Abstract
Rickettsiae are obligate intracellular alphaproteobacteria that include pathogenic species in the spotted fever, typhus, and transitional groups. The development of a standardized cell line in which diverse rickettsiae can be grown and compared would be highly advantageous to investigate the differences among and between pathogenic and nonpathogenic species of rickettsiae. Although several rickettsial species have been grown in tick cells, tick cells are more difficult to maintain and they grow more slowly than insect cells. Rickettsia-permissive arthropod cell lines that can be passaged rapidly are highly desirable for studies on arthropod-Rickettsia interactions. We used two cell lines (Aedes albopictus cell line Aa23 and Anopheles gambiae cell line Sua5B) that have not been used previously for the purpose of rickettsial propagation. We optimized the culture conditions to propagate one transitional-group rickettsial species (Rickettsia felis) and two spotted-fever-group rickettsial species (R. montanensis and R. peacockii) in each cell line. Both cell lines allowed the stable propagation of rickettsiae by weekly passaging regimens. Stable infections were confirmed by PCR, restriction digestion of rompA, sequencing, and the direct observation of bacteria by fluorescence in situ hybridization. These cell lines not only supported rickettsial growth but were also permissive toward the most fastidious species of the three, R. peacockii. The permissive nature of these cell lines suggests that they may potentially be used to isolate novel rickettsiae or other intracellular bacteria. Our results have important implications for the in vitro maintenance of uncultured rickettsiae, as well as providing insights into Rickettsia-arthropod interactions.
The genus Rickettsia comprises a diverse group of alphaproteobacteria that are largely known for causing rickettsioses. Pathogenic members of the genus occur in the spotted fever group (SFG), the typhus group (TG), and a recently designated transitional group (TRG) (10). Putatively nonpathogenic rickettsiae (herein referred to as nonpathogenic) are rickettsiae for which no vertebrate pathogenicity has been described. Unlike their pathogenic relatives, nonpathogenic Rickettsia species are not well characterized despite being widely distributed in invertebrates, vertebrates, protists, and plants (20). Yet nonpathogenic Rickettsia species may have substantial impacts on public health research as emerging infectious agents or, alternatively, as safer model organisms than their pathogenic cousins. A cell line in which these rickettsiae can be isolated and grown may aid in genomic studies of nonpathogenic rickettsiae and shed light into genetic determinants of Rickettsia-host interactions.
Additionally, the development of a standardized cell line in which diverse rickettsiae can be grown and compared would be highly advantageous to investigate the differences between pathogenic and nonpathogenic species of rickettsiae. To be optimally useful, the cell line should be permissive toward fastidious endosymbiotic species, allow rapid passaging regimens, and produce large numbers of rickettsiae for genomic or experimental studies. While several rickettsial species (e.g., Rickettsia prowazekii, R. typhi, and R. montanensis) can grow in mammalian cell lines (e.g., Vero and L929), others appear to be restricted to arthropod cell lines. Vero cells are useful for growing many species but appear to be refractory to species that grow relatively slowly, such as R. peacockii (13). Tick cells are useful for studying SFG and TRG rickettsiae but not as useful for maintaining and comparing TG rickettsial growth (13, 15, 21). Tick cells also have the disadvantage of taking up to a month between passages (13, 15).
Mosquito cells have also been used in the past to culture obligate intracellular bacteria such as Wolbachia pipientis (hereafter referred to as Wolbachia) and some rickettsiae (7, 12, 17, 21, 25). If permissive to infection, mosquito cells have many desirable characteristics, including rapid passaging (≤1 week) and the potential development of high bacterial titers. Recent attempts to maintain rickettsiae in mosquito cells have met with mixed results. Both R. felis and R. conorii were maintained successfully in the Aedes albopictus cell line C6/36 (12, 25). However, R. montanensis and R. peacockii have not been successfully maintained in two other Aedes albopictus cell lines (AeAl2 and C7/10, respectively) (13, 27).
In this study, we successfully established and propagated three rickettsial species (R. felis, R. montanensis, and R. peacockii) in two mosquito cell lines (Aedes albopictus cell line Aa23 and Anopheles gambiae cell line Sua5B). We chose these two cell lines because they had previously been shown to maintain diverse Wolbachia infections (17, 23). The three Rickettsia species were chosen based on the degree of pathogenicity and biological interest due to their transovarial transmission. R. felis is a putatively pathogenic TRG species horizontally transmitted by cat fleas (Ctenocephalides felis) to mammals and transmitted to flea offspring transovarially. R. peacockii serves as an interesting subject because it is presumed to be limited in cellular invasion capacity due to the lack of functional rickA and rompA genes (13, 26). Both R. montanensis and R. peacockii have been involved in transovarial exclusion, in which a primary rickettsial species invades a tick ovary and excludes a second species from being transmitted transovarially (3, 5, 14).
The use of these two mosquito cell lines may aid in answering questions regarding the mechanisms dictating rickettsial specificity for acarine or insect vectors and hosts. Among pathogenic rickettsiae, SFG species are associated exclusively with ticks, and TG species appear to be limited to fleas and lice (10). Nonpathogenic endosymbiotic rickettsiae have been found in association with both acarines and insects (20). The use of mosquito cell lines may allow us to explore the evolutionary implications of vertebrate pathogenicity with respect to the arthropod associate.
