Abstract
Robust genotyping methods for Lactobacillus casei are needed for strain tracking and collection management, as well as for population biology research. A collection of 52 strains initially labeled L. casei or Lactobacillus paracasei was first subjected to rplB gene sequencing together with reference strains of Lactobacillus zeae, Lactobacillus rhamnosus, and other species. Phylogenetic analysis showed that all 52 strains belonged to a single compact L. casei-L. paracasei sequence cluster, together with strain CIP107868 (= ATCC 334) but clearly distinct from L. rhamnosus and from a cluster with L. zeae and CIP103137T (= ATCC 393T). The strains were genotyped using amplified fragment length polymorphism, multilocus sequence typing based on internal portions of the seven housekeeping genes fusA, ileS, lepA, leuS, pyrG, recA, and recG, and tandem repeat variation (multilocus variable-number tandem repeats analysis [MLVA] using nine loci). Very high concordance was found between the three methods. Although amounts of nucleotide variation were low for the seven genes (π ranging from 0.0038 to 0.0109), 3 to 12 alleles were distinguished, resulting in 31 sequence types. One sequence type (ST1) was frequent (17 strains), but most others were represented by a single strain. Attempts to subtype ST1 strains by MLVA, ribotyping, clustered regularly interspaced short palindromic repeat characterization, and single nucleotide repeat variation were unsuccessful. We found clear evidence for homologous recombination during the diversification of L. casei clones, including a putative intragenic import of DNA into one strain. Nucleotides were estimated to change four times more frequently by recombination than by mutation. However, statistical congruence between individual gene trees was retained, indicating that recombination is not frequent enough to disrupt the phylogenetic signal. The developed multilocus sequence typing scheme should be useful for future studies of L. casei strain diversity and evolution.
Lactobacillus casei strains are of considerable interest in the food industry as acid-producing starter culture for milk fermentation and as maturation promoters of certain cheese specialties. In addition, L. casei has attracted intense interest as a probiotic over the last few years (37, 38). For example, L. casei strain DN-114 001 was shown to have positive effects in young children with acute diarrhea (39, 40), and the L. casei Shirota strain was shown to decrease the excretion in healthy volunteers of p-cresol, a toxic amino acid metabolite produced by intestinal bacteria (12). L. casei strains may be isolated from a wide variety of sources, including dairy products, plant products, and the urogenital and intestinal tracts of animals, including humans.
Virtually nothing is currently known about the genetic diversity and population structure of L. casei. However, knowledge of strain diversity and phylogenetic relationships would be highly relevant for understanding the evolution of ecological or biological properties of strains and for optimizing their industrial or medical exploitation. Basic but far-reaching questions about the biology of fermentative or probiotic characteristics, such as their strain specificity or evolutionary stability, cannot be answered without a proper population genetic framework. Distinguishing L. casei members is also important for identifying strains with particular phenotypic or industrial properties and for strain tracking, collection management, and traceability. Limited molecular typing data based on randomly amplified polymorphic DNA-PCR, ribotyping, pulsed-field gel electrophoresis, or insertion sequences (41, 49, 54) indicate the existence of DNA-level differences among L. casei strains. However, these methods are not considered robust for strain delineation and phylogenetic inference (2). For pathogenic bacteria, including species that are grouped together with lactobacilli in the order Lactobacillales (for example, Enterococcus faecium or Streptococcus pneumoniae), the method of choice for population genetics and standardized strain typing is multilocus sequence typing (MLST) (34). MLST consists of determining the sequence of an internal portion of a small number (most often seven) of housekeeping genes. It provides unambiguous genotype nomenclature that can easily be shared between laboratories and provides precise information on strain evolution. Although MLST is now widely used for international collaboration on strain tracking and population biology of bacterial pathogens, its application to the study of strain diversity and evolution in the field of food production microbiology is still in its infancy, with developments only for Lactobacillus plantarum and Oenococcus oeni (8, 9).
