Abstract
Recent studies have shown that the fecal indicator bacteria (FIB) currently used to indicate water quality in the coastal environment may be inadequate to reflect human viral contamination. Coliphage was suggested as a better indicator of human viral pollution and was proposed by the U.S. EPA as an alternative indicator for fecal pollution in groundwater. In this study, we investigated the occurrence and distribution of FIB, F+ coliphage, and PCR-detectable human adenovirus and enterovirus for an entire year at 15 locations around the Newport Bay watershed, an important southern California estuary for water recreation and an ecological reserve. Peak concentrations and prevalences of FIB and F+ coliphage were associated with winter storms (wet weather). Human adenoviruses and enteroviruses, however, were detected by PCR in ∼5% of samples collected in the summer (dry weather) but only once in wet weather. These results demonstrated that FIB and coliphage have similar seasonal and freshwater-to-saltwater distribution patterns, while the detection of human viruses depends on a distribution pattern that is the opposite of that of FIB and coliphage. This research suggested that coliphage and FIB share similar environmental sources, while sources of human viruses in Newport Bay are perhaps different.
Coastal recreational water quality standards in California and throughout most of the world are based on the concentration of coliforms or Enterococcus spp., known as fecal indicator bacteria (FIB). However, the adequacy of current water quality standards to indicate the presence of human viral pathogens is still questionable. For example, Marino et al. (23) conducted a study of two Mediterranean bathing beaches, using WHO/UNEP and European Community bathing water directives as the microbial water quality guidelines. They concluded that neither set of guidelines was successful for protecting the public from health hazards related to fecal contamination of bathing water (23).
FIB are from human or other warm-blooded animal waste. Studies have also shown that organic-rich soil can support the growth of Escherichia coli (references 6, 10, 14, and 28 and R. Whitman, presentation at Sustainable Beaches Conference 2005, St. Petersburg, FL). Therefore, the high concentration of FIB washed out of soil by rain may have little or no relationship to human pathogens that are associated only with human sewage. Furthermore, FIB are more susceptible to sewage treatment processes and environmental degradation than are human viruses (4). Thus, it is now recognized that the absence or low concentrations of FIB in water may not adequately reflect the absence of human viruses (11, 12, 16, 17, 24). In our previous study of southern California coastal waters, over 30% of coastal waters tested contained genomes of human adenoviruses, and the presence of these viral genomes did not correlate well with elevated levels of FIB (16). In addition, results from a preliminary investigation of Newport Bay (also referred to herein as the Bay) water quality conducted during the summers of 2000 and 2001 indicated that the occurrence of human enteroviruses did not correlate with that of FIB (R. Noble, unpublished data).
Viruses are suspected to be important causative agents of waterborne illness; however, viral diseases are hard to identify by current diagnostic techniques. The Centers for Disease Control and Prevention (7) estimated that viral infection may be the causative agent of nearly 50% of all acute gastrointestinal illnesses. Therefore, viral contamination of recreational coastal water is of particular importance and is a rising public health concern. F+ coliphage was recently proposed by the U.S. EPA as a surrogate for human viral contamination in groundwater because some groups of F+ coliphage resemble human polioviruses and other enteric viruses in terms of survival and morphology (1-3, 22). F+ coliphage are most prevalent in human sewage but may not be present in individual human feces. They were also shown to be prevalent in fecal slurry collected from animal farms but rarely isolated from animal feces (20, 21).
Newport Bay is an important estuary in southern California. To protect the public health and the beneficial uses of this valuable resource, the State Water Quality Control Board required strategies to be developed and implemented to improve Newport Bay water quality to meet recreational standards (REC-1) by 2014. Here we report an analysis of PCR-detectable human adenoviruses and enteroviruses in various locations in the Newport Bay watershed and their relationship to FIB and F+ coliphage.
MATERIALS AND METHODS
Study area.
The Newport Bay watershed encompasses a mixed urban and agriculture area of approximately 154 square miles (400 km2), with overland flows draining toward the Pacific coast into Newport Bay (Fig. 1). The principle tributary of Newport Bay is San Diego Creek, which accounts for 122 square miles (316 km2), or about 80%, of the Newport Bay watershed. Other drainage areas include the Santa Ana-Delhi Channel, the Big Canyon Wash, and some additional small tributaries.