MATERIALS AND METHODS
Rickettsial isolates and maintenance.
Prior to establishment in mosquito cells, rickettsiae were maintained in mammalian or tick cell lines. R. peacockii was maintained in DAE100 (Dermacentor andersoni) cells at 34°C in L15B300 medium and grown in plug-capped 25-cm2 culture flasks (15). R. montanensis was maintained in L929 (mouse fibroblast) cells. Infected L929 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum (FBS) and grown at 34°C with 5% CO2 in vent-capped 75-cm2 culture flasks (6). A frozen sample of R. felis (LSU strain)-infected Vero (African green monkey) cells was used for the inoculation of R. felis into mosquito cells.
Mosquito cell lines and maintenance.
Anopheles gambiae Sua5B cells (23) and Aedes albopictus Aa23 cells (17) were passaged every week in sterile Schneider's insect medium (Invitrogen Corporation, CA) supplemented with 10% FBS in plug-capped 75-cm2 cell culture flasks. Cells were removed by scraping and pelleted at 2,500 × g for 5 min. Cells were then washed and resuspended in 20 ml of fresh medium, and 5-ml aliquots were transferred into 75-cm2 flasks containing 15 ml of fresh medium. Flasks were incubated at 34°C for cells to reach confluence (2 to 3 days). Prior to infection, cell suspensions were split and incubated at room temperature (22 to 25°C) for 2 days.
Rickettsiae.
Host cell-free rickettsiae were obtained as previously described with modifications (13, 23). Rickettsia-infected cells were grown to a sufficient quantity (one 75-cm2 flask of R. montanensis-infected L929 mouse fibroblasts or R. peacockii-infected DAE100 cells). R. felis was partially purified from 1 ml of frozen R. felis-infected Vero cells. Cells were harvested by washing with fresh media until cells detached from the flask, pelleted at 2,500 × g for 3 min, resuspended in 12 ml of fresh medium, and lysed by being subjected to a vortex with sterile 3-mm borosilicate glass beads (Sigma-Aldrich, MO) for 5 min. Cell lysates were transferred aseptically into 15-ml centrifuge tubes and centrifuged at 4°C and 2,500 × g for 10 min to pellet cellular debris. The supernatants were aseptically transferred into 10-ml syringes and serially filtered through a 5-μm-pore-size Millex syringe filter (Millipore, MA) and then through a 2.7-μm-pore-size syringe filter (Whatman Inc., NJ).
Cell line inoculation.
To quantify rickettsiae, bacteria were stained with LIVE/DEAD BacLight stain according to the instructions of the manufacturer (Molecular Probes; Invitrogen Corporation, CA). Live rickettsiae were counted on a hemacytometer illuminated on a fluorescence microscope and were adjusted to a density of 107 rickettsiae/ml. The number of Aa23 or Sua5B cells was determined using a hemacytometer, and the density was adjusted to 106 cells/ml. A 500-μl rickettsial inoculum was applied to Sua5B or Aa23 cells grown in Falcon 24-well plates (Becton Dickinson) from which the medium had been removed. Plates were incubated for 2 h at room temperature, followed by the addition of 500 μl of medium/well. Plates were incubated at room temperature for 3 days, and then cells were transferred into 12.5-cm2 culture flasks. The cell suspension added to each flask was initiated from pooled cells from three replicate wells. Cells from an additional three replicate wells were used for PCR and staining with Diff-Quik Wright-Giménez stain (Dade Behring, IL). Two negative controls were included: (i) no inoculum and (ii) heat-killed rickettsiae. Each trial was replicated at least once. Inoculated cultures were sampled weekly and evaluated for infection using Diff-Quik staining and/or fluorescence in situ hybridization (FISH) and PCR. Infected cells were maintained in Schneider's insect medium-5% FBS at room temperature and passaged every 7 days for eight or more passages. Uninfected layers of Aa23 cells at 50% confluence were overlaid with infected Aa23 cells, while infected Sua5B cells were split into fresh medium.
To demonstrate that rickettsiae passaged through mosquito cells were still capable of infecting the cell lines from which they were derived, rickettsiae were partially purified from mosquito cells and applied to Vero, L929, or DAE100 cells as described above. Reinfection was confirmed by FISH, PCR, and reverse transcriptase PCR (RT-PCR). Because our primary objective was the confirmation of Rickettsia infection status, and not the establishment or maintenance of Rickettsia in mammalian cells, we confirmed infection only to passage 3.
DNA extraction and PCR.
