Abstract
Efficient production of heterologous proteins with yeasts and other eukaryotic hosts is often hampered by inefficient secretion of the product. Limitation of protein secretion has been attributed to a low folding rate, and a rational solution is the overexpression of proteins supporting folding, like protein disulfide isomerase (Pdi), or the unfolded protein response transcription factor Hac1. Assuming that other protein factors which are not directly involved in protein folding may also support secretion of heterologous proteins, we set out to analyze the differential transcriptome of a Pichia pastoris strain overexpressing human trypsinogen versus that of a nonexpressing strain. Five hundred twenty-four genes were identified to be significantly regulated. Excluding those genes with totally divergent functions (like, e.g., core metabolism), we reduced this number to 13 genes which were upregulated in the expression strain having potential function in the secretion machinery and in stress regulation. The respective Saccharomyces cerevisiae homologs of these genes, including the previously characterized secretion helpers PDI1, ERO1, SSO2, KAR2/BiP, and HAC1 as positive controls, were cloned and overexpressed in a P. pastoris strain expressing a human antibody Fab fragment. All genes except one showed a positive effect on Fab fragment secretion, as did the controls. Six out of these novel secretion helper factors, more precisely Bfr2 and Bmh2 (involved in protein transport), the chaperones Ssa4 and Sse1, the vacuolar ATPase subunit Cup5, and Kin2 (a protein kinase connected to exocytosis), proved their benefits for practical application in laboratory-scale production processes by increasing both specific production rates and the volumetric productivity of an antibody fragment up to 2.5-fold in fed-batch fermentations of P. pastoris.
Successful secretion of proteins has been accomplished with a variety of fungal hosts, including the yeasts Saccharomyces cerevisiae, Pichia pastoris, Hansenula polymorpha, and Kluyveromyces lactis and filamentous fungi like Aspergillus awamori and Trichoderma reesei. While the secretion of some proteins is readily achieved at high rates, many other proteins are secreted only at comparatively low levels (20, 24, 25). Improvement of the secretion of a recombinant protein was first attempted by random mutagenesis (1, 16). The major disadvantage of this method is usually that any positive result cannot be transferred to other strains.
It has been shown in several cases that the secretion process can be enhanced by cooverexpression of proteins which support the folding and processing of other proteins (recently reviewed in references 12 and 5). Some of these supporting factors, like protein disulfide isomerase (Pdi), have catalytic activity on the proteins, and others act by binding to the proteins and preventing them from aggregation (chaperones, e.g., BiP) or by stimulating the release of the protein to the cell exterior at a later step in the secretory pathway (Sso proteins). Another approach is to activate the secretory pathway concertedly by overexpression of the unfolded protein response (UPR) transcription factor Hac1, which induces the genes of chaperones and enzymes involved in the secretion process, among other genes, and was reported to increase the secretion of several heterologous proteins in both yeast and filamentous fungi (35, 36). While these approaches, once established, can be transferred to other strains and used for other proteins as well, their number is limited due to the lack of actual knowledge about the function of those factors supporting the secretion of other proteins.
It can be anticipated that the successful high-level secretion of a recombinant protein may be limited at a number of different steps, like folding, disulfide bridge formation, glycosylation, transport within the cell, and release from the cell. As many of these processes are still not fully understood, it can also be anticipated that there are many more proteins involved which support the secretion of a protein than is currently known. However, such helper functions cannot be predicted with the current state of the art, even when the DNA sequence of the entire genome of a host organism is available.
To understand more about the gene regulation of a host organism during protein production, we have recently performed DNA microarray hybridization experiments with P. pastoris clones expressing recombinant human trypsinogen in comparison to a nonproducing strain (29). These experiments allow a relative measure of the transcription levels of approximately one-third of all genes in P. pastoris, but they do not provide direct information on the potential of any hitherto unidentified protein to enhance secretion.
Additional analyses of the data derived from DNA hybridization have allowed us to identify potential secretion-supporting proteins, or their respective genes. To achieve this, the relative expression levels of all measured genes of trypsinogen-producing and non-trypsinogen-producing cells were compared to each other. Out of 524 differentially regulated genes, 13 potentially interesting genes involved in secretion and/or general stress response were selected for further analysis. These genes were cloned from S. cerevisiae by PCR and coexpressed in a P. pastoris strain expressing the Fab fragment of a monoclonal antibody fragment (2F5mAb) against human immunodeficiency virus type 1. By evaluating the effect of the putative helper factors on recombinant protein secretion, we could identify six novel secretion helper factors.
MATERIALS AND METHODS
Unless stated otherwise, all chemicals were purchased from VWR International, all enzymes for DNA manipulation were purchased from MBI Fermentas, KOD DNA polymerase was from Novagen, and all antisera were from Sigma.
Analysis of gene regulation due to recombinant protein expression.
Development and data acquisition of the heterologous hybridization microarray method were described in Sauer et al. (29). To gain information on transcriptional regulation due to recombinant protein production, these raw data were further analyzed. The normalized signals on each spot were compared between the different chips. All genes showing signal differences exceeding the threshold (±1.5-fold) between the trypsinogen-expressing P. pastoris strain and the nonexpressing control strain were judged as significantly regulated. Only genes regulated under expression conditions (methanol fed batch) were selected for further analysis.
Coexpression of secretion helper factors: isolation of the helper factor genes from S. cerevisiae and cloning into pGAPHis.
All the genes were amplified directly from S. cerevisiae genomic DNA by PCR with specific oligonucleotide primers. ACG was inserted directly before the start codon ATG as the Kozac sequence. The nontemplate coded restriction sites SacII and either PmlI or SfiI were added to the respective forward and backward primers. After restriction digestion of the PCR fragments of correct length (checked by agarose gel separation) with SacII and either PmlI or SfiI, these fragments were cloned into the pGAPHis vector (13) which had been digested with the respective restriction enzymes and treated with alkaline phosphatase. Additionally, the induced variant of the HAC1 gene of S. cerevisiae and the gene coding for Pdi1 were ligated into pGAPHis as described previously (13).
Construction of P. pastoris strains coexpressing 2F5 Fab and a secretion helper factor.
