Abstract
To identify the animal sources for Cryptosporidium contamination, we genotyped Cryptosporidium spp. in wildlife from the watershed of the New York City drinking water supply, using a small-subunit rRNA gene-based PCR-restriction fragment length polymorphism analysis and DNA sequencing. A total of 541 specimens from 38 species of wildlife were analyzed. One hundred and eleven (20.5%) of the wildlife specimens were PCR positive. Altogether, 21 Cryptosporidium genotypes were found in wildlife samples, 11 of which were previously found in storm runoff in the watershed, and six of these 11 were from storm water genotypes of unknown animal origin. Four new genotypes were found, and the animal hosts for four storm water genotypes were expanded. With the exception of the cervine genotype, most genotypes were found in a limited number of animal species and have no major public health significance.
Cryptosporidiosis is a significant cause of waterborne outbreaks of diarrheal diseases. Cryptosporidium oocysts are a threat to water supplies because they are resistant to chlorine disinfection, have a low infectious dose, and are excreted by almost all vertebrates (6). Although farm animals and humans have generally been considered major sources of human-pathogenic Cryptosporidium oocysts in surface water (1, 12, 13, 17, 26), wildlife has been shown to play a significant role in the overall contamination of water with Cryptosporidium spp. through aquatic activities and runoff (5, 8, 9, 11, 15, 16, 19, 20). The challenge, however, is to differentiate the human-pathogenic Cryptosporidium spp. from the non-human-pathogenic species and genotypes. The detection method implemented in the newly established Long Term 2 Enhanced Surface Water Treatment Rule (LT2ESWTR) in the United States is the Environmental Protection Agency Method 1622 (for Cryptosporidium) or 1623 (for both Cryptosporidium and Giardia), which does not differentiate Cryptosporidium species (18).
In contrast, PCR-based methods have the ability to differentiate Cryptosporidium at the species and genotype level (24). PCR tools also have the capability to assess the human-infective potential and the animal sources of Cryptosporidium oocysts in water, as different animal species are usually infected with different Cryptosporidium species and genotypes, only a few of which are human pathogens (21). Genotyping using PCR-restriction fragment length polymorphism (RFLP) analysis of the small-subunit (SSU) rRNA gene has been used successfully in assessing the source and human-infective potential of Cryptosporidium oocysts found in water (11, 15, 16, 19, 20, 22, 26).
Previous studies conducted by researchers at the Centers for Disease Control and Prevention and the New York City Department of Environmental Protection (NYCDEP) have identified at least 22 Cryptosporidium genotypes in 121 storm water samples from the Ashokan Brook, Malcolm Brook, and the N5 stream of the New York City water supply system (11, 19, 20). Almost all of these Cryptosporidium genotypes belong to those that have not been previously detected in humans or domestic animals. Results of these studies have demonstrated that molecular techniques can complement traditional detection methods by providing information on the source of contamination and human-infective potential of Cryptosporidium oocysts found in water. Several Cryptosporidium spp. were commonly found in these watersheds, including W4 (cervine genotype) from deer, W7 (muskrat genotype I) from muskrats, and the W1 genotype from an unknown animal source. Some genotypes were only found in a particular watershed. Half of the Cryptosporidium genotypes (W1, W3, W5, W6, W12, W15, and W17 to W21) found have not been attributed to known species/groups of animals (11).
In this study, Cryptosporidium spp. in fecal specimens from known wildlife species living in the watershed of the New York City water supply system were detected and genotyped by the same rRNA-based genotyping tool previously used in the analysis of storm water samples (11, 19, 20). The objectives were (i) to assess the role of wildlife in Cryptosporidium contamination in the NYCDEP watershed and to identify the remaining genotypes that could not be tracked to animal sources in the previous studies and (ii) to understand the public health significance of Cryptosporidium spp. from wildlife.
MATERIALS AND METHODS
Specimens.
A total of 541 fecal specimens were collected between September 2005 and July 2007 from 38 species of wild animals living in the NYCDEP watershed. There are three main districts to the New York City water supply, including several counties in southeastern New York State: the Catskill, Delaware, and Croton systems. Wildlife (mammal and bird) specimens were collected opportunistically within and adjacent to all three watershed districts and in New York City for the purpose of extracting an intestinal fecal sample or free-range sample (sampling of known animal latrines or bird roosting/feeding locations). The collection areas were comprised of a variety of land cover and land use categories, including reservoir buffer lands (forest, woodland, transition edge, mixed meadow/shrub), along stream corridors, and on maintained (lawns, parks) and impervious surfaces (parking lots, sidewalks, beach walls) in New York City.
Samples were collected utilizing a variety of means, including live and lethal trapping, fresh roadkill collections, and fresh fecal collections from bird roosting/feeding/loafing locations. Trap lines were set just prior to sunset and retrieved postdawn using Sherman live traps, lethal snap traps, and body-gripping traps (no. 220 and 330). Sherman traps were set in transect lines along natural or man-made boundaries such as stream corridors, fences, and stone walls, etc. Snap traps were generally placed around NYCDEP facilities. Foot-hold and body-gripping traps, used to trap beaver (Castor Canadensis), were opportunistically placed at NYCDEP-identified nuisance locations on and around the New York City reservoirs. Fish were collected from screen chambers at the reservoir locations.