DNA was extracted from cells by using the Wizard genomic DNA purification kit according to the instructions of the manufacturer (Promega Corp., WI). PCR amplification of Rickettsia rompA and of the genes encoding the 17-kDa protein and the bacterioferritin comigratory protein (bcp) was conducted on all extracted DNA by using Rickettsia genomic DNA as a positive control and a no-template negative control (Table 1). Each 25-μl reaction mixture consisted of 1 μl of template DNA, 0.4 μM concentrations of all forward and reverse primers, and Promega 2× PCR master mix. The PCR program consisted of 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min followed by a final step at 72°C for 10 min. Restriction enzyme digestion of amplified rompA fragments from each species was conducted using AluI and PstI (New England Biolabs, MA). Alu digestion was conducted by adding 2.5 μl each of AluI and buffer NEB 2 (New England Biolabs, MA) to the amplified rompA product (4 μg) and incubating for 4 h at 37°C. PstI digestion was conducted by adding 2.5 μl each of PstI, 10× bovine serum albumin, and buffer NEB 3 (New England Biolabs, MA) to the amplified rompA product (4 μg) and incubating for 4 h at 37°C. All PCR products were separated on 1% Tris-acetate-EDTA agarose gel by electrophoresis and visualized by using ethidium bromide.
TABLE 1.
Primers used in this study
| Primer | Sequence | Fragment length (bp) | Reference |
|---|---|---|---|
| Forward primer for rickettsial 17-kDa-protein gene | GCTCTTGCAACTTCTATGTT | 434 | 2 |
| Reverse primer for rickettsial 17-kDa-protein gene | CATTGTTCGTCAGGTTGGCG | 2 | |
| Forward primer for rickettsial rompA gene | ATGGCGAATATTTCTCCAAAA | 629 | 24 |
| Reverse primer for rickettsial rompA gene | GTTCCGTTAATGGCAGCATCT | 24 | |
| Forward primer for rickettsial bcp gene | CCGAAAGACGATACACCTGGATG | 313 | This study |
| Reverse primer for rickettsial bcp gene | TTTTAGCATTTACCGACCGCC | This study |
RNA extraction and RT-PCR.
RT-PCR was conducted to demonstrate the viability and growth of rickettsiae in mosquito cells after nine passages in mosquito cells or after the third passage in the source cells confirmed to be reinfected. RNA was extracted from Rickettsia-infected cells by using TRIzol (Invitrogen). RNA was treated with DNase I and purified on an RNeasy spin column (RNeasy mini kit; QIAGEN Inc, CA). RT-PCR was conducted using the SuperScript III one-step RT-PCR system with Platinum Taq DNA polymerase by following the guidelines of the manufacturer (Invitrogen). No-template and no-RT treatments were implemented as negative controls. Primers used were the gene for the bacterioferritin comigratory protein (bcp) and the rompA gene (Table 1).
Cloning and sequencing of amplified products.
PCR and RT-PCR amplicons were separated by 1% Tris-acetate-EDTA-agarose gel electrophoresis and purified using the StrataPrep DNA gel extraction kit (Stratagene, CA). Amplicons were ligated into the TOPO TA cloning vector, and Top10 chemically competent cells (Invitrogen) were transformed with the vector. Clones were selected by blue-white screening, and plasmids were extracted using the QIAprep spin miniprep kit (QIAGEN). Sequences were analyzed on an Applied Biosystems 3730XL high-throughput 96-capillary or an ABI 3100 16-capillary DNA sequencer at the University of Maryland, Baltimore, Biopolymer/Genomics Core Facility.
FISH.
General FISH to detect the presence of rickettsiae in 4% formalin-fixed cells was conducted as described by Gottlieb et al. (11). Infected cells were cytocentrifuged onto a slide (Wescor, Inc., UT) or grown overnight in Labtek chamber slides (8-well Permanox slides; Nalge Nunc International). In the latter case, the chamber walls and the gasket were removed after fixing cells for 20 min in 4% formalin-phosphate-buffered saline at room temperature. The Rickettsia-specific FISH probe was conjugated with a 5′-end 6-carboxyfluorescein label and is specific to all known rickettsial 16S rRNA genes (5′-6-carboxyfluorescein-TCCACGTCGCCGTCTTGC) (11). The hybridization probe was prepared to a final concentration of 10 pmol/ml as described by Rasgon et al. (23). Probed slides were incubated in a humid chamber at 37°C overnight. Slides were washed once in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate)-10 mM dithiothreitol, counterstained with Evans blue (0.1% in 1× phosphate-buffered saline), and incubated at 37°C for 15 min. Slides were then subjected to a series of washes as described by Rasgon et al. (23). Slides were mounted in 50 μl of ProLong (Invitrogen) containing DAPI (4′,6′-diamidino-2-phenylindole; 1 ng/ml) and observed under oil immersion (100× objective) by using an epifluorescence microscope fitted with a green (fluorescein isothiocyanate) filter, a blue (UV DAPI) filter, and a red (tetramethyl rhodamine isocyanate) filter. Pictures taken were exposed for 0.5 to 0.75 s in a 2- by 2-pixel bin at gamma 1 by using a SPOT RT digital camera (Diagnostic Instruments, Inc., MI), and images were merged with SPOT advanced imaging software (Universal Imaging, PA). Uninfected mosquito cells, Aa23 and Sua5B cells mixed with Escherichia coli that had been grown in Luria-Bertani broth overnight, and Wolbachia-infected Aa23 cells were used as negative controls to demonstrate the specificity of the probe for Rickettsia.
RESULTS
Cell growth and morphological changes after rickettsial infection.