FIG. 1.
Map of Newport Bay, California, showing sampling location sites A through O of this study.
Fifteen locations in the Newport Bay watershed were selected for this study (Fig. 1). Sites included locations with historically high concentrations of FIB, high-use recreational beaches with historically low concentrations of FIB, and harbors and marinas (http://www.ocbeachinfo.com/). Three sampling sites were located in the tributaries of Newport Bay: one in San Diego Creek (Fig. 1A) and two in the Santa Ana-Delhi Channel (Fig. 1B and C). A fourth tributary-related site was located at the mouth of Big Canyon Wash (Fig. 1D). All four tributary locations, along with 43rd Street Beach (Fig. 1E) and Harbor Patrol Beach (Fig. 1F), have historically high concentrations of FIB and are considered to be FIB “hot spots” (http://www.ocbeachinfo.com/). Recreational beach sites include North Star Beach (Fig. 1G), Newport Dunes North (Fig. 1H) and East (Fig. 1I and J), Bayshore Beach (Fig. 1K), and 43rd Street Beach. Harbor and marina sites included Balboa Yacht Basin (Fig. 1L), 10th Street Beach (Fig. 1M), Garnet Ave. Beach (Fig. 1N), and Harbor Patrol Beach. Rocky Point (Fig. 1O), near the mouth of the lower bay, is the farthest site from the influence of urban runoff. However, the frequency of boating activities and the density of bird population are high around this site.
Rainfall data were retrieved from California Irrigation Management System (CIMS) rain stations (http://www.ipm.ucdavis.edu). The Santa Ana fire station is the closest rain station covering the study area.
Sampling procedures.
Water samples were collected from each site between 6 and 9 a.m., beginning 22 May 2002 at biweekly intervals between May and September (summer) and at monthly intervals for the rest of the year. Sampling dates included weekends, holidays, and weekdays to capture the variability of water quality during high-use versus low-use periods. A total of 206 samples were collected.
At each site, water samples were taken at ankle depth in sterilized sampling bottles. Controls were used to ensure that the sampling procedure did not have carryover contamination from sample to sample. Water temperature was measured in situ by using a calibrated thermometer. Salinity was determined using refraction index. Water samples were transported within 4 h of collection to the Orange County Sanitation District laboratory for FIB analysis and to the UC Irvine laboratory for immediate processing for human viruses and F+ coliphage.
FIB analysis.
FIB, including total coliform, fecal coliform, and Enterococcus bacteria, were assayed by the state-certified microbiology laboratory for water quality testing at Orange County Sanitation District. Multitube fermentation was used for total and fecal coliform bacteria assay, and an Enterolert testing kit (IDEXX, Westbrook, ME) was used for testing for enterococci. Multitube fermentation procedures and reporting followed protocols found in Standard Methods for the Examination of Water and Wastewater (8). Enterolert procedures followed the manufacturer's protocol (IDEXX, Westbrook, ME). Previous study has shown that Enterolert testing kit results are comparable to those of traditional methods (25).
F+ coliphage assay by culture enrichment.
Two-step enrichment was carried out by using five-tube replicates to determine the most probable number (MPN) of F+ coliphage, following EPA protocol 1601. In brief, five 100-ml water samples from each site were amended with 5 ml sterile 10× trypticase soy broth medium (Difco Lab), 0.5 ml log-phase E. coli HS (pFamp)R host (ATCC 700891), and ampicillin and streptomycin in a final concentration of 15 mg/liter. Negative controls contained five tubes each with 100 ml of sterile water amended with nutrient medium, E. coli host, and antibiotics, as for the regular sample assay. The enrichment cultures were incubated at 37°C for 18 to 24 h before spot testing for the presence of F+ coliphage was performed. For spot testing, 2 μl of supernatant from the enriched culture was spotted onto preformed E. coli bacterial lawns on trypticase soy agar plates (Difco Lab). Clear spots on the bacterial lawn were scored, and the MPN value was calculated using Thomas' MPN formula (8).