The plasmids containing a helper factor gene, and an empty vector as a control, were used to transform P. pastoris strain SMD1168, already containing the expression cassettes for 2F5 Fab under the control of the GAP promoter, preselected for a high Fab secretion level. The construction of the Fab expression strains has been described in detail in Gasser et al. (13). SMD1168 is a his4 pep4 mutant. Selection was based on zeocin resistance for the antibody genes and histidine auxotrophy for the other genes. Thus, the transformed cells were cultivated on RDB agar lacking histidine (1 M sorbitol; 2 g liter−1 glucose; 1.34% yeast nitrogen base without amino acids; 0.4 mg liter−1 biotin; 0.005% l-glutamic acid, l-methionine, l-lysine, l-leucine, and l-isoleucine; 2 g liter−1 agar) for selection of His-prototrophic clones that contain the expression cassettes for the secretion helper factors.
Shake flask cultivation.
Five milliliters of YP medium (10 g liter−1 yeast extract, 20 g liter−1 peptone) containing 20 g liter−1 glycerol was inoculated with a single colony of P. pastoris selected from the RDB plates and grown overnight at 28°C. Aliquots of these cultures corresponding to a final optical density at 600 nm of 0.1 were transferred to 10 ml of main culture medium (per liter: 10 g yeast extract, 10 g peptone, 100 mM potassium phosphate buffer [pH 6.0], 13.4 g yeast nitrogen base with ammonium sulfate, 0.4 mg biotin) and incubated for 48 h at 28°C with vigorous shaking in 100-ml Erlenmeyer flasks. To stimulate recombinant protein expression, cultures with the GAP promoter were supplemented with 10 g liter−1 glucose. The same amounts of substrate were added repeatedly four times every 12 h before cells were harvested by centrifugation at 2,500 × g for 5 min at room temperature (RT) and prepared for analysis (biomass determination by measurement of optical density at 600 nm, enzyme-linked immunosorbent assay for Fab quantification in the culture supernatant).
Fed-batch fermentation.
A preculture of the individual P. pastoris strains incubated with shaking at 28°C for 24 h on YPG (per liter: 10 g yeast extract, 10 g peptone, 10 g glycerol) was used to inoculate the starting volume (1.75 liters of batch medium) of the bioreactors to a starting optical density at 600 nm of 1.0. Fermentations were carried out in 5.0-liter working volume bioreactors (Minifors, Infors, Switzerland) with a computer-based process control. Fermentation temperature was controlled at 25°C, pH was controlled at 5.0 with addition of 25% ammonium hydroxide, and the dissolved-oxygen concentration was maintained above 20% saturation by controlling the stirrer speed between 600 and 1,200 rpm, whereas the airflow was kept constant at 100 liters h−1.
The batch medium contained (per liter) 2.0 g citric acid, 12.4 g (NH4)2HPO4, 0.022 g CaCl2·2H2O, 0.9 g KCl, 0.5 g MgSO4·7H2O, 46.5 g glycerol, and 4.6 ml PTM1 trace salts stock solution. The pH was adjusted to 5.0 with 25% HCl.
The glucose fed-batch solution contained (per liter) 550 g glucose·1H2O, 10 g KCl, 6.45 g MgSO4·7H2O, 0.35 g CaCl2·2H2O, and 12 ml PTM1 trace salts stock solution.
The PTM1 trace salts stock solution contained (per liter) 6.0 g CuSO4·5H2O, 0.08 g NaI, 3.0 g MnSO4·H2O, 0.2 g Na2MoO4·2H2O, 0.02 g H3BO3, 0.5 g CoCl2, 20.0 g ZnCl2, 65.0 g FeSO4·7H2O, 0.2 g biotin, and 5.0 ml H2SO4 (95 to 98%). All chemicals for PTM1 trace salts stock solution were from Riedel-de Haën (Seelze, Germany), except for biotin (Sigma, St. Louis, MO) and H2SO4 (Merck Eurolab).
After approximately 27 h, the batch was finished and the glucose fed batch with a feed rate of 16 g h−1 was started. The fermentations were terminated at approximately 100 h. Samples were taken frequently and processed as described previously (21).
Enzyme-linked immunosorbent assay for quantification of Fab.
To determine the amount of secreted recombinantly expressed 2F5 Fab, 96-well microtiter plates (MaxiSorb, Nunc, Denmark) were coated with anti-human immunoglobulin G (Fab specific) overnight at RT (1:1,000 in phosphate-buffered saline [PBS], pH 7.4; Sigma I-5260) before serially diluted supernatants of P. pastoris cultures (starting with a 1:200 dilution in PBS-Tween 20 [0.1%] plus 1% bovine serum albumin) were applied and incubated for 2 h at RT. A Fab fragment of human immunoglobulin G (Rockland) was used as a standard protein at a starting concentration of 200 ng ml−1. After each incubation step, the plates were washed four times with PBS containing 1% Tween 20 adjusted to pH 7.4. One hundred microliters of anti-kappa light chain-alkaline phosphatase conjugate as a secondary antibody (1:1,000 in PBS-Tween plus 1% bovine serum albumin; Sigma A-3813) was added to each well and incubated for 1 h at RT. After being washed, the plates were stained with pNPP (1 mg ml−1 p-nitrophenyl phosphate in coating buffer, 0.1 N Na2CO3-NaHCO3, pH 9.6) and read at 405 nm (reference wavelength, 620 nm).
RESULTS
Identification and cloning of secretion helper factors from S. cerevisiae.
In order to identify the genes and respective proteins which play a potentially beneficial role during recombinant protein production, the gene expression pattern of a P. pastoris strain containing the gene for human trypsinogen 1 was compared to that of a nonexpressing strain by microarray analysis. The experimental procedure of the fed-batch cultivations, the microarray hybridization, and the evaluation of the obtained data were described in Sauer et al. (29).
As the genome sequence of P. pastoris has not been published so far and not many genes were characterized for P. pastoris, DNA microarrays of S. cerevisiae were used for heterologous hybridization with P. pastoris cDNA.