In evolutionarily recent bacterial groups, MLST fails to differentiate strains because nucleotide variation accumulates only at a low rate (43). For fine subtyping of these groups, methods with higher rates of evolution are needed. Methods that were shown to be useful for fine typing of homogeneous groups include multilocus variable-number tandem repeats (VNTR) analysis (MLVA) (31), single nucleotide repeat (SNR) variation (51), and clustered regularly interspaced short palindromic repeats (CRISPR) locus variation, also called spoligotyping for Mycobacterium tuberculosis (28).
The main aim of the present study was to develop an MLST scheme for L. casei and to initiate characterization of the population structure of this species. Due to the currently debated taxonomic status and relationships of L. casei and the related species Lactobacillus zeae, Lactobacillus paracasei, and Lactobacillus rhamnosus, which together are regarded as the L. casei group, we first determined the phylogenetic clustering of our strains compared to the type and reference strains of the L. casei group. MLST was then applied to the study of diversity among 52 strains. We also explored potential alternative typing methods with the hope of providing increased discrimination for some strain groups that were homogeneous based on MLST.
MATERIALS AND METHODS
Bacterial strains.
A total of 52 L. casei study strains and 11 reference strains were included (Table 1). L. casei strains (some of which were originally referred to as L. paracasei) were gathered from the collection of Danone Research, Centre de Recherche Daniel Carasso. Reference strains were obtained directly from the Collection de l'Institut Pasteur. The complete genome of strain DN-114 001 was fully sequenced (Danone Research, unpublished).
TABLE 1.
Characteristics of Lactobacillus strains studieda
| Strain code | Other name | Taxonomic designation | Source | AFLP typeb | Allele code
|
ST | rplB alleled | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fusA | ileS | lepA | leuS | pyrG | recA | recG | |||||||
| CIP102021 | L. plantarum | NA | ND | ND | ND | ND | ND | ND | ND | ND | ND | 14 | |
| CIP102840 | L. brevis | Cosmetic product | ND | ND | ND | ND | ND | ND | ND | ND | ND | 16 | |
| CIP102993 | ATCC 25598 | L. casei subsp. casei | Dairy food | ND | ND | ND | ND | ND | ND | ND | ND | ND | 6 |
| CIP103024T | ATCC 25599 | L. casei subsp. tolerans | Dairy food | ND | ND | ND | ND | ND | ND | ND | ND | ND | 11 |
| CIP103137T | ATCC 393Tc | L. casei | Dairy food | ND | ND | ND | ND | ND | ND | ND | ND | ND | 17 |
| CIP103151T | ATCC 14917 | L. plantarum | Other food | ND | ND | ND | ND | ND | ND | ND | ND | ND | 15 |
| CIP103152T | ATCC 35046 | L. animalis | Animal | ND | ND | ND | ND | ND | ND | ND | ND | ND | 12 |
| CIP103253T | ATCC 15820 | L. zeae | Cereal food | ND | ND | ND | ND | ND | ND | ND | ND | ND | 18 |
| CIP105422T | L. sakei | Other traditional food | ND | ND | ND | ND | ND | ND | ND | ND | ND | 13 | |
| CIP107868 | ATCC 334 | L. casei | Dairy food | ND | 7 | 3 | 7 | 10 | 1 | 3 | 1 | ST32 | 7 |
| CIPA157T | ATCC 7469 | L. rhamnosus | NA | ND | ND | ND | ND | ND | ND | ND | ND | ND | 19 |