Roadkill collections were conducted opportunistically during routine surveillance monitoring during predawn, postdusk, and daytime hours. Roadkill mammals were only collected prior to rigor mortis and if specimens were warm to touch, ensuring freshness, and if all viscera remained intact. All avian samples were collected off pavement and grassy surfaces using sterile syringes and plastic forceps. Direct observation of fecal elimination occurred just prior to sampling to ensure species identification and freshness of samples. Fecal samples were collected from roadkill specimens and euthanized mammals either by direct fecal extraction from the anus (externally) or the lower intestinal tract via dissection. All samples were placed in sterile plastic vials, capped, and stored in coolers with ice packs/ice for transport to the laboratory within 48 h of collection.
All specimens were identified to the species level (except Peromyscus), aged, and sexed. Animals sampled included small rodents (mice, shrews, voles, rats, and chipmunks), other rodents and mammals, birds, and fish (Table 1), with most animals being adults. Samples were initially processed in the NYCDEP laboratory, and then pellets were shipped in coolers to the laboratory at the Centers for Disease Control and Prevention for Cryptosporidium detection and genotyping.
TABLE 1.
Prevalence of Cryptosporidium genotypes in wildlife in the NYCDEP watershed
| Host | Total no. of samples | No. of positive samples | Prevalence (%) | Genotype(s) (no. positive) |
|---|---|---|---|---|
| Rodents | 263 | 86 | 32.8 | |
| Sciuridae | 48 | 18 | 37.5 | |
| Sciurus carolinensis (eastern gray squirrel) | 33 | 12 | 36.4 | W1 (5), W4 (3), W13 (1), W17 (1), C. parvum (1), C. muris (1) |
| Sciurus vulgaris (red squirrel) | 2 | 1 | 50.0 | W4 (1) |
| Glaucomys volans (southern flying squirrel) | 1 | 0 | 0.0 | |
| Tamias striatus (eastern chipmunk) | 7 | 4 | 57.1 | W4 (2), W17 (1), chipmunk II (1) |
| Marmota monax (woodchuck) | 5 | 1 | 20.0 | W4 (1) |
| Castoridae | 16 | 3 | 18.8 | |
| Castor canadensis (beaver) | 16 | 3 | 18.8 | Beaver genotype (1), W4 (2) |
| Muridae | 1 | 0 | 0 | |
| Rattus norvegicus (Norway rat) | 1 | 0 | 0 | |
| Zapodidae | 1 | 0 | 0.0 | |
| Napaeozapus insignis (woodland jumping mouse) | 1 | 0 | 0.0 | |
| Cricetidae | 193 | 65 | 33.7 | |
| Peromyscus sp. (deer mouse) | 177 | 57 | 32.2 | W3 (21), W1 (20), deer mouse genotype I (10), W4 (1), W17 (1), C. meleagridis (1), deer mouse genotypes I and II (1), W1 and deer mouse genotypes I and II (2) |
| Myodes gapperi (boreal red-backed vole) | 5 | 4 | 80.0 | W7 (4) |
| Microtus pennsylvanicus (meadow vole) | 10 | 3 | 30.0 | W15 (1), W16 (2) |
| Ondatrini zibethicus (muskrat) | 1 | 1 | 100 | W7 (1) |
| Erethizontidae | 4 | 0 | 0.0 | |
| Erethizon dorsatum (porcupine) | 4 | 0 | 0.0 | |
| Carnivores | 41 | 7 | 17.1 | |
| Mustela vison (mink) | 4 | 1 | 25.0 | Mink genotype (1) |
| Mustela erminea (ermine) | 1 | 1 | 100 | W5/W18 (1) |
| Procyon lotor (raccoon) | 21 | 4 | 19.0 | W4 (1), W13 (3) |
| Ursus americanus (black bear) | 5 | 0 | 0.0 | |
| Lontra canadensis (river otter) | 8 | 1 | 12.5 | W13 (1) |
| Mephitis mephitis (striped skunk) | 2 | 0 | 0.0 | |
| Insectivores | 5 | 2 | 40.0 | |
| Blarina brevicauda (northern short-tailed shrew) | 5 | 2 | 40.0 | W5 (2) |
| Lagomorpha | 8 | 0 | 0.0 | |
| Sylvilagus floridanus (eastern cottontail) | 8 | 0 | 0.0 | |
| Ruminants | 59 | 5 | 8.5 | |
| Odocoileus virginianus (white-tailed deer) | 59 | 5 | 8.5 | W9 (5) |
| Marsupials | 9 | 1 | 11.1 | |
| Didelphis virginiana (Virginia opossum) | 9 | 1 | 11.1 | W13 (1) |
| Birds | 100 | 10 | 10.0 | |
| Larus delawarensis (ring-billed gull) | 8 | 0 | 0.0 | |
| Larus argentatus (herring gull) | 8 | 0 | 0.0 | |
| Larus marinus (great black-backed gull) | 1 | 0 | 0.0 | |
| Phalacrocorax auritus (double-crested cormorant) | 5 | 0 | 0.0 | |
| Branta canadensis (Canada goose) | 52 | 10 | 19.2 | Goose genotype I (7), goose genotype II (2), goose genotypes II and I (1) |
| Petrochelidon pyrrhonota (cliff swallow) | 25 | 0 | 0.0 | |
| Melospiza melodia (song sparrow) | 1 | 0 | 0.0 | |
| Fish | 55 | 0 | 0.0 | |
| Perca flavescens (yellow perch) | 25 | 0 | 0.0 | |
| Alosa pseudoharengus (alewife) | 16 | 0 | 0.0 | |
| Salmo trutta (brown trout) | 5 | 0 | 0.0 | |
| Oncorhynchus mykiss (rainbow trout) | 2 | 0 | 0.0 | |
| Ameiurus natalis (yellow bullhead) | 5 | 0 | 0.0 | |
| Ambloplites rupestris (rock bass) | 1 | 0 | 0.0 | |
| Pomoxis nigromaculatus (black crappie) | 1 | 0 | 0.0 | |
| Amphibian | 1 | 0 | 0.0 | |
| Bufo americanus (American toad) | 1 | 0 | 0.0 | |
| Total | 541 | 111 | 20.5 |
Cryptosporidium detection and genotyping.