After determination of the relative expression levels of all measured genes, the genes were ordered by the relative ratios of their expression levels, and the 524 genes above the threshold (±1.5-fold regulation) were considered for further analysis. The differentially regulated genes were ranked based on their putative intracellular localizations and functions. As the DNA microarrays used for these experiments were derived from S. cerevisiae gene sequences, only putative gene functions for P. pastoris could be assigned by homology to S. cerevisiae. Out of the regulated genes, 99 had either uncharacterized or unknown gene products in S. cerevisiae and were therefore excluded. Thirty-three genes belonged to energy metabolism, 26 belonged to various cellular stress responses, 27 were involved in cell organization and biogenesis of cellular structures, and 47 were linked to the cell cycle and sporulation. By far, the biggest group of differentially regulated genes between the expression strain and the wild-type control were associated with protein metabolism, ranging from transcription, RNA and ribosome synthesis, and translation to protein processing, transport, and degradation. Out of those, genes with totally divergent functions or involvement in core metabolic activities were excluded.
Focusing on upregulated genes with potential function in the secretion machinery and/or in stress regulation, 13 potentially interesting genes were selected for further analysis, including PDI1 and HAC1, which have been shown before to have beneficial effects on recombinant protein secretion in P. pastoris (13). Additionally, the Pdi oxidase Ero1 was included in our study as it has been reported that duplication of either PDI1 or ERO1 genes led to a significant increase in the yield of human serum albumin, a disulfide-rich protein, in K. lactis (19).
The cDNAs of these genes were amplified from S. cerevisiae, as the respective P. pastoris sequences were not (and for some genes are still not) available, and transformed in a P. pastoris strain already secreting the 2F5 Fab fragment under the control of the GAP promoter.
Investigation of the effect of the secretion helper factors on heterologous protein production of recombinant 2F5 Fab in P. pastoris in shake flask cultures.
Of each of the 15 different secretion helper factor constructs, 16 individual clones were cultivated in shake flasks and compared to 16 individual clones of the control strain, which was transformed with the pGAPHis vector lacking a gene. The 2F5 Fab productivity (μg Fab per biomass and time period) was determined for all the analyzed cultures (first screening round). The six best clones of each of the constructs were then analyzed in a second screening round. Table 1 shows the mean improvements of 2F5 Fab production rate for the two screening rounds obtained by cooverexpression of the secretion helper factors relative to the control culture values.
TABLE 1.
Mean improvements of 2F5 Fab productivity obtained by cooverexpression of novel secretion helper factors relative to the control culture valuea
| Folding factor | Mean improvementb | P valuec | Statistical significanced |
|---|---|---|---|
| PDI1 | 1.7 | 0.00032 | *** |
| CUP5 | 1.7 | 0.00157 | *** |
| SSA4 | 1.6 | 0.00156 | *** |
| BMH2 | 1.5 | 0.00002 | *** |
| KIN2 | 1.5 | 0.00060 | *** |
| KAR2 | 1.5 | 0.22167 | |
| HAC1 | 1.5 | 0.00022 | *** |
| ERO1 | 1.4 | 0.00059 | *** |
| SSE1 | 1.4 | 0.00688 | *** |
| BFR2 | 1.4 | 0.03811 | ** |
| COG6 | 1.2 | 0.18851 | |
| SSO2 | 1.2 | 0.27459 | |
| COY1 | 1.2 | 0.07546 | |
| IMH1 | 1.1 | 0.26235 | |
| SEC31 | 1.0 | 0.08838 | |
| Control | 1.0 |
The previously characterized secretion helper factors PDI1, KAR2, HAC1, ERO1, and SSO2 are included for comparative reasons and highlighted in bold.
The six best clones of each tested secretion helper factor construct were compared to the six best clones of the control construct (empty pGAPHis vector). The mean improvement factor for two screening rounds is shown. The strain used in this study was P. pastoris SMD1168, containing the 2F5 Fab genes under the control of the GAP promoter, preselected for high Fab secretion prior to transformation with the pGAPHis vector for constitutive coexpression of the helper genes.
Statistical significance of the mean improvement of each tested secretion helper factor construct determined by a combined Student t test.
***, P ≤ 0.01; **, P ≤ 0.05.
As can be seen from Table 1, the secretion of the heterologous Fab fragment was increased for most of the analyzed secretion helper factors, in a range between 1.2- and 1.7-fold. Apart from the secretion helper factors already known to have a positive effect on the secretion of a heterologous protein (Pdi1, Hac1, Ero1, Kar2), cooverexpression of the novel helper factor genes CUP5, SSA4, BMH2, KIN2, SSE1, and BFR2 showed a highly significant increase in the amount of secreted 2F5 Fab, and cooverexpression of COG6 and COY1, along with the previously characterized SSO2, showed a moderate increase in the amount of secreted heterologous antibody fragments.
Effect of the novel secretion helper factors on heterologous protein production in fed-batch cultivations.
The best two clones of each construct were cultivated on a larger scale (250-ml baffled shake flask cultures) in order to select a clone for fed-batch cultivation. Clones cooverexpressing the genes for the newly identified secretion helper factors CUP5, SSA4, BMH2, KIN2, SSE1, and BFR2 were chosen, in addition to PDI1 as a reference and the control strain containing no additional helper factor.
Fed-batch cultivations on the laboratory scale were performed to evaluate the feasibility of the novel secretion helper factors for production and to verify their beneficial role on heterologous protein secretion under more-defined conditions. As expected, in the control strain the final titer of 2F5 Fab (47.27 mg liter−1) was comparable to previous results obtained with nonengineered P. pastoris (13, 21).