Viral concentration.
For human virus detection, 500 ml of water from each site was concentrated ∼1,000-fold to a final volume of ∼500 μl, using a Centricon Plus-80 ultrafiltration system with a 100-kDa molecular mass cut-off membrane (Millipore Inc.). To determine the rate of viral recovery with this concentration system, bacteriophage φHSIC, isolated from the Sand Island Canal, Hawaii (18), was seeded into a subset of Newport Bay samples before processing. Portions of concentrate (100 μl) were serially diluted to determine the titer of φHSIC before and after concentration and were used to calculate viral recovery rates. Since φHSIC was not found in the Newport Bay water, there was no interference for this phage titration. Concentrates were stored frozen at −80°C and used for human virus assay by PCR at a later date.
Detection of viruses by PCR.
Water concentrates were subjected to extraction of viral nucleic acid, using the method originally developed by Boom et al. (5) with minor modifications (16). A comparison of this extraction method with that of the QIAmp viral RNA mini-kit (QIAGEN Inc., CA) showed no significant differences in viral nucleic acid recovery and removal of PCR inhibition from water concentrates (S. C. Jiang et al., unpublished data).
Primers used for specific amplification of the enteroviruses were 5′-CCTCCGGCCCCTGAATG-3′ and 5′-ACCGGATGGCCAATCCAA-3′, which target the 5′ untranslated region (R. DeLeon, Y. S. C. Shieh, R. S. Baric, and M. D. Sobey, presentation at the Water Quality Conference, San Diego, CA, 1990). The procedure for reverse transcription PCR (RT-PCR) of enterovirus followed the protocol developed by Tsai et al. (31), with a modification of the total reaction volume. Amplification products were further confirmed by probing with an internal oligonucleotide probe (5′-TACTTTGGGTGTCCGTGTTTC-3′) after Southern transfer of amplicons to a charged nylon membrane (MSI, Inc.) as previously described (17).
For adenovirus detection, a nested PCR protocol was used as previously described by Pina et al. (27). The outer primers used were 5′-GCCGCAGTGGTCTTACATGCACATC-3′ and 5′-CAGCACGCCGCGGATGTCAAAGT-3′, which yielded a 301-bp amplicon of the hexon gene. The nested primers used were 5′-GCCACCGAGACGTACTTCAGCCTG-3′ and 5′-TTGTACGAGTACGCGGTATCCTCGCGGTC-3′, which yielded a 143-bp amplicon (27). Viral nucleic acid extracts (1 to 3 μl) were used as the template for a nested PCR or an RT-PCR assay of each virus.
Statistical analysis.
All statistical analyses were performed using SPSS software (SPSS, Inc. Chicago, IL). The presence/absence data for virus were presented as the frequency of detection for temporal and spatial analysis. The mean values of temperature, salinity, and FIB were used for the analyses. Pearson correlation and partial correlation, controlling for temperature, salinity, and season, were used to test the relationship among FIB, coliphage, and human viruses. An independent-samples t test was used to make seasonal (wet/dry) and site (tributary/bay) comparisons. For the seasonal comparison, May to September was considered summer, which was also referred to as dry weather because the precipitation level during summer was <0.5 in. for the entire season. October to April was considered winter for this study, which was also referred to as wet weather, with >7 in. of precipitation. In all analyses, a P value of less than 0.05 was considered significant.
RESULTS
Environmental parameters.