After washing the specimens twice in distilled water, genomic DNA was extracted from 0.2 ml of specimens using a FastDNA SPIN kit for soil (BIO 101, Carlsbad, CA) and eluted in 100 μl of reagent-grade water as described previously (10). Cryptosporidium oocysts presented in the specimens were genotyped initially by nested PCR amplification of an approximate 830-bp fragment of the SSU rRNA gene and RFLP analysis of the secondary PCR products using restriction enzymes SspI and VspI (11). All secondary PCR products were sequenced to confirm the genotype identification. Each fecal specimen was analyzed at least twice by the PCR-RFLP technique, using 2 μl of the DNA solution per PCR. DNA of Cryptosporidium serpentis was used as the positive control in all SSU rRNA-based PCR-RFLP analyses. To neutralize residual PCR inhibitors in the extracted DNA, 400 ng/μl of nonacetylated bovine serum albumin (Sigma-Adrich, St. Louis, MO) was used in all primary PCRs.
Sequence analysis.
After being purified using Montage PCR filters (Millipore, Bedford, MA), the secondary PCR products were sequenced directly with secondary PCR primers using an ABI BigDye Terminator v. 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and the manufacturer-suggested procedures. Sequences were read on an ABI3130 genetic analyzer (Applied Biosystems). Sequence accuracy was confirmed by two-directional sequencing and sequencing of at least two PCR products from each positive specimen. Nucleotide sequences obtained were aligned with reference Cryptosporidium sequences using the ClustalX 1.81 package (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/) and the default setting. Phylogenetic analysis was carried out to assess the relationship between parasites in animals and known Cryptosporidium spp. Neighbor-joining trees were constructed using the TreeCon package (http://www.psb.rug.ac.be/bioinformatics/psb/Userman/treeconw.html), based on the evolutionary distances calculated by the Kimura two-parameter model. An SSU rRNA sequence of Eimeria tenella (GenBank accession no. AF026388) was used as the outgroup. The reliability of various clusters was evaluated by the bootstrap method with 1,000 replicates.
Nucleotide sequence accession numbers.
Unique partial SSU rRNA sequences obtained from wildlife during the study were deposited in the GenBank database under accession numbers EF641009 to EF641030, EU096237, and EU096238.
RESULTS
Wildlife sampled.
A total of 541 fecal specimens from wild animals were analyzed for Cryptosporidium spp. by PCR. The animals examined included mammals (385 specimens from 23 species), birds (100 specimens from seven species), fish (55 specimens from seven species), and an amphibian (one specimen). Most of the mammal specimens were from rodents (263 specimens), with some from ruminants (59 specimens from white-tailed deer), carnivores (38 specimens from five species), insectivores (five specimens from northern short-tailed shrew), lagomorphs (eight specimens from eastern cottontails), and marsupials (nine specimens from Virginia opossum). Within rodents, 13 species of animals were sampled. However, over two-thirds of the rodent specimens (177) were from deer mouse (Peromyscus sp.) (Table 1).
Prevalence of Cryptosporidium genotypes in wildlife.
Cryptosporidium spp. were detected in 111 (20.5%) studied animals (Table 1). Most of the positive animals (101/111) were mammals, with only 10 from birds and none from fish. Within mammals, rodents had the highest prevalence of Cryptosporidium, with a prevalence of 32.8%. Most of the high Cryptosporidium prevalence in rodents was attributable to the high infection rate in the well-sampled deer mice (57/177 or 32.2%), even though high prevalence (37.5%) of Cryptosporidium was also seen in the squirrel family (18/48) (Table 1). The only ruminants studied, the white-tailed deer, had a much lower infection rate (5/59 or 8.5%).
Cryptosporidium genotypes in wildlife.