| D573 | DN-114 001 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 |
| D6.1 | L. casei | Dairy food | ND | 1 | 1 | 1 | 1 | 1 | 1 | 2 | ST2 | 1 | |
| D629 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D631 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D635 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D636 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D637 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D639 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D640 | L. casei | Dairy food | 19 | 5 | 4 | 5 | 5 | 1 | 1 | 4 | ST14 | 4 | |
| D641 | L. casei | Dairy food | 19 | 5 | 4 | 5 | 5 | 1 | 1 | 1 | ST15 | 4 | |
| D642 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D643 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D644 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D645 | L. casei | Dairy food | 21 | 4 | 5 | 6 | 6 | 1 | 1 | 5 | ST16 | 5 | |
| D647 | L. casei | Other traditional food | 11 | 4 | 2 | 8 | 2 | 1 | 1 | 6 | ST21 | 6 | |
| D648 | DSM 3173 | L. casei | Fermented fruits | 1 | 7 | 3 | 7 | 9 | 1 | 1 | 7 | ST22 | 7 |
| D655 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D656 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D657 | ATCC 27092 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 |
| D658 | ATCC 27139 | L. casei | NA | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 |
| D659 | L. casei | Dairy food | A/14 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | ST3 | 1 | |
| D660 | L. casei | Dairy food | 8 | 4 | 2 | 3 | 3 | 1 | 1 | 3 | ST9 | 8 | |
| D661 | L. casei | Dairy food | 22 | 4 | 5 | 6 | 6 | 1 | 2 | 5 | ST17 | 5 | |
| D662 | L. casei | Dairy food | 8 | 4 | 2 | 3 | 3 | 1 | 1 | 3 | ST9 | 8 | |
| D664 | DSM 2649 | L. casei | Cereal | 20 | 5 | 2 | 4 | 4 | 1 | 1 | 4 | ST11 | 4 |
| D666 | L. casei | Other traditional food | 16 | 4 | 3 | 9 | 5 | 1 | 1 | 8 | ST23 | 9 | |
| D667 | L. casei | Cereal | 12 | 5 | 1 | 1 | 1 | 3 | 1 | 2 | ST24 | 1 | |
| D669 | L. casei | Dairy food | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 | |
| D671 | ATCC 334 | L. casei | Dairy food | 2 | 7 | 4 | 7 | 10 | 1 | 3 | 1 | ST25 | 7 |
| D679 | L. casei | Other traditional food | 6 | 6 | 3 | 7 | 8 | 1 | 1 | 1 | ST19 | 7 | |
| D685 | L. casei | Other traditional food | 17 | 2 | 6 | 5 | 11 | 1 | 1 | 1 | ST26 | 10 | |
| D686 | L. casei | Cereal | 9 | 2 | 3 | 9 | 3 | 1 | 1 | 8 | ST27 | 10 | |
| D692 | BL23 | L. casei | NA | A/14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 |
| D693 | L. casei | Other traditional food | 10 | 4 | 7 | 5 | 3 | 1 | 1 | 1 | ST28 | 8 | |
| D694 | L. casei | Other traditional food | 18 | 4 | 2 | 3 | 1 | 1 | 1 | 3 | ST8 | 8 | |
| D695 | L. casei | Other traditional food | 4 | 7 | 8 | 7 | 8 | 1 | 1 | 1 | ST29 | 7 | |
| D696 | L. casei | Dairy food | 5 | 6 | 3 | 7 | 7 | 1 | 1 | 1 | ST18 | 7 | |
| D697 | L. casei | Dairy food | 5 | 6 | 3 | 7 | 7 | 1 | 1 | 1 | ST18 | 7 | |
| D698 | L. casei | Other traditional food | 13 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | ST6 | 1 | |
| D699 | L. casei | Other traditional food | 15 | 8 | 2 | 10 | 12 | 1 | 4 | 9 | ST30 | 10 | |
| SB3847 | R.1 | L. casei | Human | A | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 | 1 |