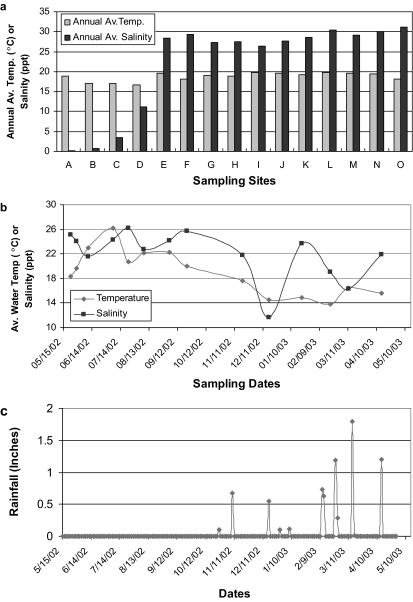
Figure 2 shows the seasonal and spatial distribution of average temperatures and salinity values for all study sites. There were no significant differences in water temperature among sampling sites located in the Bay (Fig. 2a), while tributary sites have slightly lower annual average temperatures than the Bay sites. In contrast, there were clear seasonal differences in water temperature. During summer, the water temperature averaged 22°C, approximately 5 to 7°C higher than in winter (Fig. 2b). Salinity at tributary sites averaged less than 3 parts per thousand (ppt), while Bay site salinity values ranged from 27 to 31 ppt (Fig. 2a). Seasonal salinity differences reflect seasonal rainfall, with salinity averaging 24 ppt during May through September (summer), while significant dips in salinity were found in December and March, which corresponded with rainstorms (Fig. 2b). Precipitation records indicated <0.5 in. of cumulative rainfall during summer, while the cumulative precipitation was >7 in. in winter. The first salinity depression was measured within 24 h after the rain, while the second salinity depression was measured 2 weeks after a series of storms and immediately before the largest storm of the season (Fig. 2c).
FIG. 2.
Average temperature and salinity by sampling sites (a) and dates (b) and the precipitation record for the area (c). The average water temperature is expressed as °C, and salinity is expressed as ppt.
FIB and F+ coliphage.
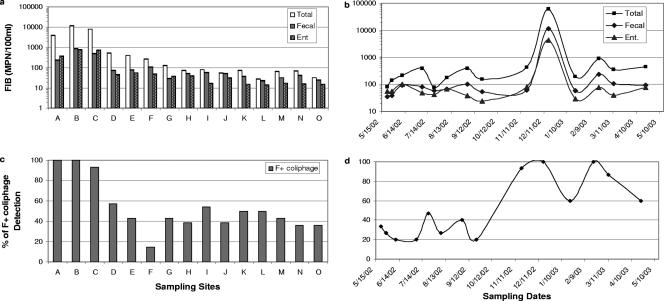
All three FIB (total coliform, fecal coliform, and Enterococcus) counts were significantly higher (t test, P < 0.01 for each type of bacteria) at tributary sites than at Bay sites (Fig. 3a). However, seasonal peaks of FIB concentration were associated with storms. Two peaks of elevated FIB were found following major winter storms, while concentrations fluctuated by 1 log unit during summer (Fig. 3b). Total and fecal coliform and enterococcus concentrations were significantly correlated (with partial correlation analysis controlling for temperature, salinity, and season; P < 0.01) and were inversely correlated with salinity (Pearson correlation, P < 0.01). However, an independent-samples t test revealed no significant differences for data binned for wet (winter, Oct. to April) and dry (summer, May to Sept.) weather (P > 0.05).
FIG. 3.
Distribution of mean FIB MPN value by sampling site (a) and sampling date (b) in Newport Bay. Similar distribution patterns of F+ coliphage detection frequency were also observed by sampling site (c) and sampling date (d). Error bars show the 95% confidence interval for FIB. Total, total coliform bacteria; Fecal, fecal coliform bacteria; Ent, Enterococcus.
Similar patterns in F+ coliphage detection were also observed (Fig. 3c and d). Since no dilution was made for the multiple tube enrichment assay, the upper detection limit for F+ coliphage was an MPN of 1.79/100 ml. The concentration of F+ coliphage exceeded this MPN upper detection limit in most samples tested. Therefore, only the presence or absence of the coliphage was used. The frequency of detection instead of the mean concentration of coliphage was used for data analysis. Results showed that F+ coliphage was most frequently detected at tributary sites (t test, P < 0.01) and was found at all sites at two samplings following storms. The frequency of F+ coliphage detection was significantly higher in the winter season than in the summer (t test, P < 0.01). As for FIB, coliphage detections were negatively correlated with salinity (Pearson correlation, P < 0.01).
Adenovirus and enterovirus.
