 | 0.031 | 0.65 | 0.06 | <0.001 | 0.10 |
Estimated from fecal counts and taking into account the slurry preparation.
SD refers to the standard deviation in counts across volunteers.
SED, standard error of the difference.
Analyzed as two-way ANOVA with volunteer as random effect and substrate, pH, and their interaction as fixed effects.
Relationship between lactate utilization and bacterial groups.
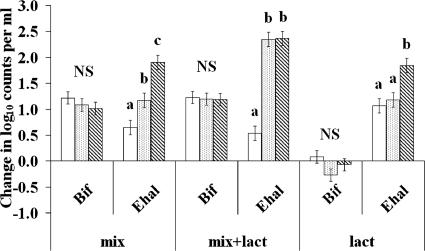
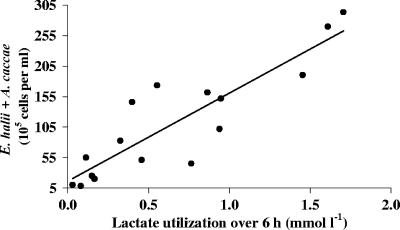
The initial populations of the Eubacterium hallii group (estimated with the Ehal1469 probe) were low and variable (0.04 to 0.31% of total bacteria). Nevertheless, the abundance of this bacterial group increased substantially during all incubations, especially at pH 5.9 and 6.4 (Table 4), and more so when both carbohydrates and lactate were present (P = 0.028; Fig. 2). Another related butyrate-producing, lactate-utilizing bacterium, Anaerostipes caccae (estimated with the Acac194 probe), was also present at low abundance in the fecal samples (<0.3%), and this showed a smaller increase during incubation. When the two lactate-utilizing bacterial groups were combined, a positive linear relationship was observed with lactate utilization at 6 and 24 h (77 and 69% of variance accounted for lactate utilization at 6 and 24 h, respectively; P < 0.001) at the higher pH values (5.9 and 6.4; Fig. 3). Lactate utilization was small or negligible at pH 5.2 (except on lactate alone), and these values were not included. Stimulation of E. hallii numbers following lactate addition, which was particularly evident at pH 5.9 and 6.4 in medium containing the polysaccharide mix, was accompanied by increased butyrate, while acetate decreased at pH 5.9 when both polysaccharides and lactate were present. Overall, a positive linear relationship was observed between butyrate production and the abundance of lactate-utilizing bacteria (with 44% of variance accounted for, P < 0.001).
FIG. 2.
Change in log10 counts per ml in bifidobacteria (Bif) and Eubacterium hallii (Ehal) groups, detected using the probes Bif164 and Ehal1469, in 24-h batch culture containing a mixture of polysaccharides (mix), lactate alone (lact), or the mixture of carbohydrates and lactate together (mix+lact) and incubated at three different pHs: 5.2 (open bars), 5.9 (dotted bars), and 6.4 (striped bars). Data were analyzed as two-way ANOVA with random effect for volunteer and fixed effect for substrate, pH, and their interaction. Error bars indicate standard errors of the means obtained from ANOVA. NS, not significant; different letters indicate statistically significant differences (P < 0.05).
FIG. 3.
Relationship between final abundance of lactate-utilizing bacteria (Eubacterium hallii and Anaerostipes caccae groups, detected using Ehal1469 and Acac194 probes) and lactate utilization over 6 h in batch cultures inoculated with fecal slurries from four different volunteers and incubated for 24 h with lactate at two different pHs (5.9 and 6.4) and in the presence or absence of a mixture of carbohydrates. (Seventy-seven percent of variance was accounted for; P was <0.001 for effect of lactate utilization at 6 h, based on linear regression of bacterial count on lactate utilization.)