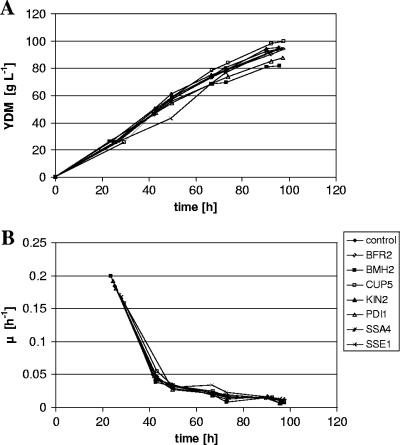
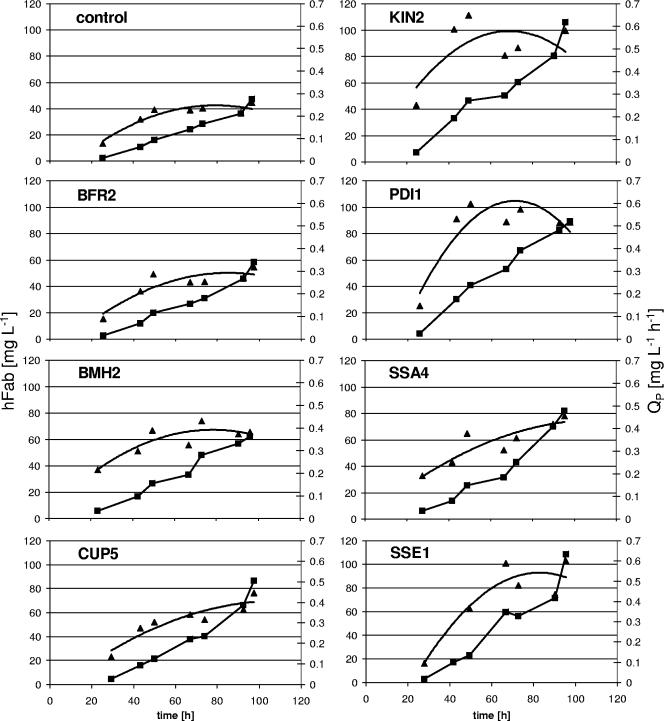
Figure 1 shows the development of biomass concentration and specific growth rate over time for all the different strains, while the accumulation of 2F5 Fab in the supernatant and the volumetric productivity (QP) over time are plotted in Fig. 2 for each of the strains individually. Apparently, the biomass concentrations as well as the specific growth rates reach comparable levels in all the analyzed strains, whether containing a gene encoding a secretion helper factor or not (Fig. 1). As can also be seen in Table 2, the biomass yield coefficients do not deviate significantly between the secretion-engineered strains and their vector control strain.
FIG. 1.
Kinetics of growth-related parameters of the secretion-engineered P. pastoris and the control strain in fed-batch cultures. Development of biomass concentration (g liter−1 yeast dry mass [YDM]) (A) and specific growth rates (μ) (B) are shown for the P. pastoris expression strains (SMD1168 secreting 2F5 Fab under the control of the GAP promoter) coexpressing the novel secretion helper factors and the control strain expressing only the antibody fragment.
FIG. 2.
Increased production of secreted antibody fragments in P. pastoris coexpressing novel secretion helper factors in fed-batch cultures. The development of product titer (rectangles with solid black lines) and QP (triangles with trend lines) are shown for each of the secretion-engineered strains and their parental strain (control) individually.
TABLE 2.
Comparison of secretion-engineered P. pastoris strains and their parental strain in fed-batch cultivationsa
| Strainb | Mean qP (mg g−1 h−1) | Mean μ (h−1) | QP maximum (mg liter−1 h−1) | YDM (g liter−1) | hFab (mg liter−1) | Total YDM (g) | Total product (mg hFab) | YP/X (mg g−1) | Y′X/S |
|---|---|---|---|---|---|---|---|---|---|
| Control | 0.0085 | 0.0249 | 0.2585 | 94.2 | 47.3 | 220.2 | 66.9 | 0.36 | 0.46 |
| BFR2 | 0.0105 | 0.0245 | 0.3172 | 94.2 | 58.4 | 219.5 | 83.0 | 0.44 | 0.42 |
| BMH2 | 0.0141 | 0.0223 | 0.4303 | 82.0 | 62.2 | 185.9 | 95.0 | 0.57 | 0.39 |
| CUP5 | 0.0143 | 0.0265 | 0.4443 | 99.8 | 86.5 | 229.8 | 112.5 | 0.60 | 0.46 |
| KIN2 | 0.0185 | 0.0243 | 0.6491 | 95.3 | 106.0 | 220.2 | 144.9 | 0.78 | 0.47 |
| PDI1 | 0.0187 | 0.0236 | 0.5968 | 87.6 | 89.2 | 201.1 | 134.5 | 0.74 | 0.43 |
| SSA4 | 0.0154 | 0.0252 | 0.4562 | 93.9 | 81.8 | 211.1 | 110.1 | 0.62 | 0.48 |
| SSE1 | 0.0211 | 0.0254 | 0.5982 | 94.6 | 108.5 | 212.9 | 152.1 | 0.81 | 0.48 |
Abbreviations: μ, average growth rate; YDM, final biomass concentration; hFab, final product concentration; YP/X, product yield; Y′X/S, observed biomass yield coefficient (g YDM per g glucose). Total YDM is calculated as biomass concentration times culture volume; total product is determined as hFab volume in the free culture volume (total volume as deducted by the YDM).
The control is P. pastoris SMD1168, expressing only the 2F5 Fab fragment under the control of the GAP promoter, whereas BFR2, BMH2, CUP5, KIN2, PDI1, SSA4, and SSE1 are coexpressing the respective S. cerevisiae genes identified as secretion helper factors.
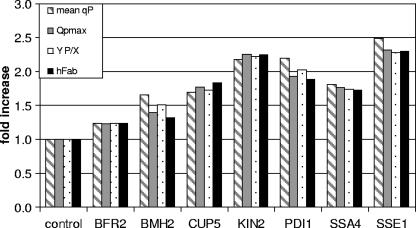
Mean specific product formation rates (qP), on the other hand, which are given in Table 2, are enhanced in all the analyzed cases by cooverexpression of the secretion helper factors, thereby raising 2F5 Fab titers in the culture supernatants as well as QP levels compared to those in the control strain (Fig. 2). While an additional copy of BFR2 leads to a moderate increase of 20% in the measured parameters, overexpression of regulator protein BMH2, the vacuolar ATPase subunit CUP5, or the chaperone SSA4 improves process yields up to 1.7-fold (as illustrated in Fig. 3). PDI1 overexpression, which was used as a positive (previously established) control in our study, raised 2F5 Fab titers to 89.2 mg liter−1 (1.9-fold increase) and resulted in a 2.3-fold improvement in QP. As depicted in Fig. 3, these results were even topped by overexpression of the newly identified secretion helper factors KIN2 and SSE1, where heterologous protein secretion was enhanced 2.2- and 2.3-fold, respectively, mounting up to 108.5 mg liter−1 Fab in the supernatant. A 2.3-fold increase in qP led to a 2.5-fold improvement in QP. All the fermentation parameters are summarized in Table 2, whereas the improvements in terms of total QP, qP, and the antibody fragment titers of the six novel secretion helper factors and Pdi1 compared to the control culture value are presented in Fig. 3.