| SB3864 | DSM 5622 | L. paracasei | NA | ND | 6 | 3 | 7 | 8 | 2 | 1 | 1 | ST20 | 7 |
| SB3865 | DSM 8741 | L. paracasei | Human | ND | 4 | 2 | 3 | 2 | 1 | 1 | 3 | ST7 | 6 |
| SB3866 | DSM 8742 | L. paracasei | Human | ND | 3 | 3 | 5 | 4 | 1 | 1 | 4 | ST12 | 4 |
| SB3879 | DSM 20012 | L. paracasei | Dairy food | H | 1 | 1 | 1 | 1 | 1 | 1 | 2 | ST2 | 1 |
| SB3880 | ATCC 393Tc | L. casei | Dairy food | C | 3 | 3 | 5 | 4 | 1 | 1 | 4 | ST12 | 4 |
| SB3883 | DSM 4905 | L. paracasei | Human | D | 4 | 2 | 3 | 2 | 1 | 1 | 3 | ST7 | 6 |
| SB3884 | DSM 5457 | L. paracasei | NA | E | 1 | 3 | 5 | 4 | 1 | 1 | 4 | ST13 | 4 |
| SB3885 | DSM 20006 | L. paracasei | Beer | I | 2 | 1 | 1 | 1 | 1 | 1 | 1 | ST4 | 1 |
| SB3886 | DSM 20008 | L. paracasei | Dairy food | J | 4 | 2 | 4 | 4 | 1 | 1 | 4 | ST10 | 4 |
| SB3887 | DSM 20020 | L. paracasei | Human | F | 3 | 1 | 1 | 1 | 1 | 1 | 1 | ST5 | 1 |
| SB3888 | R.3 | L. paracasei | Human | G | 1 | 9 | 9 | 5 | 1 | 1 | 8 | ST31 | 10 |
NA, not available; ND, not determined.
AFLP types are labeled with numbers or letters depending on the batch in which they were analyzed. The only common type is labeled A/14.
This strain is the official type strain of L. casei but is phylogenetically distinct from most L. casei/L. paracasei strains.
The gene rplB is not used in the MLST scheme.
AFLP.
Lactobacillus casei strains were grown overnight at 37°C in 2 ml of MRS broth. Bacteria were pelleted by centrifugation and resuspended in 500 μl of TES (1 mM EDTA, 10 mM Tris [pH 8], 250 g/liter sucrose). Cells were again pelleted by centrifugation, suspended in 300 μl of TES and 90 μl of enzyme solution (lyzozyme [360 mg/ml] and mutanolysin [1,400 U/ml]), and incubated for 2 h at 37°C. Three hundred microliters of saline solution (NaCl [150 mM] and EDTA [10 mM]) and 40 μl of 20% (wt/vol) sodium dodecyl sulfate were added. DNA was purified by the phenol-chloroform method (45). Amplified fragment length polymorphism (AFLP) data were obtained from KeyGene (Wageningen, The Netherlands) using restriction enzymes NlaIII and TacI and six combinations of the eight following primers: N02 (5-AGACTGCGTACACATGC-3), N03 (5′-AGACTGCGTACACATGG-3′), N04 (5′-AGACTGCGTACACATGT-3′), T11 (5′-GATGAGTCCTGACCGAAA-3′), T14 (5′-GATGAGTCCTGACCGAAT-3′), T15 (5′-GATGAGTCCTGACCGACA-3′), T23 (5′-GATGAGTCCTGACCGATA-3′), and T25 (5′-GATGAGTCCTGACCGATG-3′). These primers were used in the following combinations: N02 and T23, N03 and T11, N03 and T14, N03 and T23, N04 and T15, and N04 and T25. For primers whose names start with “N” and “T,” the selective nucleotides were the last two and three nucleotides, respectively. AFLP profiles that did not differ by more than one band were defined as belonging to the same AFLP type.
MLST.
Lactobacillus casei strains were grown at 30°C overnight in 10 ml of MRS broth. The cells were pelleted by centrifugation and resuspended in 500 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) solution containing 15 mg/ml of lysozyme (Sigma, Germany) and 15 μl of mutanolysine (5 U/μl). Cells were incubated overnight at 37°C and then lysed by adding 150 μl of 25% (wt/vol) sodium dodecyl sulfate and 150 μl of proteinase K (20 mg/ml) (Sigma).