Uninfected Aa23 and Sua5B cells were adherent, formed filopodia, and were spindle shaped. For all media tested, infected mosquito cells incubated in medium with 5% FBS at 34°C clumped and detached, which ultimately led to cell death. The use of 10% FBS at 34°C prevented cell death, but cell growth was rapid, and infections were sometimes lost. Cells grown at room temperature with 5% FBS grew more slowly, and persistent to semipersistent infections were successfully established. We therefore used room temperature for the incubation of cells postinfection and supplemented the media with 5% FBS for inoculated cells in all subsequent inoculations. For Aa23 cells, we were able to split Rickettsia-infected cells, but after two passages, the infected cells attached poorly. Infected Aa23 cells were therefore added to a 50% confluent layer of uninfected cells to establish semipersistent infections unless otherwise specified. R. montanensis- and R. peacockii-infected Sua5B cells could be persistently maintained without the addition of new cells. R. felis-infected Sua5B cells were subcultured in a manner similar to Aa23 cells due to the vigorous nature of R. felis infections.
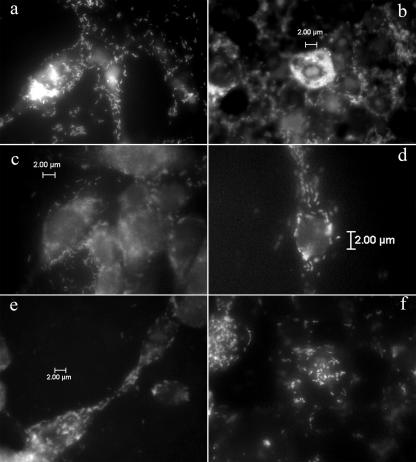
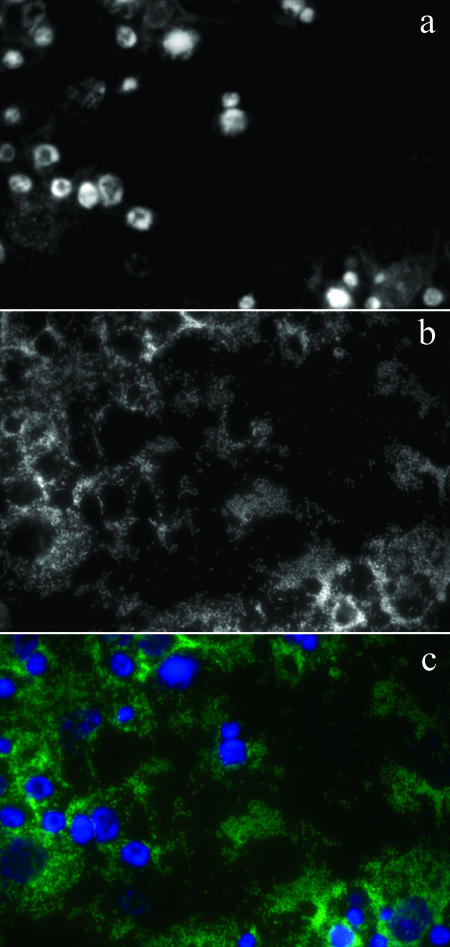
Rickettsial morphology varied with growth stage, an observation previously made by Wisseman et al. (28) and Wisseman and Waddell (29). Log-phase rickettsiae were dividing, often appearing bacillary (Fig. 1a and e) or forming short chains (Fig. 1c). Stationary-phase rickettsiae were smaller coccobacilli (Fig. 1b and f). We observed rickettsiae surrounding the cell nuclei (Fig. 1b and d), dispersed throughout the cytoplasm (Fig. 1), located within filopodia (Fig. 1e), and positioned in cell-to-cell bridges between adjacent cells (Fig. 1a and d) that were grown on Labtek chamber slides. Because we did not perform confocal microscopy, we could not discern whether rickettsiae had in fact entered the nuclei. However, we observed rickettsiae surrounding cell nuclei (stained with DAPI) and localizing in the nuclear region (Fig. 2). In later stages of infection, extracellular rickettsiae were also observed (Fig. 2).
FIG. 1.
FISH detection of R. montanensis and R. peacockii in Anopheles gambiae cell line Sua5B and Aedes albopictus cell line Aa23. (a and b) R. peacockii in Sua5B cells, shown dividing and forming short chains (a) and appearing as coccobacilli in several heavily infected Sua5B cells, especially surrounding the nucleus (b); (c) R. peacockii in Aa23 cells (note division and the formation of short chains); (d) R. montanensis in Sua5B cells; (e and f) R. montanensis in Aa23 cells, appearing bacillary in the filopodia and cytoplasm (e) and stationary (as coccobacilli) in Aa23 cytoplasm (f).
FIG. 2.
Microscopic detection of R. felis in Anopheles gambiae cell line Sua5B. (a) DAPI image; (b) FISH image; (c) color-merged image of FISH and DAPI images of R. felis in Anopheles gambiae cell line Sua5B. The Rickettsia-specific 16S rRNA FISH probe fluoresced bright green; DAPI fluoresced blue and bound to DNA.