FIG. 3.
Improvements in antibody fragment secretion yields by overexpression of novel helper factors in yeast. The increases (n-fold) of qP (striped bars), maximum QP (QPmax) (gray bars), the product yield coefficient YP/X (dotted bars), and the final antibody fragment titer (black bars) during fed-batch cultivations are displayed for the secretion-optimized P. pastoris strains and their parental strain.
DISCUSSION
Novel secretion helper factors for the production of heterologous proteins in yeasts could be identified by mining transcriptional regulation data from a trypsinogen-expressing strain and a wild-type P. pastoris strain hybridized to S. cerevisiae cDNA microarrays (29). As we aimed to determine general secretion helpers with potential benefits for more than just one specific heterologous protein and expression system, a reporter different from human trypsinogen was evaluated in order to preclude protein specific effects. The Fab fragment of the anti-human immunodeficiency virus antibody 2F5 was chosen because it has been shown previously that secretion levels were increased significantly in P. pastoris strains engineered for the coexpression of PDI1 or the UPR transcription factor Hac1 (both derived from S. cerevisiae) (13).
The human antibody fragment is a heterodimer (heavy chain and light chain) containing several disulfide bonds and shares with trypsinogen the fate of being partially intracellularly retained although targeted for secretion in P. pastoris, thereby inducing the UPR pathway and indicating a bottleneck in the secretory pathway (13, 15).
Thirteen potentially beneficial proteins, or their respective genes, were selected from 524 differentially regulated genes, based on their functions and intracellular locations in S. cerevisiae (Table 3). They are supposed to be connected to either the protein secretory pathway or general stress responses and include already known folding helpers, such as the endoplasmic reticulum (ER) chaperone BiP/Kar2, protein disulfide isomerase Pdi1, Pdi oxidase Ero1, and a protein acting later in the secretory pathway, Sso2, in addition to novel putative helper proteins.
TABLE 3.
Changes in transcript levels of P. pastoris secretion- and stress-related genes due to recombinant protein secretion
| Common name | S. cerevisiae ORF designation | trp/wt ratioa | Descriptionb |
|---|---|---|---|
| BFR2 | YDR299W | 1.7 | Essential protein possibly involved in secretion; multicopy suppressor of sensitivity to brefeldin A |
| BMH2 | YDR099W | 1.6 | 14-3-3 protein isoform; binds proteins and DNA, involved in regulation of many processes, including exocytosis and vesicle transport |
| COG6 | YNL041C | 2.1 | Component of the conserved oligomeric Golgi complex (Cog1p through Cog8p), a cytosolic tethering complex that functions in protein trafficking to mediate fusion of transport vesicles to Golgi compartments |
| COY1 | YKL179C | 1.9 | Golgi membrane protein with similarity to mammalian CASP; genetic interactions with GOS1, involved in tethering of transport vesicles and in the organization of the Golgi stack |
| CUP5 | YEL027W | 4.3 | Proteolipid subunit of the V-ATPase V0 sector (subunit c; dicyclohexylcarbodiimide binding subunit); required for vacuolar acidification and important for copper and iron metal ion homeostasis |
| ERO1 | YML130C | #NV | Pdi oxidase; glycoprotein required for oxidative protein folding in the ER, protein thiol-disulfide exchange |
| HAC1 | YFL031W | #NV | bZIP transcription factor that regulates the unfolded protein response, via UPRE binding, and membrane biogenesis |
| IMH1 | YLR309C | 2.1 | Protein involved in vesicular transport; mediates transport between an endosomal compartment and the Golgi; contains a Golgi localization (GRIP) domain |
| KAR2 | YJL034W | 2.0 | Binding protein BiP, an ATPase involved in protein import into the ER, acts as a chaperone to mediate protein folding in the ER; regulates the unfolded protein response, involved in ER quality control and ER-associated degradation |
| KIN2 | YLR096W | 2.1 | Serine/threonine protein kinase involved in regulation of exocytosis; localizes to the cytoplasmic face of the plasma membrane |
| PDI1 | YCL043C | 2.9 | Protein disulfide isomerase catalyzes the formation and isomerization of disulfide bonds during the folding of secretory proteins |
| SEC31 | YDL195W | 2.4 | COPII coat of secretory pathway vesicle component (p150), involved in protein transport from endoplasmic reticulum to Golgi, structural molecule |
| SSA4 | YER103W | 3.4 | Cytoplasmic member of 70-kDa heat shock protein family, chaperone activity, involved in cotranslational targeting and translocation of nascent polypeptides into the ER |
| SSE1 | YPL106C | 2.2 | Chaperonine in the HSP110 subclass of the HSP70 family, nuclease exchange factors for HSP70 chaperones, ATPase of the Hsp90 chaperone complex; binds unfolded proteins; localized to the cytoplasm |
| SSO2 | YMR183C | 2.1 | Plasma membrane t-SNARE involved in fusion of secretory vesicles at the plasma membrane; syntaxin homolog that is functionally redundant with Sso1 |
Ratios of the average gene expression levels of the P. pastoris expression strain X33+pPICZαB_trypsinogen (trp) and the nonexpressing control X33 (wt) during methanol-induced recombinant protein expression in fed batch fermentations, hybridized to S. cerevisiae cDNA microarrays. #NV indicates genes that gave no signal in the microarray experiment.
The functions of the putative secretion helper factors are derived from the S. cerevisiae homologs of the P. pastoris genes (adapted from the Saccharomyces Genome Database [http://www.yeastgenome.org]).