DISCUSSION
The aim of this investigation was to establish the impact of nutritional and environmental factors (pH) on lactate metabolism by the bacterial communities found in the human colon. In the batch culture incubations employed, the set pH remained relatively stable over 24 h but the initial pH had a major effect on the fermentation products. A reduction in initial pH from 6.4 to 5.9 resulted in decreased production of propionate relative to butyrate, an effect previously reported in pH-controlled fermentor experiments (52). In the current study, at the lowest pH of 5.2, which is within the range reported for human intestinal contents from colitis patients (44) and close to the values observed in healthy individuals (8, 44), butyrate and propionate formation were nearly zero, although there was still acetate accumulation. Only at this low pH, however, was net lactate formation observed after 24 h.
Two explanations might account for the net lactate production at pH 5.2. First, there could be an increase in lactate production, associated with lactate-producing bacteria competing more successfully for the carbohydrate substrates. Second, a reduction in lactate utilization might also cause lactate accumulation. In terms of the first explanation, bifidobacteria can be important lactate producers in the colon (18). In the presence of polysaccharides, bifidobacterial numbers increased here by at least sixfold, independently of pH, and represented 42 to 73% of total bacteria after 24 h of incubation at pH 5.2. Bifidobacteria can produce only l-lactate (30), however, so the d-lactate detected at pH 5.2 must be due to activity of other lactate producers, including Lactobacillus spp., Faecalibacterium prausnitzii, and Bacteroides spp. (11, 34). In addition, many other bacterial groups have the ability to produce lactate as one of their fermentation products under some nutritional conditions (4). The kinetic measurements at 6 h showed that lactate production (Flin) occurred at all pHs investigated but was greatest at pH 5.9. Previous studies have shown that lactate production by bifidobacteria, in pure culture, is stimulated at a slightly acidic pH (5).
As lactate formation was independent of pH, then the lack of net lactate accumulation at higher pHs (both 5.9 and 6.4) must be due to enhanced lactate utilization by other bacteria. This was confirmed by the stable-isotope studies, with lactate utilization (Flout) considerably greater at pH 5.9 or 6.4 than at pH 5.2. At pH 5.2, however, the rate of lactate utilization was less than the rate of production and hence accumulation occurred for all volunteers.
Isotope 13C fractionation was observed when lactate was the only substrate at pH 5.2, in agreement with previous observations for microbial metabolism (6, 33). Lactate formation and utilization therefore may be underestimated due to this isotope effect. At the higher pH, however, both labeled and unlabeled lactate were fully metabolized after 24 h and so the overall general conclusions are still valid.
There are several possible routes for lactate utilization by gut bacteria, and studies with stable isotopes have indicated that butyrate and propionate are both formed from lactate in human fecal incubations (7, 42). In the current study, the dominant route of propionate formation was via the acrylate pathway, similar to that observed for organisms such as Megasphaera elsdenii, known to play an important role in the rumen (9). Lower rates of synthesis were observed via the alternative randomizing pathway through succinate (21). Lactate conversion to butyrate was probably via acetyl coenzyme A as described recently for bacteria related to E. hallii and A. caccae (5, 13). Nutrient supply and pH, however, affected the metabolic fate of lactate. At pH 5.9 in the presence of polysaccharides, butyrate production dominated in all four donors, while at pH 6.4 significant conversion to propionate also occurred. In the absence of polysaccharides, lactate was converted to both propionate and butyrate, with propionate the major product at pH 6.4. This difference between the presence and absence of polysaccharides probably reflects the physiology and ecology of the different lactate utilizers. For example, E. hallii populations increased in all incubations, often by more than 100-fold, and are probably responsible for most of the butyrate formation from lactate (Fig. 3). Which groups of bacteria were responsible for conversion to propionate is less clear but might involve a subpopulation of the clostridial cluster IX or XIVa groups.