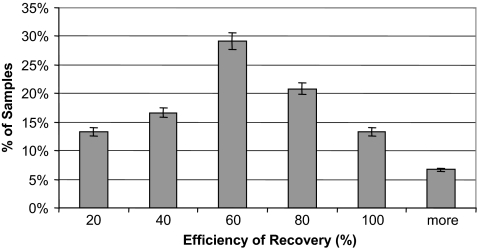
Since a high efficiency of viral recovery during viral concentration is critical for downstream detection, we first evaluated the viral recovery rates using a Centricon Plus-80 centrifugal ultrafiltration system (Fig. 4). Over 70% of samples had a recovery rate for φHSIC of greater than 60%. The phage, a Siphoviridae family member used for the seeding study, is larger than enterovirus but similar to adenovirus in head size. The recovery rate for enterovirus may be less than that indicated by the phage seeding study. In addition, the rate of recovery varied between sites and seasons. However, there is no clear trend of recovery rate by site or by season based on the current data. The variability may be attributed to virus binding to suspended solids (the total suspended solids effect) or adhering to the membrane filter surface. In an attempt to increase the viral recovery rate, a second round of elution using a glycine buffer (pH 9.0) was performed for selected samples, but this second round of elution recovered less than 15% of remaining virus (data not shown). Combining a second elution with the first elution would dilute the viral concentration. Therefore, a second round of elution was not routinely performed during sample processing. Due to the variability of the recovery rate, the recovery experiment data provided only understanding of the conservative nature of human virus detection. The information was not used to compute the human virus detection rate.
FIG. 4.
Frequency of viral recovery rates using a Centricon Plus-80 ultrafiltration system. The viral recovery rate is determined by seeding bacteriophage φHSIC. Error bars show 95% confidence intervals.
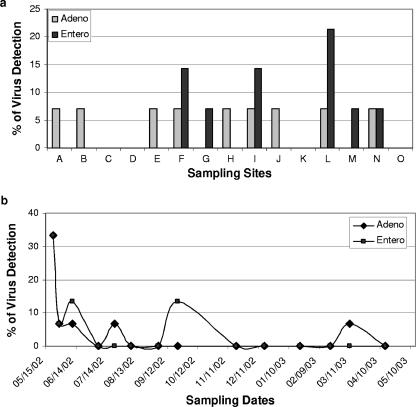
Results of the human virus assay by nested PCR and RT-PCR showed that on average, 4.3% and 4.8% of all samples were positive for human adenoviruses and enteroviruses, respectively. There were no statistical differences in the frequency of viral detection by sites due to the overall low frequency of viral signal (Fig. 5a). However, the frequency of viral detection showed a seasonal trend (Fig. 5b). All positive samples, except one, were found in summer. Only a single sample, collected on 11 March 2003, was positive for human adenovirus in the storm season (Fig. 5b). The seasonal detection rate for adenovirus and enteroviruses, however, was not statistically different between the wet and the dry seasons (t test, P > 0.05), again due to the overall low rate of viral detection.
FIG. 5.
Detection of human adenovirus (Adeno) and enterovirus (Entero) by sampling site (a) and sampling date (b).
Relationship of viruses and indicators.
The partial correlation analysis, controlling for temperature, salinity, and sampling date, showed that the seasonal detection of human adenovirus and enterovirus is negatively correlated to that of FIB and coliphage. However, these correlations were statistically insignificant (P > 0.05). Similarly, no statistical relationship can be identified by the sampling site between human viruses and FIB and coliphage.
DISCUSSION
Concentrations of FIB in Newport Bay displayed a strong seasonal pattern. This is in agreement with ∼30 years of historical monitoring data collected by the Orange County Health Care Agency (26). Peak FIB concentrations were associated with winter storms, suggesting that watershed-wide urban and agriculture storm runoff may be the major source of bacteria input into the Bay. This is consistent with the spatial distribution data that showed high levels of FIB in tributaries. Surbeck et al. (29) concluded that it was unlikely that a high load of FIB during storms originated from sewage because the loading of FIB calculated from storms far exceeded the total capacity of sewage treatment plants located within the watershed. They suggested a “mud puddle hypothesis,” in which FIB from animal feces, agriculture, and urban runoff persisted in mud puddles, which were collected with rain during storms.