All PCR products were digested with restriction enzymes SspI and VspI to differentiate Cryptosporidium spp. in wildlife. Multiple banding patterns were seen for both SspI and VspI. The combination of SspI and VspI RFLP identified at least 10 restriction patterns, with most of them not previously seen in humans and domestic animals (data not shown). PCR products of all positive specimens were sequenced at least twice to identify Cryptosporidium genotypes. Altogether, 21 Cryptosporidium genotypes were identified in New York wildlife. They included 11 of the 22 Cryptosporidium genotypes previously found in storm water in the NYCDEP watershed: W1, W3, W4, W5, W7, W9, W13, W15, W16, W17, and W18. Previously, animal sources for six of the Cryptosporidium genotypes in storm water were not known (11, 19): W1 (deer mouse genotype III) from deer mice and eastern gray squirrels; W3 (deer mouse genotype IV) from deer mice; W5 (shrew genotype) from northern short-tailed shrews and one ermine; W15 (vole genotype) from a meadow vole; W17 (chipmunk genotype I) from one chipmunk, eastern gray squirrel, and deer mouse each; and W18 from one ermine. The known animal hosts for four of the storm water genotypes were expanded, with W4 (cervine genotype) seen in three eastern gray squirrels, two eastern chipmunks, two beavers, and one each of the red squirrel, woodchuck, deer mouse, and raccoon. These are in addition to known W4 hosts from previous studies such as deer, sheep, mouflon sheep, blesboks, nyalas, lemurs, and humans. W7 (muskrat genotype I) was seen in four boreal red-backed voles in addition to muskrats, W13 (skunk genotype) was seen in one eastern gray squirrel, river otter, and Virginia opossum each, in addition to skunks and raccoons, and W16 was seen in two meadow voles in addition to muskrats (Table 2). Six established Cryptosporidium genotypes previously not seen in New York City watershed storm water were found in wildlife in this study, including goose genotypes I and II from Canada geese, a deer mouse genotype (renamed deer mouse genotype I in this study) from deer mice, Cryptosporidium parvum in an eastern gray squirrel, Cryptosporidium meleagridis in a deer mouse, and Cryptosporidium muris in an eastern gray squirrel. In addition, four new Cryptosporidium genotypes were found in this study, including the mink genotype in a mink, beaver genotype in a beaver, deer mouse genotype II in deer mice, and chipmunk genotype II in an eastern chipmunk (Table 2).
TABLE 2.
Cryptosporidium genotypes found in the NYCDEP watershed and their animal sources
| Water genotype namea | Common genotype name | Previous known hosts | Hosts in this studyb |
|---|---|---|---|
| W1 | Deer mouse genotype III | Unknown | Deer mouse (21), eastern gray squirrel (5) |
| W2 | Opossum genotype I | Opossum | None |
| W3 | Deer mouse genotype IV | Unknown | Deer mouse (21) |
| W4 | Cervine genotype | Deer, sheep, lemur, mouflon sheep, blesbok, nyala, human | Eastern gray squirrel (3), eastern chipmunk (2), beaver (2), red squirrel (1), woodchuck (1), deer mouse (1), raccoon (1) |
| W5c | Shrew genotype | Wildebeest | Northern short-tailed shrew (2) |
| W6c | None | Unknown | Ermine (1) |
| W7 | Muskrat genotype I | Muskrat | Boreal red-backed vole (4), muskrat (1) |
| W8 | Opossum genotype II | Opossum | None |
| W9 | Deer genotype | White-tailed deer | White-tailed deer (5) |
| W10 | C. baileyi | Bird | None |
| W11 | Snake genotype | Snake | None |
| W12 | None | Unknown | None |
| W13 | Skunk genotype | Skunk and raccoon | Eastern gray squirrel (1), raccoon (1), opossum (1), river otter (1) |
| W14 | C. hominis | Human, cattle, sheep, dugong | None |
| W15 | Vole genotype | Unknown | Meadow vole (1) |
| W16 | Muskrat genotype II | Muskrat | Meadow vole (2) |
| W17 | Chipmunk genotype I | Unknown | Eastern chipmunk (1), eastern squirrel (1), deer mouse (1) |
| W18c | None | Unknown | Ermine (1) |
| W19 | None | Unknown | None |
| W20 | None | Unknown | None |
| W21 | None | Unknown | None |
| W22 | C. galli | Finch | None |
| W23d | None | Unknown | None |
| W24e | Fox genotype | Fox | None |
| None | Deer mouse genotype I | Deer mouse | Deer mouse (13) |
| None | Beaver genotypef | Beaver (1) | |
| None | Deer mouse genotype IIf | Deer mouse (3) | |
| None | Mink genotypef | Mink (1) | |
| None | Chipmunk genotype II | Eastern chipmunk (1) | |
| None | Goose genotype I | Canada goose | Canada goose (8) |
| None | Goose genotype II | Canada goose | Canada goose (3) |
| None | C. meleagridis | Bird, human, dog | Deer mouse (1) |
| None | C. parvum | Ruminant, human, horse, mouse, raccoon dog | Eastern gray squirrel (1) |
| None | C. muris | Mouse, rat, Japanese field mouse, Siberian chipmunk, camel, hyrax, ringed seal, cat, bilby, cynomolgus monkey, human | Eastern squirrel (1) |
Numbers in parentheses are the numbers of samples positive for each genotype or species.
May be one genotype.
A new genotype in a sample (8784-4) collected from a Delaware watershed.
Originally misidentified as W16 in a storm sample collected from the N5 basin (8650-2) in Jiang et al. (11).