DNA extraction was performed using the Wizard genomic DNA purification kit (Promega, Madison, WI). In order to design primers suitable for PCR amplification of all L. casei strains (Table 2), we optimized primers suggested to be useful for a wide range of bacterial groups (46). Optimization was achieved by reducing degeneracy in the original primers, using the genome sequence data of strain DN-114 001. The eight loci for which PCR amplification was successful corresponded to gene elongation factor EF-2 (fusA), isoleucyl-tRNA synthetase (ileS), GTP-binding protein LepA (lepA), leucyl-tRNA synthetase (leuS), CTP synthetase (pyrG), recombinase A (recA), ATP-dependent DNA helicase (recG), and 50S ribosomal protein L2 (rplB). These genes are widely separated on the chromosome sequence of strain DN-114 001 (Table 2), excepted for rplB and fusA, whose start codons are only 5,744 nucleotides apart. PCR conditions for all amplification reactions were as follows: initial denaturation at 94°C for 5 min; 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30s; and final extension at 72°C for 5 min. PCR products were purified by ultrafiltration (Millipore), and nucleotide sequences were obtained using the PCR primers and BigDye Terminator v3.1 chemistry (Applied Biosystems, Foster City, CA) on an ABI 3700 apparatus (Applied Biosystems, Foster City, CA). Sequence traces were edited and stored using BioNumerics version 4.6 (Applied-Maths, St. Maartens-Latem, Belgium). For reliability, the quality of the chromatogram traces was checked and the sequences were repeated until every nucleotide in the consensus sequence was supported by at least two sequence chromatogram traces.
TABLE 2.
Primers used for MLST
| Locus | Forward primer | Reverse primer | Locationa |
|---|---|---|---|
| fusA | 5′-CCG TAA TAT CGG GAT CAT GGC TCA CAT CGA-3′ | 5′-CAA CAA CAT CTG AAC ACC CTT GTT-3′ | 2476902-2476240 |
| ileS | 5′-TCC TGG TTG GGA TAC TCA CGG-3′ | 5′-AGG AAC CGG AAT CGA ACC ACA CAT C-3′ | 1258650-1259009 |
| lepA | 5′-CAT CGC CCA CAT TGA TCA CGG GAA-3′ | 5′-CAT ATG CAG CAA GCC TAA GAA CCC-3′ | 1556848-1556300 |
| leuS | 5′-GGG ACG GTT GTT GCA AAC GAA GAA GT-3′ | 5′-CGG TTC ACC CCA ATA ACG CT-3′ | 859623-860264 |
| pyrG | 5′-GGG GTC GTA TCG TCA TTG GGT AAA GG-3′ | 5′-GGA ATG GCA ATG ATT CGA TAT CGC CAA-3′ | 2553308-2552964 |
| recA | 5′-CCG GAA AGT TCC GGC AAA ACA AC-3′ | 5′-CGC GAC CAC CTG GTG TCG TTT C-3′ | 900181-900495 |
| recG | 5′-AGG CGA TGT TGG GAG CGG TAA AAC-3′ | 5′-GTG TTC GGG GAA TAG GCG TCG C-3′ | 1614495-1614154 |
| rlpB | 5′-CAA CAG TTA AAG CAA TCG AAT ACG ATC C-3′ | 5′-CAC CAC CAC CAT GCG GGT GAT C-3′ | 2469701-2469336 |
Coordinates of the MLST sequence template for the complete genome of strain ATCC 334 (GenBank accession no. CP000423).
MLVA.
In order to identify tandem repeats in the L. casei genome, we used the program Tandem Repeat Finder (3) with our unpublished genome data for strain DN-114 001. Initially, 15 potential VNTR loci were selected based on criteria known to maximize polymorphism: shorter repeat unit length, higher number of repeats, and higher degree of sequence conservation among the repeats. PCR primers flanking the tandem repeats were designed, and nine VNTR loci (Table 3) were selected based on successful PCR amplification. PCR amplification was performed in 50-μl volumes. VNTR loci were amplified in separate PCRs using the following program: 5 min at 94°C, followed by 35 cycles of 30 s at 93°C, 30 s at 55°C, and 30 s at 72°C, and a final step of 7 min at 72°C. After PCR, samples were diluted 1:20 in water and 1 μl of the diluted samples was mixed with 1 μl of Rox labeled GENEFLO625 DNA ladder (EURx, Gdansk, Poland) and 12 μl of formamide. After heat denaturation for 5 min at 95°C, fragments were separated using an ABI 3700 apparatus by using the standard GeneScan module. GeneScan chromatograms were imported into BioNumerics version 4.6 (Applied-Maths, St. Maartens-Latem, Belgium) to perform sizing and to calculate the repeat number.