F+ coliphage also displayed a temporal and spatial distribution pattern that was similar to FIB. Most striking was the 100% detection of coliphage from all sites in Newport Bay following winter storms. This suggested that F+ coliphage in Newport Bay were brought in by storm water, perhaps from the same source as FIB. Long et al. (20, 21) showed that although F+ coliphage were prevalent in sewage and animal fecal lagoons, they were rare in animal feces. They suggested that the high prevalence of coliphage in lagoons and sewage was a result of coliphage replication in these environments. The similar seasonal signatures of F+ coliphage and FIB in Newport Bay suggested that perhaps coliphage in storm runoff also fits the “mud puddle hypothesis.” Animal waste lagoons and stagnant small puddles in the watershed may provide an environment for the generation of coliphage. Our current knowledge of viruses in the environment suggests that the phages are never far behind their hosts (for a review see reference 32). It is logical that coliphages are closely associated with their hosts in the environment. Environmental conditions that promote the proliferation of coliform bacteria (reference 15 and R. Whitman presentation, 2005) may also permit the replication of their phages. Additional studies are necessary to provide validation of coliphage replication in the environment.
In contrast to FIB and coliphage, PCR-positive human viral genomes were more frequently found in dry weather (summer) than in wet weather (winter) in Newport Bay. One possible explanation for this may be the seasonal variability of enteric virus shedding to the environment. Tani et al. (30) reported enteroviruses were more prevalent during summer months. Alternatively, the positive detection may be due to heightened anthropogenic activities during summer. Random small localized sources, such as small sewage line problems and leaks or illegal discharges of vessel waste tanks by recreational boaters, leaks of vessel waste pump-out stations, and dock and marina wash-down activities, may have contributed to the human viral contamination in the Bay. However, this explanation was not supported by the City of Newport Beach sewer line and vessel waste pump-out station maintenance program, which inspects and prevents sewage leakage on a routine basis. A recent investigation of vessel waste disposal by boaters at two marinas in Newport Beach also demonstrated little or no impact of vessel waste on water quality, based on monitoring of FIB (15). However, the investigation did not consider viral contamination of the Newport Bay water.
Since PCR detects only genetic material, this assay cannot distinguish infectious viruses from the presence of fragments of viral genome. In addition, PCRs are sensitive to inhibitors in environmental samples, which may produce false-negative assay results. False negatives can also result from the loss of viral target during viral concentration, purification, and limited sample volume.
The occurrence of viral genome detected by PCR was not positively correlated either with FIB or with F+ coliphage. The lack of positive correlation between human viruses and FIB was not surprising since several previous studies had shown similar results (13, 19). The explanation for this lack of correlation was thought to be due to differential decay of human viruses and FIB in the environment. However, it was interesting to observe, in this study, an opposite seasonal trend for FIB and the PCR-detectable viral signals. One interpretation of this finding may be that FIB and viral signals detected by PCR were from different sources. Peak loads of FIB from Newport Bay were from watershed-wide storm runoff, which may include upstream agriculture (manure), urban animal waste (dogs, cats), and native animal waste (rabbits, coyotes, etc.) harboring high concentrations of FIB but not necessarily human waste. These high levels of FIB of nonhuman waste origin mask the relationship between FIB and human viruses of human waste origin. Alternatively, our ability to detect virus via PCR was significantly hampered by the presence of high levels of PCR inhibitors in the storm water. The storm water also dilutes the minute source of human waste contamination in the watershed, which reduced our ability to detect the viral signal. We have shown that the addition of storm water extracts significantly inhibited PCR efficiency in a previous study (29).