In shake flask cultures, the newly identified helper factors improved heterologous protein secretion as efficiently as or even more efficiently than known candidates. In particular, overproduction of subunit c of the yeast vacuolar (H+)-ATPase (V-ATPase) V0 domain Cup5 was able to stimulate Fab secretion as much as Pdi1 (1.7-fold increase in relative Fab productivity). V-ATPases belong to a family of ATP-dependent proton pumps that acidify several organelles, including the yeast central vacuole, endosomes, the Golgi network, and secretory vesicles (8, 9). The V0 domain is an integral membrane structure of five subunits responsible for transporting protons across the membrane. Assembly of the V0 domain is not possible in the absence of Cup5. V-ATPase function is important for many processes, including endocytosis, protein processing, degradation and targeting, and coupled transport across the vacuolar membrane (10). However, so far we cannot draw a conclusion as to how the functions of Cup5 influence recombinant protein secretion. Eide et al. (6) report a role for V-ATPase in detoxification of copper, iron metabolism, and mitochondrial function.
The cytosolic chaperone Ssa4 is involved in the targeting of nascent proteins to the ER membrane prior to the cotranslational translocation of the proteins into the ER lumen, whereas the 14-3-3 protein encoded by BMH2 was identified to participate in multiple steps of vesicular trafficking, especially in protein exit from the ER and forward trafficking of multimeric cell surface membrane proteins (23). Kin2 and the closely related protein Kin1 are two serine/threonine protein kinases localized at the cytoplasmic side of the plasma membrane. According to Elbert et al. (7), the catalytic activity of Kin2 is essential for its function in regulation of exocytosis by phosphorylation of the plasma membrane t-SNARE Sec9, a protein acting at the final step of exocytosis. By overexpression of any of these three genes, similar levels of improvement (1.5-fold) in heterologous protein secretion could be achieved as in strains overexpressing KAR2 or HAC1.
The flavoenzyme Ero1, which is required for oxidation of protein dithiols in the ER and acts as a specific oxidant of protein disulfide isomerase (Pdi), has been shown to increase human serum albumin yields in K. lactis (19) and scFv in S. cerevisiae (38). Overexpression of ERO1 in P. pastoris led to a 1.4-fold increase in 2F5 Fab secretion in shake flask cultivations, similarly to the novel factors SSE1 and BFR2. Their impact on protein secretion is not as obvious: the chaperonines of the Sse/Hsp110 subclass of the Hsp70 family only assist in folding by binding to nascent peptides and holding them in a folding-competent state; however, they cannot actively promote folding reactions. Even less is known about the function of Bfr2, which has been isolated as a multicopy suppressor of the drug brefeldin A, a fungal metabolite that perturbs the protein flux into the Golgi apparatus and the structure of the Golgi apparatus itself (4).
Although the yeast syntaxin homologs Sso1 and Sso2, which are necessary for the fusion of secretory vesicles to the plasma membrane by acting as t-SNAREs (soluble N-ethylmaleimide-sensitive factor attachment protein receptors), have been reported to increase the amounts of secreted proteins in S. cerevisiae on average twofold (17, 31), in our study the improvement was rather moderate (20%). However, Ruohonen et al. (26) concluded that the enhancing effect of SSO overexpression correlated with the amount of Sso proteins, which were up to 20 times overproduced in their experiments (due to multiple copies of the SSO gene), while we integrated only one additional copy of S. cerevisiae SSO2 into the P. pastoris genome.
Similar enhancements (1.2-fold) in relative Fab productivity were achieved by the overexpression of three other putative secretion helper factors connected to the Golgi vesicle-mediated transport, more precisely COG6, COY1, and IMH1. COG6 belongs to one of eight genes coding for the conserved oligomeric Golgi (COG) complex, an eight-subunit peripheral Golgi protein that is engaged in membrane trafficking and synthesis of glycoconjugates. Moreover, the COG complex is not only necessary for maintaining normal Golgi structure and function but also directly involved in retrograde vesicular transport within the Golgi apparatus and retrograde traffic from the endosome to the Golgi apparatus (recently reviewed in reference 33). The product of the IMH1/SYS3 gene is a member of the peripheral membrane golgins involved in vesicular transport between a prevacuolar, endosome-like compartment and the late Golgi apparatus (32). While the molecular function of Coy1, a protein identified by similarity to mammalian CASP (14), is not established yet, it seems to play a role in Golgi vesicle transport through interaction with Gos1. Gos1 is a SNARE protein commonly used as a marker of later compartments of the Golgi apparatus in S. cerevisiae (22). Ungar et al. (33) suggested that, at least in mammalian cells, the homologs of latter proteins represent a subset of Golgi membrane proteins that are transported by COG complex-dependent vesicles. It may be speculated that overproduction of these Golgi transport network-associated proteins facilitates the retrieval of intra-Golgi enzymes, thereby stimulating the overall rate of protein processing (e.g., glycosylation), but that the effects on the heterologous antibody fragments are not as profound, as the bottleneck is elsewhere down the secretory pathway.
No effect on the productivity of 2F5 Fab could be seen when SEC31 was overexpressed. SEC31 encodes an essential phosphoprotein component of the COPII (coat protein complex II) coat of secretory pathway vesicles, in complex with Sec13 (27). This is most probably due to the fact that the Sec31/Sec13 subcomplex has a mainly structural role and needs the other two main components of the COPII complex to form the specialized cargo vesicles from the ER to the Golgi apparatus (reviewed in reference 3).
In most cases, the rate-limiting step in the eukaryotic secretion pathway has been identified to be the exit of proteins from the ER to the Golgi apparatus (30). However, when the results obtained in our coexpression experiment are examined in more detail, it becomes obvious that interfering with folding and translocation into the ER results in an increased flux into the secretory pathway and thus improves heterologous protein productivity (up to 1.7-fold) but is still limited as it just shifts the bottlenecks.
Consequently, combinations of the individual helper factors may overcome these limitations. The combination of Pdi1 and its oxidase Ero1 seems especially logical, as disulfides generated de novo within Ero1 are transferred to Pdi and then to substrate proteins by dithiol-disulfide exchange reactions (11). Additionally, auxiliary factors in the early steps of the secretory pathway (e.g., translocation into the ER, folding and disulfide bonding in the ER, the ER quality control mechanism) should be merged with enhanced rates of posttranslational protein processing and transport by overexpression of Golgi network and vesicular elements.