New genotypes found in this study.
Intragenotypic variations in Cryptosporidium genotypes.
Sequence heterogeneity was observed in deer mouse genotypes I, W4, W5, and W7 seen in this study. These variations included nucleotide substitutions, deletions, and insertions (Fig. 1). As previously reported (accession no. AY737592 and AY737593), two types of sequences were seen for W4 which differed from each other in the presence of an AT deletion in some sequences and were both seen in one specimen (no. 12355). Likewise, two types of sequences were seen in deer mouse genotype I which differed from each other in the presence of a TTT deletion in some sequences and were both seen in one specimen (no. 13077). The W5 sequence obtained in the study from the ermine (no. 14966) was identical to the W5 previously seen in storm water (accession no. AY737594), but other W5 sequences had a T-to-C nucleotide change and some also had an A and a C deletion. Compared to the W7 (muskrat genotype I) previously seen in muskrats in Maryland (AY120904), the W7 sequences from voles all had a G-to-A nucleotide change and a TTT insertion (a few with a TTTTT insertion) and a TT deletion in two T-repeat regions downstream. Some of the vole sequences differed from each other in the change of AT to TA proceeding to the first T-repeat region, and both types of sequences were seen in one specimen (no. 11715) (Fig. 1). Nevertheless, phylogenetic analysis showed that these intragenotypic variable sequences clustered together for each genotype.
FIG. 1.
Intragenotypic diversity in the sequences of the SSU rRNA genes of some Cryptosporidium genotypes seen in this study. Dots denote nucleotide identity to the reference sequence from GenBank. Dashes denote sequence deletions.
Phylogenetic relationship of Cryptosporidium spp. in wildlife.
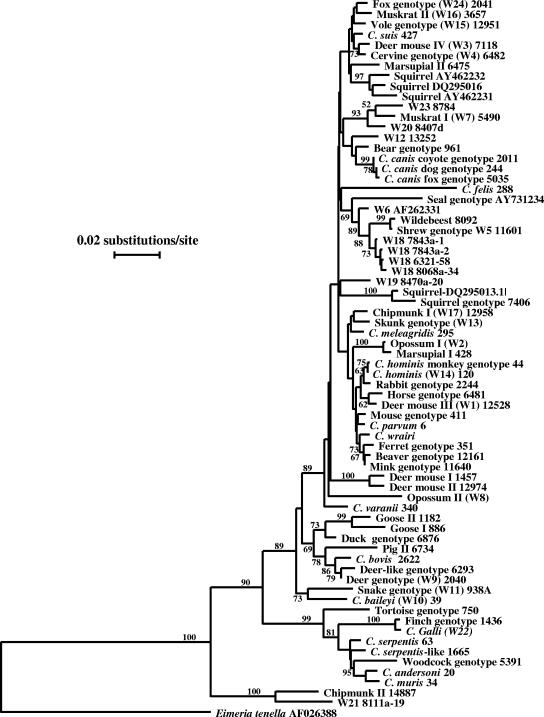
A multiple-sequence alignment in the range of 193 to 1,043 residues of Cryptosprodium hominis (GenBank accession no. AF093489), which included the most polymorphic region of the SSU rRNA gene, was used in the confirmation of genotype identification and the assessment of the phylogenetic relationship among Cryptosporidium genotypes found in the study. With the exception of two genotypes, most Cryptosporidium genotypes found in the study clustered in the intestinal group (Fig. 2). Within this, most of the genotypes were grouped in the Cryptosporidium cluster containing C. parvum, C. hominis, C. wrairi, C. meleagridis, C. varanii, C. canis, and C. felis. Three genotypes (deer genotype and goose genotypes I and II), however, were placed in other clusters within the intestinal Cryptosporidium group (Fig. 2).
FIG. 2.
Phylogenetic relationship among various Cryptosporidium genotypes in wildlife in the NYCDEP watershed and known Cryptosporidium species and genotypes, as inferred by a neighbor-joining analysis of the SSU rRNA sequences. Numbers on branches are percent bootstrapping values (>50) using 1,000 replicates.
Within the two genotypes placed outside the intestinal Cryptosporidium group, one was a C. muris variant (one C-to-T nucleotide change, one TAT insert, and one A insert) and was placed in the gastric group. The other one, the chipmunk genotype II, had high sequence homology to a genotype (W21) previously identified in a NYCDEP storm water sample and was placed outside the intestinal and gastric groups of Cryptosporidium spp. in the phylogenetic analysis (Fig. 2).
DISCUSSION
In this study, the overall Cryptosporidium prevalence in 541 wild animals in the watersheds of New York City water supplies was 20.5%. The infection rate, however, varied greatly among different animals. Among animals with reasonable numbers of specimens, rodents had the highest infection rate (32.8% in 236 animals) and fish had the lowest infection rate (0% in 55 animals), with ruminants (white-tailed deer) and birds in between (8.5% in 59 deer and 10.0% in 100 birds). Within birds, Cryptosporidium was only seen in Canada geese (19.2% in 52 geese). Altogether, 21 Cryptosporidium genotypes were found. As most animals with large numbers of samples had multiple Cryptosporidium genotypes, more Cryptosporidium genotypes could have been found if the sample sizes for some animal species were increased.