Coliphage was suggested as a better indicator for human virus because it shares decay characteristics that are similar to those of human viruses (1, 2, 11, 22). Several studies, including our own study of the southern California coast, showed a correlation between coliphage and PCR-detectable human viral genome (16). Colford et al. (9) also showed positive correlation, although weak, between F+ coliphage and recreational illness in a large epidemiological investigation in Mission Bay, California. However, few studies have quantified the seasonal change of coliphage prevalence and their relationship with human viruses on a watershed-wide scale. Perhaps the correlation between coliphage and human viral genome in previous studies was a reflection of decay characteristics between coliphage and human viruses when the sources of these two organisms were identical, while the seasonal decoupling of coliphage and human viruses in a watershed perhaps indicated multiple sources of input of these organisms. The similar seasonal patterns of coliphage and FIB imply that it was unlikely that the source of coliphage during the storm season in Newport Bay was of human origin.
Acknowledgments
Funding for this research was provided by the Santa Ana Regional Water Quality Control Board through the City of Newport Beach.
The fecal indicator bacteria data were kindly provided by Charles McGee of Orange County Sanitation District. We thank Linda Candelaria, Amanda Carr, Chris Crompton, Donna Ferguson, Larry Honeybourne, Dave Kiff, Monica Mazur, Charlie McGee, Douglas Moore, and Jack Skinner of the Newport Bay fecal coliform TMDL Technical Advisory Committee for helpful discussions. We also thank the Orange County Coast Keepers for assistance with sample collection, Sam Choi from UC Irvine for assistance with sample processing and statistical analysis, and Stuart Goong of County of Orange for editing the manuscript.
Footnotes
Published ahead of print on 24 August 2007.
REFERENCES
- 1.Allwood, P. B., Y. S. Malik, C. W. Hedberg, and S. M. Goyal. 2003. Survival of F-specific RNA coliphage, feline calicivirus, and Escherichia coli in water: a comparative study. Appl. Environ. Microbiol. 69:5707-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arraj, A., J. Bohatier, H. Laveran, and O. Traore. 2005. Comparison of bacteriophage and enteric virus removal in pilot scale activated sludge plants. J. Appl. Microbiol. 98:516-524. [DOI] [PubMed] [Google Scholar]
- 3.Atherholt, T., E. Feerst, B. Hovendon, J. Kwak, and J. D. Rosen. 2003. Evaluation of indicators of fecal contamination in groundwater. J. Am. Water Works Assoc. 95:119-131. [Google Scholar]
- 4.Berg, G. 1983. Viral pollution of the environment. CRC Press, Boca Raton, FL.
- 5.Boom, R., C. Sol, M. Beld, J. Weel, J. Goudsmit, and P. Wertheim-Van Dillen. 1999. Improved silica-guanidiniumthiocyanate DNA isolation procedure based on selective binding of bovine alpha-casein to silica particles. J. Clin. Microbiol. 37:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byappanahalli, M. N., and R. S. Fujioka. 1998. Evidence that tropical soil environment can support the growth of Escherichia coli. Water Sci. Technol. 38:171-174. [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1993. Water-related disease outbreaks, 1991-1992. Morb. Mortal. Wkly. Rep. 42:1-22. [Google Scholar]
- 8.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC.