The aim of the present study was, however, to discover single novel secretion helper factors with the ability to increase heterologous protein productivity in fungal hosts. In order to verify the results obtained in shake flasks, the novel helper factors were tested along with Pdi1 as a positive reference in fed-batch fermentations. Compared to that of a control strain (containing the empty vector only), specific 2F5 Fab productivity could be improved in all of the analyzed coexpression strains, ranging from 1.2-fold in the case of BFR2 to 2.5-fold when SSE1 is overexpressed. Unlike for previous studies, where overexpression of helper proteins was reported to impair growth of the engineered hosts (2, 34), we cannot see any significant difference in mean specific growth rates for either of the strains in our experiments during the batch. As can be seen in Fig. 1, the development of biomass over time for the vector control strain is similar to that for the secretion-optimized strains.
Taken together, increased productivity and unaffected growth rates give rise to increased titers of antibody fragments in the culture supernatants, thereby enhancing QP (Fig. 2). Optimization of QP is crucial for the reduction of overall production process costs, as described by Werner (39). Apart from Bfr2 (1.2-fold increase), all the other novel secretion helpers improved QP more than 1.5 times, whereby the chaperonine Sse1 and the protein kinase Kin2 are especially outstanding (2.3-fold increase). Overexpression of the vacuolar protease subunit Cup5, the chaperone Ssa4, and the regulator protein Bmh2 resulted in QP elevation similar to that for the reference protein Pdi1 (1.7 to 1.8-fold, all summarized in Fig. 3).
The process of protein folding and subsequent secretion is rather complex, with many interacting participants. Due to this tight interdependence of each of the participating factors, increasing the rate of one step can lead to rate limitation of the following one, which can then become the bottleneck of the expression system. Therefore, additional studies will be required to investigate potential synergetic actions of combinations of the individual secretion helper factors in the future. Interestingly, some of the genes identified as novel secretion helper factors in our study appear to be induced during B-cell differentiation (28, 37). The gradual increase of cytosolic chaperones and resident ER proteins, such as BiP and Pdi, upon B-cell proliferation can be easily explained by their task of becoming professional secretors of large amounts of immunoglobulins. More recently, Lee and coworkers reported the induction of subunits of the COG complex and of the vacuolar ATPase upon stimulation through the B-cell antigen receptor (18), indicating that the responses to high-level protein production are evolutionary conserved from yeasts to mammalian cells. This can give rise to the mutual exploitation of beneficial knowledge about secretory components.
Acknowledgments
This work was supported by the Austrian Research Promotion Agency (program FHplus) and Polymun Scientific GmbH, Vienna, Austria.
We thank Stefanie Müller for her excellent technical assistance with fermentation.
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Archer, D., D. Jeenes, and D. Mackenzie. 1994. Strategies for improving heterologous protein production from filamentous fungi. Antonie Leeuwenhoek 65:245-250. [DOI] [PubMed] [Google Scholar]
- 2.Bao, W., and H. Fukuhara. 2001. Secretion of human proteins from yeast: stimulation by duplication of polyubiquitin and protein disulfide isomerase genes in Kluyveromyces lactis. Gene 272:103-110. [DOI] [PubMed] [Google Scholar]
- 3.Bickford, L., E. Mossessova, and J. Goldberg. 2004. A structural view of the COPII vesicle coat. Curr. Opin. Struct. Biol. 14:147-153. [DOI] [PubMed] [Google Scholar]
- 4.Chabane, S., E. Gachet, and F. Képès. 1998. Over-expression of the yeast BFR2 gene partially suppresses the growth defects induced by Brefeldin A and by four ER-to-Golgi mutations. Curr. Genet. 33:21-28. [DOI] [PubMed] [Google Scholar]
- 5.Conesa, A., P. J. Punt, N. van Luijk, and C. A. van den Hondel. 2001. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33:155-171. [DOI] [PubMed] [Google Scholar]
- 6.Eide, D., J. Bridgham, Z. Zhao, and J. Mattoon. 1993. The vacuolar H(+)-ATPase of Saccharomyces cerevisiae is required for efficient copper detoxification, mitochondrial function, and iron metabolism. Mol. Gen. Genet. 241:447-456. [DOI] [PubMed] [Google Scholar]
- 7.Elbert, M., G. Rossi, and P. Brennwald. 2005. The yeast par-1 homologs kin1 and kin2 show genetic and physical interactions with components of the exocytic machinery. Mol. Biol. Cell 16:532-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forgac, M. 1989. Structure and function of vacuolar class of ATP-driven proton pumps. Physiol. Rev. 69:765-796. [DOI] [PubMed] [Google Scholar]
- 9.Forgac, M. 1998. Structure, function and regulation of the vacuolar (H+)-ATPases. FEBS Lett. 440:258-263. [DOI] [PubMed] [Google Scholar]
- 10.Forgac, M. 2000. Structure, mechanism and regulation of the clathrin-coated vesicle and yeast vacuolar H(+)-ATPases. J. Exp. Biol. 203:71-80. [DOI] [PubMed] [Google Scholar]
- 11.Frand, A. R., and C. A. Kaiser. 1998. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell 1:161-170. [DOI] [PubMed] [Google Scholar]
- 12.Gasser, B., and D. Mattanovich. 2007. Antibody production with yeasts and filamentous fungi: on the road to large scale? Biotechnol. Lett. 29:201-212. [DOI] [PubMed] [Google Scholar]
- 13.Gasser, B., M. Maurer, J. Gach, R. Kunert, and D. Mattanovich. 2006. Engineering of Pichia pastoris for improved production of antibody fragments. Biotechnol. Bioeng. 94:353-361. [DOI] [PubMed] [Google Scholar]