Of the 21 Cryptosporidium genotypes found in wildlife in the NYCDEP watershed, 19 of them were intestinal. The finding of the only gastric species in this study, C. muris, in only one animal is surprising, especially when one considers the fact that a large number of several species of rodents was sampled. The result is nevertheless in agreement with the previous finding of Cryptosporidium genotypes in storm runoff samples collected from the same watershed, where 20 of the 22 Cryptosporidium genotypes were intestinal (11, 19). One Cryptosporidium genotype previously found in a storm water sample from the watershed, W21, clustered outside the known intestinal and gastric Cryptosporidium species/genotypes, even though a BLAST search of the GenBank database clearly established its identity of Cryptosporidium (11). Interestingly, the chipmunk genotype II found in this study had high sequence homology to the W21 genotype at the 3′ and 5′ ends of the SSU rRNA gene and formed a cluster together outside the intestinal and gastric Cryptosporidium species/genotypes (Fig. 2). The biological uniqueness of these two Cryptosporidium genotypes warrants further studies.
Results of this study support the previous conclusion that most Cryptosporidium spp. from wildlife are host adapted in nature (25, 27-29). Thus, most Cryptosporidium genotypes in this study were found in a few related animals, such as deer mouse genotypes I, II, and IV (W3) in deer mice, muskrat genotypes I (W7) and II (W16) in muskrats and voles, a deer genotype (W9) in deer, a vole genotype (W15) in voles, chipmunk genotype I (W17) in chipmunks and squirrels, and goose genotypes I and II in Canada geese. Results of phylogenetic analysis also support the suggestion of host adaptation and parasite-host coevolution of Cryptosporidium spp., because some related animal species have related Cryptosporidium genotypes. For example, it was previously shown that (i) opossum genotype I in Virginia opossum (the American marsupial) is related to but distinct from the marsupial genotype I seen in kangaroos and koalas in Australia; (ii) the monkey genotype has only minor sequence differences from C. hominis seen in humans; (iii) the ruminant parasites deer genotypes (in deer), deer-like genotype (in cattle), and Cryptosporidium bovis (in cattle and sheep) are related to each other; and (iv) distinct C. canis genotypes are seen in dogs, coyotes, and foxes (27) (Fig. 2). In this study, the newly found mink genotype is related to the ferret genotype with only minor sequence differences (with 5 bp of substitutions and 3 bp of deletions), and they cluster together in the phylogenetic tree (Fig. 2). Minor differences are present between muskrats and voles in the sequences of muskrat genotype I or II. The C. muris sequence obtained from an eastern gray squirrel had small differences from sequences obtained previously from other rodents and mammals. In some hosts, even though multiple genotypes are present, these Cryptosporidium genotypes are related to each other, such as deer genotypes I and II in deer mice, goose genotypes I and II in Canada geese, and the various squirrel genotypes seen in squirrels in California (Fig. 2).
Whether deer mouse genotypes I and II are two true Cryptosporidium genotypes remains to be determined. Even though there is greater than 1% difference between these two types of sequences, which is above the extent of difference among many established Cryptosporidium species, a few deer mouse genotype I sequences were also seen in animals with deer mouse genotype II. In fact, many PCR products of genotype I were difficult to sequence. It was previously shown that some Cryptosporidium spp. such as C. felis and marsupial genotype I have very different copies of the SSU rRNA gene, which is responsible for the existence of mixed RFLP profiles and difficulties in sequencing some PCR products (23). In this study, the existence of heterogeneous copies of the SSU rRNA gene was also seen in the multiple products in one specimen each of C. parvum, cervine genotype (W4), and muskrat genotype I (W7), even though the sequence differences were all minor. It is possible that at least some of the divergent Cryptosporidium sequences obtained from squirrels in California (2) were from heterogeneous copies of the SSU rRNA gene. In contrast, the goose genotypes I and II were usually seen in different specimens from Canada geese (29).
Interestingly, the only ermine specimen examined in the study produced both W5 and W18 sequences in repeated analyses. Because the sequence differences (eight or nine nucleotide changes) between W5 and W18 are restricted to the hypervariable region of the gene (Fig. 1), both types of sequences were found in the same animal, and phylogenetically they clustered together (Fig. 2), it is very possible that W5 and W18 represent sequences from heterogeneous copies of the rRNA gene of the same Cryptosporidium genotype (shrew genotype). Whether W6 is in fact also the shrew genotype (W5) remains to be determined, as it is also phylogenetically related to W5 and W18 and differs from W5 and W18 only in the hypervariable regions of the SSU rRNA gene (Fig. 1 and Fig. 2). It also remains to be determined whether the shrew genotype (of W5 and W18 sequences) is a true parasite of ermines, as its finding in the one ermine could be from an infected rodent ingested by the animal.