R. felis.
Both Aa23 and Sua5B cells supported R. felis growth. Aa23 cells inoculated with R. felis clumped, lysed, and released rickettsiae extracellularly. Similarly, rickettsiae grew to high numbers in Sua5B cells, and extracellular rickettsiae were also observed (Fig. 2b and c). We observed condensed and degraded nuclei of infected cells with DAPI staining (Fig. 2a) and high numbers of extracellular rickettsiae (Fig. 2b). The numbers of rickettsiae were high (more than 100 per cell by passage 9), and even with 1:10 dilutions of R. felis-infected cells with uninfected monolayers, cells had to be passaged within 1 to 2 weeks or infected cells would die. Close to 100% of cells were infected within 7 days of passaging. Partially purified R. felis cells from Aa23 and Sua5B cells also induced cell detachment and nuclear degradation in DAE100 cells (grown at 34°C) and L929 cells (grown at 34°C with 5% CO2). Molecular confirmation of infection was achieved by restriction digestion of PCR-amplified rompA and by RT-PCR (Table 2). We sequenced PCR products of the 17-kDa-protein gene, rompA, and bcp from both mosquito cell lines infected with R. felis and RT-PCR products of bcp from R. felis-infected cells (Table 2). R. felis was able to reinfect L929 cells when partially purified from mosquito cells at passage 9 (Table 2).
TABLE 2.
Infection statuses of arthropod cell lines
| Cell line | Rickettsial species | No. of passages | Infection status detected by:
|
Infectivity in originating cellsf (cell line) | ||||
|---|---|---|---|---|---|---|---|---|
| FISHa | PCRb | RE digestionc | RT-PCRd | Sequencinge | ||||
| Anopheles gambiae Sua5B | R. peacockii | 9 | + | + | + | + | + | + (DAE) |
| R. montanensis | 15 | + | + | + | + | + | + (L929) | |
| R. felis LSU | 9 | + | + | + | + | + | + (Vero) | |
| Aedes albopictus Aa23 | R. peacockii | 9 | + | + | + | + | + | + (DAE) |
| R. montanensis | 15 | + | + | + | + | + | + (L929) | |
| R. felis LSU | 9 | + | + | + | + | + | + (Vero) | |
A Rickettsia-specific FISH probe was used to detect infections microscopically.
Rickettsial genes amplified at each passage were rompA, the 17-kDa-protein gene, and/or bcp.
PstI and AluI restriction enzyme (RE) digestion of each species was used to confirm species-specific restriction patterns.
RT-PCR was used to amplify rickettsial bcp or rompA transcripts.
Sequencing of PCR and RT-PCR products of bcp and rompA was conducted.
Infections were confirmed by partially purifying rickettsiae from mosquito cells after the ninth passage, inoculating the originating cell line (L929, Vero, or DAE100, as indicated), and then assaying for rickettsial presence by FISH, PCR, and RT-PCR after three passages. +, infective in originating cell line.
R. montanensis.
Both Aa23 and Sua5B cells supported the growth of R. montanensis. Initial responses to rickettsial infection by Aa23 and Sua5B cells were similar to those observed when cells were infected with R. felis. Counts of rickettsiae ranged from 0 to more than 50 per cell, with at least 80% of cells being infected. Because infections in Sua5B cells could be maintained persistently, cells were maintained by splitting infected cells into new flasks. Molecular confirmation of infection was achieved by restriction digestion of PCR-amplified rompA and by RT-PCR. We sequenced PCR products of the 17-kDa-protein gene, rompA, and bcp from both mosquito cell lines infected with R. montanensis (Table 2). Additionally, we sequenced RT-PCR products of rompA from R. montanensis-infected Sua5B cells and bcp from R. montanensis-infected Aa23 cells after passage 9 (Table 2). R. montanensis was able to reinfect L929 and Vero cells (both grown at 34°C with 5% CO2) when partially purified from mosquito cells at passage 9 (Table 2).
R. peacockii.
R. peacockii was maintained most efficiently in mosquito cells at room temperature in 5% FBS with semipersistent passaging for Aa23 cells or persistent passaging for Sua5B cells. While initial exposure to R. peacockii caused many cells in both lines to detach and round up, subsequent passages resulted in good R. peacockii growth without significant cell death. Counts of rickettsiae ranged from 0 to more than 50 per cell, with at least 80% of cells being infected. Molecular confirmation of R. peacockii infection was achieved by restriction digestion of PCR-amplified rompA and by RT-PCR (Table 2). We sequenced PCR products of the 17-kDa-protein gene, rompA, and bcp from both mosquito cell lines infected with R. peacockii and RT-PCR products of bcp from R. peacockii-infected cells (Table 2). R. peacockii was able to reinfect DAE100 cells when partially purified from mosquito cells at passage 9 (Table 2). R. peacockii was detected in L929 cells in the third passage by using RT-PCR, although it was lost after the second passage in Vero cells (both grown at 34°C with 5% CO2).