- 14.Gillingham, A., A. Pfeifer, and S. Munro. 2002. CASP, the alternatively spliced product of the gene encoding the CCAAT-displacement protein transcription factor, is a Golgi membrane protein related to giantin. Mol. Biol. Cell 13:3761-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hohenblum, H., B. Gasser, M. Maurer, N. Borth, and D. Mattanovich. 2004. Effects of gene dosage, promoters, and substrates on unfolded protein stress of recombinant Pichia pastoris. Biotechnol. Bioeng. 85:367-375. [DOI] [PubMed] [Google Scholar]
- 16.Lang, C., and A. Looman. 1995. Efficient expression and secretion of Aspergillus niger RH5344 polygalacturonase in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 44:147-156. [DOI] [PubMed] [Google Scholar]
- 17.Larsson, S., P. Cassland, and L. Jönsson. 2001. Development of a Saccharomyces cerevisiae strain with enhanced resistance to phenolic fermentation inhibitors in lignocellulose hydrolysates by heterologous expression of laccase. Appl. Environ. Microbiol. 67:1163-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, J. A., R. S. Sinkovits, D. Mock, E. L. Rab, J. Cai, P. Yang, B. Saunders, R. C. Hsueh, S. Choi, S. Subramaniam, and R. H. Scheuermann. 2006. Components of the antigen processing and presentation pathway revealed by gene expression microarray analysis following B cell antigen receptor (BCR) stimulation. BMC Bioinformatics 7:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lodi, T., B. Neglia, and C. Donnini. 2005. Secretion of human serum albumin by Kluyveromyces lactis overexpressing KlPDI1 and KlERO1. Appl. Environ. Microbiol. 71:4359-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macauley-Patrick, S., M. L. Fazenda, B. McNeil, and L. M. Harvey. 2005. Heterologous protein production using the Pichia pastoris expression system. Yeast 22:249-270. [DOI] [PubMed] [Google Scholar]
- 21.Maurer, M., M. Kuhleitner, B. Gasser, and D. Mattanovich. 2006. Versatile modeling and optimization of fed batch processes for the production of secreted heterologous proteins with Pichia pastoris. Microb. Cell. Fact. 5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNew, J., J. Coe, M. Søgaard, B. Zemelman, C. Wimmer, W. Hong, and T. Söllner. 1998. Gos1p, a Saccharomyces cerevisiae SNARE protein involved in Golgi transport. FEBS Lett. 435:89-95. [DOI] [PubMed] [Google Scholar]
- 23.Michelsen, K., T. Mrowiec, K. Duderstadt, S. Frey, D. Minor, M. Mayer, and B. Schwappach. 2006. A multimeric membrane protein reveals 14-3-3 isoform specificity in forward transport in yeast. Traffic 7:903-916. [DOI] [PubMed] [Google Scholar]
- 24.Porro, D., M. Sauer, P. Branduardi, and D. Mattanovich. 2005. Recombinant protein production in yeasts. Mol. Biotechnol. 31:245-259. [DOI] [PubMed] [Google Scholar]
- 25.Punt, P. J., N. van Biezen, A. Conesa, A. Albers, J. Mangnus, and C. van den Hondel. 2002. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 20:200-206. [DOI] [PubMed] [Google Scholar]
- 26.Ruohonen, L., J. Toikkanen, V. Tieaho, M. Outola, H. Soderlund, and S. Keranen. 1997. Enhancement of protein secretion in Saccharomyces cerevisiae by overproduction of Sso protein, a late-acting component of the secretory machinery. Yeast 13:337-351. [DOI] [PubMed] [Google Scholar]
- 27.Salama, N., J. Chuang, and R. Schekman. 1997. Sec31 encodes an essential component of the COPII coat required for transport vesicle budding from the endoplasmic reticulum. Mol. Biol. Cell 8:205-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salonen, J. M., L. Valmu, G. Ronnholm, N. Kalkkinen, and M. Vihinen. 2006. Proteome analysis of B-cell maturation. Proteomics 6:5152-5168. [DOI] [PubMed] [Google Scholar]
- 29.Sauer, M., P. Branduardi, B. Gasser, M. Valli, M. Maurer, D. Porro, and D. Mattanovich. 2004. Differential gene expression in recombinant Pichia pastoris analysed by heterologous DNA microarray hybridisation. Microb. Cell. Fact. 3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shuster, J. 1991. Gene expression in yeast: protein secretion. Curr. Opin. Biotechnol. 2:685-690. [DOI] [PubMed] [Google Scholar]
- 31.Toikkanen, J., L. Sundqvist, and S. Keränen. 2004. Kluyveromyces lactis SSO1 and SEB1 genes are functional in Saccharomyces cerevisiae and enhance production of secreted proteins when overexpressed. Yeast 21:1045-1055. [DOI] [PubMed] [Google Scholar]
- 32.Tsukada, M., E. Will, and D. Gallwitz. 1999. Structural and functional analysis of a novel coiled-coil protein involved in Ypt6 GTPase-regulated protein transport in yeast. Mol. Biol. Cell 10:63-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ungar, D., T. Oka, M. Krieger, and F. M. Hughson. 2006. Retrograde transport on the COG railway. Trends Cell Biol. 16:113-120. [DOI] [PubMed] [Google Scholar]
- 34.Vad, R., E. Moe, K. Saga, A. Kvinnsland, and T. Oyen. 1998. High-level production of human parathyroid hormone (hPTH) by induced expression in Saccharomyces cerevisiae. Protein Expr. Purif. 13:396-402. [DOI] [PubMed] [Google Scholar]
- 35.Valkonen, M., M. Penttilä, and M. Saloheimo. 2003. Effects of inactivation and constitutive expression of the unfolded-protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valkonen, M., M. Ward, H. Wang, M. Penttilä, and M. Saloheimo. 2003. Improvement of foreign-protein production in Aspergillus niger var. awamori by constitutive induction of the unfolded-protein response. Appl. Environ. Microbiol. 69:6979-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Anken, E., E. P. Romijn, C. Maggioni, A. Mezghrani, R. Sitia, I. Braakman, and A. J. Heck. 2003. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity 18:243-253. [DOI] [PubMed] [Google Scholar]
- 38.Wentz, A. E., and E. V. Shusta. 2007. A novel high-throughput screen reveals yeast genes that increase secretion of heterologous proteins. Appl. Environ. Microbiol. 73:1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werner, R. G. 2004. Economic aspects of commercial manufacture of biopharmaceuticals. J. Biotechnol. 113:171-182. [DOI] [PubMed] [Google Scholar]