As expected, the host specificity of Cryptosporidium spp. is not strict and there are apparent exceptions to the hypothesis of host adaptation. The most noticeable example is the cervine genotype (W4), which was found in this study in multiple species of rodents (beaver, eastern gray squirrel, red squirrel, chipmunk, and woodchuck) as well as in a raccoon. Previously, it was also found in various ruminants (white-tailed deer, sheep, mouflon sheep, blesbok, and nyala) and primates (human and lemur) (Table 2). The generalist nature of the host specificity of the parasite and habitat sharing are probably responsible for the wide occurrence of the cervine genotype in animals. The cervine genotype was the most common and the only year-round Cryptosporidium in storm water in the NYCDEP watershed (11, 19, 20). A few other Cryptosporidium species/genotypes also had more-broad host specificity; the deer mouse genotype III (W1) was found in a few eastern gray squirrels in addition to deer mice, the skunk genotype was found in one striped skunk, one squirrel, one opossum, and one river otter, and C. parvum was found in one eastern gray squirrel. The latter is now generally considered mostly a parasite of ruminants and humans, but it has been found occasionally in a few other species of animals such as horses, mice, raccoons, and dogs (21). Thus in the NYCDEP watershed, interspecies transmission of Cryptosporidium mostly occurs in the cervine genotype.
Among the 21 Cryptosporidium genotypes found in this study, only C. parvum, C. meleagridis, C. muris, cervine (W4), and chipmunk I (W17) genotypes have been found in humans. The former three were each found in only one animal and there were minor SSU rRNA sequence differences in C. parvum and C. muris between eastern gray squirrels and humans (one or two nucleotide changes depending on the copies of the gene for C. parvum, and one nucleotide substitution and four nucleotide insertions/deletions for C. muris). Even though the cervine genotype was commonly seen in many animals and in storm water, this Cryptosporidium sp. has been found in only a few human cases around the world (24). Likewise, the chipmunk genotype I has only been reported in two persons in Wisconsin (7). Thus, wildlife in the NYCDEP watershed is unlikely to be a major contributor of human-pathogenic Cryptosporidium spp. in the source water. A similar conclusion was previously made regarding Cryptosporidium spp. in reptiles and Canada geese and in aquatic mammals in the watershed in Maryland (25, 28, 29). This is in contrast to previous suggestions by others (3, 4, 14). The latter, however, was mostly based on the erroneous assumption that C. parvum commonly infects various species of mammals and on dated nomenclature for Cryptosporidium species and genotypes. Even though most Cryptosporidium genotypes in wildlife in this study were grouped in phylogenetic analysis in the cluster containing Cryptosporidium types infectious to humans, one should not conclude that all genotypes within that cluster are infectious to humans. In fact, there are many species and genotypes within this cluster that have never been found in humans. It is likely that only very subtle genetic differences are required to change host specificity; therefore, genotypes can be very similar yet not infect the same hosts.
Results of this study demonstrate that genotyping tools could provide information on the host specificity and the human-infective potential of Cryptosporidium oocysts in wildlife and on the animal source of contamination in water. Wildlife may contribute to Cryptosporidium contamination in the water but may not have major public health significance because they are generally infected with non-human-pathogenic species and genotypes. Nevertheless, watershed protection programs should attempt to control pathogen inputs from wildlife in addition to anthropogenic and agricultural sources, and more attention should be directed to studying the transport of pathogens from wildlife to water and to monitoring pathogens in watersheds deemed protected or pristine.
Acknowledgments
This study was supported in part by a grant from the NYCDEP and the New York State Department of Environmental Conservation through Safe Drinking Water Act funding. Yaoyu Feng was further supported by a grant from the Program for New Century Excellent Talents in Chinese Universities (NCET-05-0382) and an Emerging Infectious Diseases Fellowship from the Centers for Disease Control and Prevention and the Association of Public Health Laboratories.
We thank Tom Baudanza of NYCDEP and Robert Stranges of HDR, P.C., for sampling support and Don Culjack, Alex Maestri, and Deb Schwarz for technical support.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 24 August 2007.
REFERENCES
- 1.Atwill, E. R., M. D. Pereira, L. H. Alonso, C. Elmi, W. B. Epperson, R. Smith, W. Riggs, L. V. Carpenter, D. A. Dargatz, and B. Hoar. 2006. Environmental load of Cryptosporidium parvum oocysts from cattle manure in feedlots from the central and western United States. J. Environ. Qual. 35:200-206. [DOI] [PubMed] [Google Scholar]
- 2.Atwill, E. R., R. Phillips, M. D. Pereira, X. Li, and B. McCowan. 2004. Seasonal shedding of multiple Cryptosporidium genotypes in California ground squirrels (Spermophilus beecheyi). Appl. Environ. Microbiol. 70:6748-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajer, A., S. Caccio, M. Bednarska, J. M. Behnke, N. J. Pieniazek, and E. Sinski. 2003. Preliminary molecular characterization of Cryptosporidium parvum isolates of wildlife rodents from Poland. J. Parasitol. 89:1053-1055. [DOI] [PubMed] [Google Scholar]
- 4.Bednarska, M., A. Bajer, K. Kulis, and E. Sinski. 2003. Biological characterisation of Cryptosporidium parvum isolates of wildlife rodents in Poland. Ann. Agric. Environ. Med. 10:163-169. [PubMed] [Google Scholar]
- 5.Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fayer, R., C. A. Speer, and J. P. Dubey. 1997. The general biology of Cryptosporidium, p. 1-41. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, FL.