DISCUSSION
While several arthropod cell lines have been used to support rickettsial growth, the two cell lines described herein have not been used for Rickettsia maintenance before this study. Further, both cell lines not only supported rickettsial growth but were also permissive toward the most fastidious species of the three tested, R. peacockii. We chose to use the Aa23 cell line because it was readily available and has previously supported several diverse Wolbachia strains (9). Consequently, the Aa23 cell line may conceivably serve as an arena for studies of intergenus interactions between Anaplasmataceae (namely, Wolbachia, Ehrlichia, and Anaplasma species) and Rickettsia. Alternatively, the Aa23 cell line could be used to compare and contrast the different lifestyles of intracellular bacteria (endosymbionts versus pathogens and membrane-bound bacteria versus those free in the cytoplasm, etc.) and explore interbacterial interactions. We chose to use Sua5B cells because the Sua5B cell line promises to be a particularly useful model system for Rickettsia-arthropod interactions. Since the genome sequence of Anopheles gambiae has been published, microarrays are readily available to study differential gene expression. Aguilar et al. and Dimopoulos and colleagues have conducted several excellent microarray-based experiments investigating the immune and stress responses of Sua5B cells to exposure to bacteria, fungi, and Plasmodium species (1, 8). Hence, while the Sua5B cell line is useful as a maintenance tool for diverse rickettsiae, it is also a potential model system for vector-Rickettsia interactions.
To our knowledge, neither R. montanensis nor R. peacockii has been maintained previously in mosquito cell systems. A prior attempt to culture R. montanensis in Aedes albopictus AeAl2 cells was unsuccessful, although attachment did occur (27). Similarly, R. peacockii was not maintained in Aedes albopictus C7/10 cells, although it grew well in several tick species and one lepidopteran line (Trichoplusia ni) (13). Differences in Aedes albopictus cell line characteristics may explain why both R. peacockii and R. montanensis were able to grow in the particular Aedes albopictus cell line we used but did not grow in either of the cell lines used by Kurtti et al. and Uchiyama (13, 27).
The culturing of R. peacockii in mosquito cells is significant because R. peacockii is considered to be deficient in the abilities to invade and grow in mammalian cells. Unlike other SFG species, R. peacockii does not polymerize actin owing to the deletion of a rickA gene (26). Nor does R. peacockii express the outer membrane protein encoded by rompA, although it does possess the rompA gene and transcripts have been detected previously (4, 26). R. peacockii is not known to adhere to tick cells, and tick cells exposed to R. peacockii have not been observed to exhibit phagocytic behavior, suggesting that R. peacockii may have a disadvantage in entering host cells (15). Both Sua5B and Aa23 cell lines have hemocyte-like characteristics and are capable of phagocytosis (8, 17). Since many rickettsiae enter cells by inducing endocytosis, it is conceivable that R. peacockii could infect mosquito cells that exhibit phagocytic behavior provided that R. peacockii could avoid digestion. Based on RT-PCR and FISH visualization of R. peacockii after nine passages, it would appear that R. peacockii somehow avoids digestion and grows in the cytoplasm of mosquito cells. Somewhat paradoxically, R. peacockii was transcriptionally active in L929 cells after the third passage. It remains to be seen if R. peacockii infections can be sustained for several more passages in L929 or other mammalian cell lines.
Previous studies have demonstrated the successful maintenance of R. felis and R. conorii in the Aedes albopictus cell line C6/36 (12, 25). We therefore anticipated that R. felis would grow in Aa23 cells. Similarly, R. felis infection of Sua5B cells was also expected as Sua5B cells have previously supported the growth of an intracellular endosymbiont Wolbachia species (21). The R. felis LSU strain we used was originally derived from colonies of R. felis-infected cat fleas and has been transovarially maintained in these fleas for several years. The strain may therefore have characteristics of an adapted flea colony rather than those of a wild-type strain. We have recently acquired the Pedreira strain of R. felis and are currently maintaining it in Aa23 and Sua5B cells (seven passages to date). Again, this outcome was not surprising since the Pedreira strain was originally isolated from wild-caught R. felis-infected cat fleas and maintained in Aedes albopictus C6/36 cells (12).
R. felis falls phylogenetically into a transitional group between the spotted fever and typhus groups and is transmitted by fleas (10). The use of an insect cell line to study this TRG rickettsial pathogen may provide insight into the evolution of Rickettsia-vector interactions and perhaps host jumps from acarine to insect vectors or visa versa. Additionally, we may be able to explain why mosquitoes are not infected with Rickettsia spp. but many are infected with Wolbachia. For example, if infection-dependent gene expression in mosquitoes elicited by Wolbachia differs from that elicited by exposure to Rickettsia spp., this finding may explain why Rickettsia spp. have not become established in mosquitoes. Furthermore, the use of the R. felis-insect interactions may shed light into what genes and/or environmental cues cause rickettsial species to exist as endosymbionts, reproductive parasites, or pathogens.