Bacteroides and bacteria belonging to clostridial clusters XIVa and XIVb (estimated with the Erec482 probe) are usually the most abundant bacterial groups in both continuous fermentors (52) and fecal samples (15). Numbers of Bacteroides increased in the absence of lactate only at pH 6.4, consistent with the previously observed sensitivity of human colonic Bacteroides to reduced pH (52). The increase in numbers of bacteria comprising clostridial clusters XIVa and XIVb in incubations at pH 5.9 and 6.4 can be explained mainly by E. hallii proliferation. Interestingly, the population of bifidobacteria showed a significant increase here in the presence of polysaccharides for all three pH conditions. This contrasts with poor competitiveness in continuous-culture systems with the same polysaccharide substrates (52) and may reflect the capacity of the bifidobacteria for rapid rates of growth at the high nutrient availability during the early stage of batch culture incubation. Notably, lactate production is associated with rapid growth but relatively low growth efficiency (48).
In summary, at pH 5.9 and 6.4 fecal bacteria rapidly utilized lactate produced from incubations that favored the growth of lactate-producing Bifidobacterium species. E. hallii-related bacteria appeared to play a major role in conversion of lactate to butyrate in the presence of fermentable polysaccharides, while production of propionate via the acrylate pathway by unknown species was also important in the absence of carbohydrates and the presence of lactate. The main conclusion from our work is that lactate accumulated at pH 5.2 because production was maintained but utilization was reduced markedly. Such differential pH sensitivity in production and utilization of lactate creates a potential for imbalance in net accumulation at low pH and could ultimately result in further lactate accumulation and in acidosis, which may be realized with certain gut disorders or dietary intakes and lead to adverse consequences. Further research is necessary to determine the relevance in vivo of lactate-utilizing butyrate-producing bacteria, such as E. hallii, and to devise possible therapies to prevent lactate accumulation in certain gut disorders.
Acknowledgments
The Rowett Research Institute and Biomathematics and Statistics Scotland are supported by the Scottish Executive Environment and Rural Affairs Department. A. Belenguer received financial support from the Spanish Ministry of Education and Science.
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Al Jassim, R. A. M., P. T. Scott, A. L. Trebbin, D. Trott, and C. C. Pollitt. 2005. The genetic diversity of lactic acid producing bacteria in the equine gastrointestinal tract. FEMS Microbiol. Lett. 248:75-81. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avivi-Green, C., S. Polak-Charcon, Z. Madar, and B. Schwartz. 2000. Apoptosis cascade proteins are regulated in vivo by high intracolonic butyrate concentration: correlation with colon cancer inhibition. Oncol. Res. 12:83-95. [DOI] [PubMed] [Google Scholar]
- 4.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, and H. J. Flint. 2000. Phylogenetic relationships of dominant butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belenguer, A., S. H. Duncan, A. G. Calder, G. Holtrop, P. Louis, G. E. Lobley, and H. J. Flint. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 72:3593-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blair, N., A. Leu, E. Muñoz, J. Olsen, E. Kwong, and D. Des Marais. 1985. Carbon isotopic fractionation in heterotrophic microbial metabolism. Appl. Environ. Microbiol. 50:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourriaud, C., R. J. Robins, L. Martin, F. Kozlowski, E. Tenailleau, C. Cherbut, and C. Michel. 2005. Lactate is mainly fermented to butyrate by human intestinal microfloras but inter-individual variation is evident. J. Appl. Microbiol. 99:201-212. [DOI] [PubMed] [Google Scholar]
- 8.Brown, R. L., J. A. Gibson, G. E. Sladen, B. Hicks, and A. M. Dawson. 1974. Effects of lactulose and other laxatives on ileal and colonic pH as measured by a radiotelemetry device. Gut 15:999-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Counotte, G. H. M., R. A. Prins, H. A. M. Janssen, and M. J. A. Debie. 1981. Role of Megasphaera elsdenii in the fermentation of [2-13C]lactate in the rumen of dairy cattle. Appl. Environ. Microbiol. 42:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings, J. H. 1995. Short chain fatty acids, p. 101-130. In G. R. Gibson and G. T. Macfarlane (ed.), Human colonic bacteria. CRC Press, Boca Raton, FL.