TABLE 3.
Selected VNTR loci and primers used for PCR amplification
| VNTR locus | Chromosomal locationa | Consensus size (bp) | Copy no.a | Forward primer | Reverse primer |
|---|---|---|---|---|---|
| VNTR2 | 278137-278405 | 15 | 18 | 5′-Hex-ATG GCG GTT ACG TGC CAG AAC G | 5′-TCA GGC GTC GTA GTC TGC ACA G |
| VNTR3 | 278225-278424 | 18 | 12 | 5′-Hex-GCA ACG GCT TCG AAC GCA GCT G | 5′-CAG GCG CAG CAC TGG ACT CAG GCG |
| VNTR4 | 362596-362658 | 9 | 7 | 5′-Hex-GTT ACG TAT TCA TAT ACT GAT CAA G | 5′-CGC GGC AAA CGT GTT CCC GAA TTG |
| VNTR5 | 583072-583190 | 9 | 13 | 5′-Hex-GAC AAT AGC GAT GAT ACG GCT AGC G | 5′-AAG TAC CAT TAT GTG ACA GCG |
| VNTR9 | 1339400-1339559 | 6 | 25 | 5′-Hex-TCG CGA GTC TGA CAA GAC TGA TG | 5′-CAC TTC TCA AAT TGG TTA GGC AGA C |
| VNTR10 | 1966577-1966679 | 15 | 7 | 5′-Hex-ATC ATG ACG ATC AAA CTC ACC C | 5′-GAT CAC GAC TCT GAA GAA GAG C |
| VNTR12 | 2625703-2625879 | 18 | 9 | 5′-Hex-GTC GAA GCC ATC ATC AGC TTC C | 5′-TGA AGC AGC CGG CTT TGA AGG C |
| VNTR13 | 2628427-2628848 | 24 | 18 | 5′-Hex-CCG ACT CAT CGG CTG ACT ACT TG | 5′-ATC CAG GTG CTA AGC CTT CGA C |
| VNTR14 | 2760423-2760527 | 6 | 18 | 5′-Hex-TCA TCT GGG TCG TCA TCA CTG TC | 5′-GAG GCA CCA CGT CGT AAG AAG C |
Based on the genome sequence of strain DN-114 001.
Ribotyping.
The automated-device RiboPrinter microbial characterization system (Qualicon, Wilmington, DE) was used for ribotyping. Standard reagents were used in all steps of the analysis. The methods involves the release of DNA, EcoRI digestion of chromosomal DNA, separation of the resulting fragments by agarose gel electrophoresis, and transfer onto a nitrocellulose membrane, followed by hybridization using as a probe the rrnB rRNA operon sequence from Escherichia coli (50).
Amplification and sequencing of the CRISPR locus.
CRISPRs are a family of DNA direct repeats found in many prokaryotic genomes (27). The CRISPR region was amplified with primers 482F (5′-CCAGGGTTCAAATAAGTTATTAATCGC-3′) and 483R (5′-TTTAAGTGCCAGAGACTTTTCGTCGG-3′), which targeted the region flanking the unique CRISPR locus found in the genome of strain DN-114 001. PCR amplification conditions were as follows: initial denaturation at 94°C for 2 min; 30 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 30s; and a final extension at 72°C for 5 min. Nucleotide sequencing was performed from the two ends using the PCR primers.
Amplification and sequencing of SNRs.