Because these two cell lines were supportive of R. peacockii growth, it is conceivable that they can be used to isolate and maintain the growing number of newly described nonpathogenic rickettsiae from a diverse range of invertebrate and protozoan hosts. The significance of nonpathogenic rickettsiae extends beyond ecological or evolutionary interests and may have implications relevant to public health. First, nonpathogenic rickettsiae may serve as sources of emerging vertebrate pathogens. Perlman et al. suggested that since the majority of rickettsial diversity lies in species associated with invertebrates, ancestral rickettsiae are associated with invertebrates and only secondarily develop into vertebrate pathogens by rare horizontal transmission events in evolutionary time (20). Second, rickettsiae commonly referred to as nonpathogenic may in fact be pathogenic rickettsiae whose pathogenicity was not immediately apparent upon discovery. R. parkeri is one such example, having been considered nonpathogenic until 60 years after its discovery (18). Likewise, R. canadensis and R. montanensis have been referred to as being of unknown pathogenicity (19, 22). Third, some nonpathogenic rickettsiae appear to affect the epidemiology of pathogenic rickettsiae through competition for the same tick niche. The classic example of this relationship is the geographic limitation of R. rickettsii to D. andersoni populations on the west side of the Bitterroot Valley of Montana, presumably through transovarial exclusion by the closely related nonpathogenic R. peacockii (13, 15, 16). Last, nonpathogenic rickettsiae may serve as useful model systems and provide vital clues into the genetic components of rickettsial pathogenicity. Thus, the availability of these cell lines that will potentially support diverse Rickettsia species of various degrees of pathogenicity for rickettsial maintenance is highly advantageous for the study of nonpathogenic species.
The three Rickettsia species grown in these two cell lines vary in genetic characteristics and the degree of infectivity in eukaryotic cells. It is often difficult to make conclusions about their behavior without considering the cell line in which they were grown as a variable of their pathogenicity (or lack thereof). The use of a standard cell line such as Aa23 or Sua5B to grow all three species would remove the variation due to cell line differences as a factor. Further, it would be ideal to have a cell line that can support the growth of both insect-associated and tick-associated rickettsiae for studying broader questions of Rickettsia-host interactions. We propose that since both Aa23 and Sua5B cells support diverse rickettsiae of various degrees of pathogenicity derived from insect and tick sources, they are excellent candidates for future studies into the evolution of rickettsial pathogenicity and the coevolution of Rickettsia species with their arthropod vectors and/or hosts.
Acknowledgments
We thank N. Ammerman and M. Beier (University of Maryland, Baltimore) for support in BSL3; N. Ammerman for R. felis strain LSU in Vero cells; K. Macaluso (Louisiana State University) for R felis strain LSU; J. Sacci and R. Maag (University of Maryland, Baltimore) for microscopy and imaging advice; J. Rasgon (Johns Hopkins University) for W. pipientis, Aa23, and Sua5B cells; and U. Munderloh (University of Minnesota) for DAE100 cells and R. peacockii-infected DAE100 cells. We also thank J. Rasgon, J. Gillespie (University of Maryland, Baltimore), M. S. Rahman (University of Maryland, Baltimore), and M. Beier for critical review of the manuscript. Lastly, we thank the two anonymous reviewers for their valuable input.
The work presented here is supported by NIH/NIAID grants AI017828 and AI063534.
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Aguilar, R., A. E. Jedlicka, M. Mintz, V. Mahairaki, A. L. Scott, and G. Dimopoulos. 2005. Global gene expression analysis of Anopheles gambiae responses to microbial challenge. Insect Biochem. Mol. Biol. 35:709-719. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, B. E., R. L. Regnery, G. M. Carlone, T. Tzianabos, J. E. McDade, Z. Y. Fu, and W. J. Bellini. 1987. Sequence analysis of the 17-kilodalton-antigen gene from Rickettsia rickettsii. J. Bacteriol. 169:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad, A. F., and C. B. Beard. 1998. Rickettsial pathogens and their arthropod vectors. Emerg. Infect. Dis. 4:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldridge, G. D., N. Y. Burkhard, J. A. Simser, T. J. Kurtti, and U. G. Munderloh. 2004. Sequence and expression analysis of the ompA gene of Rickettsia peacockii, an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni. Appl. Environ. Microbiol. 70:6628-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgdorfer, W., S. F. Hayes, and A. J. Mavros. 1981. Nonpathogenic rickettsiae in Dermacentor andersoni: a limiting factor for the distribution of Rickettsia rickettsii, p. 585-594. In W. Burgdorfer and R. L. Anaker (ed.), Rickettsiae and rickettsial diseases. Academic Press, New York, NY.