- 11.Duncan, S. H., G. L. Hold, H. J. M. Harmsen, C. S. Stewart, and H. J. Flint. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52:2141-2147. [DOI] [PubMed] [Google Scholar]
- 12.Duncan, S. H., K. P. Scott, A. G. Ramsay, H. J. M. Harmsen, G. W. Welling, C. S. Stewart, and H. J. Flint. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl. Environ. Microbiol. 69:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan, S. H., P. Louis, and H. J. Flint. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 70:5810-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan, S. H., G. Holtrop, G. E. Lobley, G. Calder, C. S. Stewart, and H. J. Flint. 2004. Contribution of acetate to butyrate formation by human faecal bacteria. Br. J. Nutr. 91:915-923. [DOI] [PubMed] [Google Scholar]
- 15.Duncan, S. H., A. Belenguer, G. Holtrop, A. M. Johnstone, G. E. Lobley, and H. J. Flint. 2007. Reduced dietary intake of carbohydrate, by obese subjects, results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 73:1073-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, et al. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint, H. J. 2006. The significance of prokaryote diversity in the human gastrointestinal tract, p. 65-90. In N. A. Logan, H. M. Lappin-Scott, and P. C. F. Oyston (ed.), Prokaryotic diversity: mechanisms and significance. SGM symposium, vol. 66. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 18.Florent, C., B. Flourie, A. Leblond, M. Rautureau, J. J. Bernier, and J. C. Rambaud. 1985. Influence of chronic lactulose ingestion on the colonic metabolism of lactulose in man (an in vivo study). J. Clin. Investig. 75:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill, C. I. R., and I. R. Rowland. 2002. Diet and cancer: assessing the risk. Br. J. Nutr. 88(Suppl. 1):S73-S87. [DOI] [PubMed] [Google Scholar]
- 21.Gottschalk, G. 1979. Bacterial metabolism. Springer-Verlag, New York, NY.
- 22.Guarner, F., and J. R. Malagelada. 2003. Gut flora in health and disease. Lancet 361:512-519. [DOI] [PubMed] [Google Scholar]
- 23.Harmsen, H. J. M., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashizume, K., T. Tsukahara, K. Yamada, H. Koyama, and K. Ushida. 2003. Megasphaera elsdenii JCM1772T normalizes hyperlactate production in the large intestine of fructooligosaccharide-fed rats by stimulating butyrate production. J. Nutr. 133:3187-3190. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture methods. Microbiol. Immunol. 46:535-548. [DOI] [PubMed] [Google Scholar]
- 26.Hino, T., K. Shimada, and T. Maruyama. 1994. Substrate preference in a strain of Megasphera elsdenii, a ruminal bacterium, and its implications in propionate production and growth competition. Appl. Environ. Microbiol. 60:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 28.Hold, G. L., A. Schwiertz, R. I. Aminov, M. Blaut, and H. J. Flint. 2003. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 69:4320-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hove, H., I. Norgard Andersen, and P. B. Mortensen. 1994. Fecal dl-lactate concentrations in 100 gastrointestinal patients. Scand. J. Gastroenterol. 29:255-259. [DOI] [PubMed] [Google Scholar]
- 30.Knol, J., P. Scholtens, C. Kafka, J. Steenbakkers, S. Groß, K. Helm, M. Klarczyk, H. Schöpfer, H. M. Böckler, and J. Wells. 2005. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J. Pediatr. Gastroenterol. Nutr. 40:36-42. [DOI] [PubMed] [Google Scholar]
- 31.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lay, C., M. Sutren, V. Rochet, K. Saunier, J. Dore, and L. Rigottier-Gois. 2005. Design and validation of 16S rRNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ. Microbiol. 7:933-946. [DOI] [PubMed] [Google Scholar]