- 7.Feltus, D. C., C. W. Giddings, B. L. Schneck, T. Monson, D. Warshauer, and J. M. McEvoy. 2006. Evidence supporting zoonotic transmission of Cryptosporidium in Wisconsin. J. Clin. Microbiol. 44:4303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heitman, T. L., L. M. Frederick, J. R. Viste, N. J. Guselle, U. M. Morgan, R. C. Thompson, and M. E. Olson. 2002. Prevalence of Giardia and Cryptosporidium and characterization of Cryptosporidium spp. isolated from wildlife, human, and agricultural sources in the North Saskatchewan River Basin in Alberta, Canada. Can. J. Microbiol. 48:530-541. [DOI] [PubMed] [Google Scholar]
- 9.Jellison, K. L., H. F. Hemond, and D. B. Schauer. 2002. Sources and species of Cryptosporidium oocysts in the Wachusett Reservoir watershed. Appl. Environ. Microbiol. 68:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, J., K. A. Alderisio, A. Singh, and L. Xiao. 2005. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl. Environ. Microbiol. 71:1135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang, J., K. A. Alderisio, and L. Xiao. 2005. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 71:4446-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nichols, R. A., B. M. Campbell, and H. V. Smith. 2006. Molecular fingerprinting of Cryptosporidium oocysts isolated during water monitoring. Appl. Environ. Microbiol. 72:5428-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ono, K., H. Tsuji, S. K. Rai, A. Yamamoto, K. Masuda, T. Endo, H. Hotta, T. Kawamura, and S. Uga. 2001. Contamination of river water by Cryptosporidium parvum oocysts in western Japan. Appl. Environ. Microbiol. 67:3832-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perz, J. F., and S. M. Le Blancq. 2001. Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Appl. Environ. Microbiol. 67:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruecker, N. J., N. Bounsombath, P. Wallis, C. S. Ong, J. L. Isaac-Renton, and N. F. Neumann. 2005. Molecular forensic profiling of Cryptosporidium species and genotypes in raw water. Appl. Environ. Microbiol. 71:8991-8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruecker, N. J., S. L. Braithwaite, E. Topp, T. Edge, D. R. Lapen, G. Wilkes, W. Robertson, D. Medeiros, C. W. Sensen, and N. F. Neumann. 2007. Tracking host sources of Cryptosporidium spp. in raw water for improved health risk assessment. Appl. Environ. Microbiol. 72:3945-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward, P. I., P. Deplazes, W. Regli, H. Rinder, and A. Mathis. 2002. Detection of eight Cryptosporidium genotypes in surface and waste waters in Europe. Parasitology 124:359-368. [DOI] [PubMed] [Google Scholar]
- 18.Weintraub, J. M. 2006. Improving Cryptosporidium testing methods: a public health perspective. J. Water Health 4(Suppl. 1):23-26. [PubMed] [Google Scholar]
- 19.Xiao, L., K. Alderisio, J. Limor, M. Royer, and A. A. Lal. 2000. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 66:5492-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao, L., K. A. Alderisio, and J. Jiang. 2006. Detection of Cryptosporidium oocysts in water: effect of the number of samples and analytic replicates on test results. Appl. Environ. Microbiol. 72:5942-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao, L., R. Fayer, U. Ryan, and S. J. Upton. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao, L., A. A. Lal, and J. Jiang. 2004. Detection and differentiation of Cryptosporidium oocysts in water by PCR-RFLP. Methods Mol. Biol. 268:163-176. [DOI] [PubMed] [Google Scholar]
- 23.Xiao, L., J. R. Limor, L. Li, U. Morgan, R. C. Thompson, and A. A. Lal. 1999. Presence of heterogeneous copies of the small subunit rRNA gene in Cryptosporidium parvum human and marsupial genotypes and Cryptosporidium felis. J. Eukaryot. Microbiol. 46:44S-45S. [PubMed] [Google Scholar]
- 24.Xiao, L., and U. M. Ryan. 2004. Cryptosporidiosis: an update in molecular epidemiology. Curr. Opin. Infect. Dis. 17:483-490. [DOI] [PubMed] [Google Scholar]
- 25.Xiao, L., U. M. Ryan, T. K. Graczyk, J. Limor, L. Li, M. Kombert, R. Junge, I. M. Sulaiman, L. Zhou, M. J. Arrowood, B. Koudela, D. Modry, and A. A. Lal. 2004. Genetic diversity of Cryptosporidium spp. in captive reptiles. Appl. Environ. Microbiol. 70:891-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao, L., I. M. Sulaiman, U. M. Ryan, L. Zhou, E. R. Atwill, M. L. Tischler, X. Zhang, R. Fayer, and A. A. Lal. 2002. Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int. J. Parasitol. 32:1773-1785. [DOI] [PubMed] [Google Scholar]
- 28.Zhou, L., R. Fayer, J. M. Trout, U. M. Ryan, F. W. Schaefer III, and L. Xiao. 2004. Genotypes of Cryptosporidium species infecting fur-bearing mammals differ from those of species infecting humans. Appl. Environ. Microbiol. 70:7574-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou, L., H. Kassa, M. L. Tischler, and L. Xiao. 2004. Host-adapted Cryptosporidium spp. in Canada geese (Branta canadensis). Appl. Environ. Microbiol. 70:4211-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]