- 9.Colford, J. M., T. J. Wade, K. C. Schiff, C. C. Wright, J. F. Griffith, S. K. Sandhu, S. Burns, M. Sobsey, G. Lovelace, and S. B. Weisberg. 2007. Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemiology 18:27-35. [DOI] [PubMed] [Google Scholar]
- 10.Fujioka, R., C. Sian-Denton, M. Borja, J. Castro, and K. Morphew. 1999. Soil: the environmental source of Escherichia coli and enterococci in Guam's streams. J. Appl. Microbiol. 85:S83-S89. [DOI] [PubMed] [Google Scholar]
- 11.Fujioka, R. S., and B. S. Yoneyama. 2002. Sunlight inactivation of human enteric viruses and fecal bacteria. Water Sci. Technol. 46:291-295. [PubMed] [Google Scholar]
- 12.Grabow, W. O. K., M. B. Taylor, and J. C. de Villiers. 2001. New methods for the detection of viruses: call for review of drinking water quality guidelines. Water Sci. Technol. 43:1-8. [PubMed] [Google Scholar]
- 13.Griffin, D. W., C. J. Gibson, E. K. Lipp, K. Riley, J. H. Paul, and J. B. Rose. 1999. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl. Environ. Microbiol. 65:4118-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishii, S., W. B. Ksoll, R. E. Hicks, and M. J. Sadowsky. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong, Y., S. B. Grant, S. Ritter, A. Pednekar, L. Candelaria, and C. Winant. 2005. Identifying pollutant sources in tidally mixed systems: case study of fecal indicator bacteria from marinas in Newport Bay, southern California. Environ. Sci. Technol. 39:9083-9093. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, S., R. Noble, and W. Chu. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, S. C., and W. Chu. 2004. PCR detection of pathogenic viruses in southern California urban rivers. J. Appl. Microbiol. 97:17-28. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, S. C., C. A. Kellogg, and J. H. Paul. 1998. Characterization of marine temperate phage-host systems isolated from Mamala Bay, Oahu, Hawaii. Appl. Environ. Microbiol. 64:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipp, E. K., S. A. Farrah, and J. B. Rose. 2001. Assessment and impact of microbial fecal pollution and human enteric pathogens in a coastal community. Mar. Pollut. Bull. 42:286-293. [DOI] [PubMed] [Google Scholar]
- 20.Long, S. C., S. S. El-Khoury, S. J. G. Oudejans, M. D. Sobsey, and J. Vinje. 2005. Assessment of sources and diversity of male-specific coliphages for source tracking. Environ. Eng. Sci. 22:367-377. [Google Scholar]
- 21.Long, S. C., E. Shafer, F. C. Arango, and D. Siraco. 2003. Evaluation of three source tracking indicator organisms for watershed management. J. Water Supply Res. Technol. AQUA 52:565-575. [Google Scholar]
- 22.Maillard, J. Y. 1996. Bacteriophages: a model system for human viruses. Lett. Appl. Microbiol. 23:273-274. [DOI] [PubMed] [Google Scholar]
- 23.Marino, F. J., M. A. Morinigo, E. Martinezmanzanares, and J. J. Borrego. 1995. Microbiological epidemiologic-study of selected marine beaches in Malaga (Spain). Water Sci. Technol. 31:5-9. [Google Scholar]
- 24.Meng, Q. S., and C. P. Gerba. 1996. Comparative inactivation of enteric adenoviruses, poliovirus, and coliphage by ultraviolet irradiation. Water Res. 30:2665-2668. [Google Scholar]
- 25.Noble, R. T., S. B. Weisberg, M. K. Leecaster, C. D. McGee, K. Ritter, K. O. Walker, and P. M. Vainik. 2003. Comparison of beach bacterial water quality indicator measurement methods. Environ. Monit. Assess. 81:301-312. [PubMed] [Google Scholar]
- 26.Pednekar, A. M., S. B. Grant, Y. Jeong, Y. Poon, and C. Oancea. 2005. Influence of climate change, tidal mixing, and watershed urbanization on historical water quality in Newport Bay, a saltwater wetland and tidal embayment in southern California. Environ. Sci. Technol. 39:9071-9082. [DOI] [PubMed] [Google Scholar]
- 27.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solo-Gabriele, H. M., M. A. Wolfert, T. R. Desmarais, and C. J. Palmer. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Surbeck, C. Q., S. C. Jiang, J. H. Ahn, and S. B. Grant. 2006. Flow fingerprinting fecal pollution and suspended solids in stormwater runoff from an urban coastal watershed. Environ. Sci. Technol. 40:4435-4441. [DOI] [PubMed] [Google Scholar]
- 30.Tani, N., Y. Dohi, N. Kurumatani, and K. Yonemasu. 1995. Seasonal distribution of adenoviruses, enteroviruses and reoviruses in urban river water. Microbiol. Immunol. 39:577-580. [DOI] [PubMed] [Google Scholar]
- 31.Tsai, Y. L., M. D. Sobsey, L. R. Sangermano, and C. J. Palmer. 1993. Simple method of concentrating enteroviruses and hepatitis A virus from sewage and ocean water for rapid detection by reverse transcriptase-polymerase chain reaction. Appl. Environ. Microbiol. 59:3488-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]