- 33.Londry, K. L., and D. J. Des Marais. 2003. Stable carbon isotope fractionation by sulfate-reducing bacteria. Appl. Environ. Microbiol. 69:2942-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macfarlane, G. T., and G. R. Gibson. 1991. Co-utilization of polymerized carbon sources by Bacteroides ovatus grown in a two-stage continuous culture system. Appl. Environ. Microbiol. 57:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macfarlane, G. T., and G. R. Gibson. 1997. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine, p. 269-318. In R. I. Mackie and B. A. White (ed.), Gastrointestinal microbiology, vol. 1. Chapman and Hall, London, United Kingdom. [Google Scholar]
- 36.Macfarlane, G. T., S. Hay, and G. R. Gibson. 1989. Influence of mucin on glycosidase, protease and arylamidase activities of human gut bacteria grown in a 3-stage continuous culture system. J. Appl. Bacteriol. 66:407-417. [DOI] [PubMed] [Google Scholar]
- 37.Macfarlane, G. T., G. R. Gibson, and J. H. Cummings. 1992. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72:57-64. [DOI] [PubMed] [Google Scholar]
- 38.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 39.Mariadason, J. M., G. A. Corner, and L. H. Augenlicht. 2000. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 60:4561-4572. [PubMed] [Google Scholar]
- 40.McNeil, N. I. 1984. The contribution of the large intestine to energy supplies in man. Am. J. Clin. Nutr. 39:338-342. [DOI] [PubMed] [Google Scholar]
- 41.Miyazaki, K., J. C. Martin, R. Marinsek-Logar, and H. J. Flint. 1997. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis B14). Anaerobe 3:373-381. [DOI] [PubMed] [Google Scholar]
- 42.Morrison, D. J., W. G. Mackay, C. A. Edwards, T. Preston, B. Dodson, and L. T. Weaver. 2006. Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br. J. Nutr. 96:570-577. [PubMed] [Google Scholar]
- 43.Mortensen, P. B., and M. R. Clausen. 1996. Short chain fatty acids in the human colon: relation to gastrointestinal health and disease. Scand. J. Gastrointestinol. 31(Suppl. 216):132-148. [DOI] [PubMed] [Google Scholar]
- 44.Nugent, S. G., D. Kumar, D. S. Rampton, and D. F. Evans. 2001. Intestinal luminal pH in inflammatory bowel disease: possible determinants and implications for therapy with aminosalicylates and other drugs. Gut 48:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Padan, A., D. Zilberstein, and S. Shuldiner. 1981. pH homeostasis in bacteria. Biochim. Biophys. Acta 650:151-166. [DOI] [PubMed] [Google Scholar]
- 46.Pryde, S. E., S. H. Duncan, G. L. Hold, C. S. Stewart, and H. J. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133-139. [DOI] [PubMed] [Google Scholar]
- 47.Richardson, A. J., A. G. Calder, C. S. Stewart, and A. Smith. 1989. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett. Appl. Microbiol. 9:5-8. [Google Scholar]
- 48.Russell, J. B., and R. J. Wallace. 1997. Energy-yielding and energy-consuming reactions, p. 246-282. In P. N. Hobson and C. S. Stewart (ed.), The rumen microbial ecosystem. Blackie, London, United Kingdom.
- 49.Seeliger, S., P. H. Janssen, and B. Schink. 2002. Energetics and kinetics of lactate fermentation to acetate and propionate via methylmalonyl-CoA or acrylyl-CoA. FEMS Microbiol. Lett. 211:65-70. [DOI] [PubMed] [Google Scholar]
- 50.Slyter, L. L. 1976. Influence of acidosis on rumen function. J. Anim. Sci. 43:910-929. [DOI] [PubMed] [Google Scholar]
- 51.Vernia, P., R. Caprilli, G. Latella, F. Barbetti, F. M. Magliocca, and M. Cittadini. 1988. Fecal lactate and ulcerative colitis. Gastroenterology 95:1564-1568. [DOI] [PubMed] [Google Scholar]
- 52.Walker, A. W., S. H., Duncan, E. C. M. Leitch, M. W. Child, and H. J. Flint. 2005. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl. Environ. Microbiol. 71:3692-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson, A. J. M. 2006. An overview of apoptosis and the prevention of colorectal cancer. Crit. Rev. Oncol. Hematol. 57:107-121. [DOI] [PubMed] [Google Scholar]