- 6.Chen, S.-M., V. L. Popov, H.-M. Feng, J. Wen, and D. H. Walker. 1995. Cultivation of Ehrlichia chaffeensis in mouse embryo, Vero, BGM, and L929 cells and study of Ehrlichia-induced cytopathic effect and plaque formation. Infect. Immun. 3:647-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cory, J., and C. E. Yunker. 1974. Rickettsial plaques in mosquito cell monolayers. Acta Virol. 18:512-513. [PubMed] [Google Scholar]
- 8.Dimopoulos, G., G. K. Christophides, S. Meister, J. Schultz, K. P. White, C. Barillas-Mury, and F. C. Kafatos. 2002. Genome expression analysis of Anopheles gambiae: responses to injury, bacterial challenge, and malaria infection. Proc. Natl. Acad. Sci. USA 99:8814-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobson, S. L., E. J. Marsland, Z. Veneti, K. Bourtzis, and S. L. O'Neill. 2002. Characterization of Wolbachia host cell range via the in vitro establishment of infections. Appl. Environ. Microbiol. 68:656-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillespie, J. J., M. S. Beier, M. S. Rahman, N. C. Ammerman, J. M. Shallom, A. Purkayastha, B. S. Sobral, and A. F. Azad. 2007. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS ONE 2:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb, Y., M. Ghanim, E. Chiel, D. Gerling, V. Portnoy, S. Steinberg, G. Tzuri, A. R. Horowitz, E. Belausov, N. Mozes-Daube, S. Kontsedalov, M. Gershon, S. Gal, N. Katzir, and E. Zchori-Fein. 2006. Identification and localization of a Rickettsia sp. in Bemesia tabaci (Homoptera: Aleyrodidae). Appl. Environ. Microbiol. 72:3646-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horta, M. C., M. B. Labruna, E. L. Durigon, and T. T. S. Schumaker. 2006. Isolation of Rickettsia felis in the mosquito cell line C6/36. Appl. Environ. Microbiol. 72:1705-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurtti, T. J., J. A. Simser, G. D. Baldridge, A. T. Palmer, and U. G. Munderloh. 2005. Factors influencing in vitro infectivity and growth of Rickettsia peacockii (Rickettsiales: Rickettsiaceae), an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni (Acari, Ixodidae). J. Invertebr. Pathol. 90:177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macaluso, K. R., D. E. Sonenshine, S. M. Ceraul, and A. F. Azad. 2002. Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia. J. Med. Entomol. 39:809-813. [DOI] [PubMed] [Google Scholar]
- 15.Munderloh, U. G., Y. Liu, M. Wang, C. Chen, and T. J. Kurtti. 1994. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J. Parasitol. 80:533-543. [PubMed] [Google Scholar]
- 16.Niebylski, M. L., M. E. Schrumpf, W. Burgdorfer, E. R. Fisher, K. L. Gage, and T. G. Schwan. 1997. Rickettsia peacockii sp. nov, a new species infecting wood ticks, Dermacentor andersoni, in western Montana. Int. J. Syst. Bacteriol. 47:446-452. [DOI] [PubMed] [Google Scholar]
- 17.O'Neill, S. L., M. M. Pettigrew, S. P. Sinkins, H. R. Braig, T. G. Andreadis, and R. B. Tesh. 1997. In vitro cultivation of Wolbachia pipientis in an Aedes albopictus cell line. Insect Mol. Biol. 6:33-39. [DOI] [PubMed] [Google Scholar]
- 18.Paddock, C. D., J. W. Sumner, J. A. Comer, S. R. Zaki, C. S. Goldsmith, J. Goddard, S. L. F. McLellan, C. L. Tamminga, and C. A. Ohl. 2004. Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clin. Infect. Dis. 38:805-811. [DOI] [PubMed] [Google Scholar]
- 19.Parola, P., C. D. Paddock, and D. Raoult. 2005. Tick-borne rickettsioses around the world: emerging diseases challenging Old World concepts. Clin. Microbiol. Rev. 18:719-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perlman, S. J., M. S. Hunter, and E. Zchori-Fein. 2006. The emerging diversity of Rickettsia. Proc. R. Soc. Lond. B 273:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pornwiroon, W., S. S. Pourciau, L. D. Foil, and K. R. Macaluso. 2006. Rickettsia felis from cat fleas: isolation and culture in a tick-derived cell line. Appl. Environ. Microbiol. 72:5589-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raoult, D., and V. Roux. 1997. Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 10:694-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasgon, J. L., X. Ren, and M. Petridis. 2006. Can Anopheles gambiae be infected with Wolbachia pipientis? Insights from an in vitro system. Appl. Environ. Microbiol. 72:7718-7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roux, V., P.-E. Fournier, and D. Raoult. 1996. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J. Clin. Microbiol. 34:2058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rovery, C., P. Renesto, N. Crapoulet, K. Matsumoto, P. Parola, H. Ogata, and D. Raoult. 2005. Transcription response of Rickettsia conorii exposed to temperature variation and stress starvation. Res. Microbiol. 156:211-218. [DOI] [PubMed] [Google Scholar]
- 26.Simser, J. A., M. S. Rahman, S. M. Dreher-Lesnick, and A. F. Azad. 2005. A novel and naturally occurring transposon, ISRpe1 in the Rickettsia peacockii genome disrupting the rickA gene involved in actin-based motility. Mol. Microbiol. 58:71-79. [DOI] [PubMed] [Google Scholar]
- 27.Uchiyama, T. 2005. Growth of typhus group and spotted fever group rickettsiae in insect cells. Ann. N. Y. Acad. Sci. 1063:215-221. [DOI] [PubMed] [Google Scholar]
- 28.Wisseman, C. L., Jr., E. A. Edlinger, A. D. Waddell, and M. R. Jones. 1976. Infection cycle of Rickettsia rickettsii in chicken embryo and L-929 cells in culture. Infect. Immun. 14:1052-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wisseman, C. L., Jr., and A. D. Waddell. 1975. In vitro studies on rickettsia-host cell interactions: intracellular growth cycle of virulent and attenuated Rickettsia prowazekii in chicken embryo cells in slide chamber cultures. Infect. Immun. 11:1391-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]