SNRs are short sequence stretches of the same nucleotide base. Four loci with mononucleotide repeats of 9 or 10 nucleotides were selected from the genome sequence of strain DN-114 001 (unpublished), and PCR primers flanking the SNR were designed. Two loci were successfully amplified and sequenced with primers SNR2F (5′-GTG TTG CTA ATT GCA TCG TCA CG) and SNR2R (5′-TTC ACG ATG GTC GGC TTG TCT GG) and primers SNR4F (5′-GGA CTG CCA TCA ACA CTG TCG) and SNR4R (5′-CCA TAT CGC ACG ATG ACA TGG). Locus SNR2 is located on position 2453190, whereas SNR4 is located at position 3063640, on the genome sequence of strain DN-114 001. Both loci are intergenic.
Data analysis.
For each MLST locus, an allele number was given to each distinct sequence variant, and a distinct sequence type (ST) number was attributed to each distinct combination of alleles at the seven genes. Minimum spanning tree analysis was performed using the software program BioNumerics v4.6 (Applied-Maths, Sint Maartens-Latem, Belgium). Neighbor-joining tree analysis was performed using MEGA v3.1 (30) or SplitsTree v4b06 (26). Recombination tests were performed using RDP2 (36). Nucleotide diversity was calculated using DNAsp v4 (44). To test for phylogenetic congruence among the genes, the 31 distinct STs were used. Neighbor-joining trees were generated using PAUP* v4 (http://paup.csit.fsu.edu/index.html) for each gene individually and for the concatenated sequence of the eight genes. Using the method of Feil et al. (16), for each gene, the differences in log likelihood were computed, using PAUP* software, between the tree for that gene and the trees constructed using the other genes, with branch lengths optimized. These differences were compared to those obtained for 100 randomly generated trees. The recombination rate during the diversification of clonal lineages was computed according to the previously described principle (18, 23) that considers, between single-locus variants, allelic changes with more than one nucleotide change as resulting from recombination, whereas changes with a single nucleotide difference are considered to result from mutation. Possible recombination events introducing a single nucleotide change were excluded. The relative contributions of recombination on allelic and nucleotide changes were computed according to the above-described principle using the program MultiLocus Analyzer (S. Brisse, unpublished).
For MLVA data, each allele was coded using the deduced number of repeats. Each unique allelic combination of the repeat numbers at the nine VNTR loci was considered as a new repeat type (RT). Negative amplifications (see Results) were not taken into account for pairwise profile comparison. Hence, VNTR profiles with one or more missing loci were associated with the profile composed of the same values at the other loci. To keep track of the missing information, we coded these profiles with a delta suffix followed by the locus number that was missing (for example, RT1Δ9 corresponds to RT1 except that locus VNTR-9 was not amplified). The relatedness between the different STs or RTs was investigated using BioNumerics software by the minimum spanning tree method (48).
Nucleotide sequence accession numbers.
The rplB sequences generated in this study are available from the GenBank/EMBL databases under the accession numbers AM502819 to AM502835. Sequence data for the other genes are available through our MLST web site (www.pasteur.fr/mlst) and were also deposited in GenBank/EMBL, under the accession numbers EU030989 to EU031043.
RESULTS AND DISCUSSION
Confirmation of identification using rplB gene sequencing.
The genus Lactobacillus is the largest group among the Lactobacteriaceae and contains more than 100 species (11). The species Lactobacillus zeae, Lactobacillus rhamnosus, Lactobacillus casei, and Lactobacillus paracasei are phylogenetically and phenotypically closely related and are regarded together as the L. casei group. The taxonomic status of these species and identification of strains as these species are still matters of debate (13, 14), mostly because the type strain of L. casei, strain ATCC 393, is divergent compared to most L. casei/L. paracasei strains and appears related to L. zeae. We will hereafter follow the common practice that uses L. casei to refer to strains that are regarded as either L. casei or L. paracasei, including the reference strain ATCC 334, and will not designate with the L. casei name the strains of the L. zeae-ATCC 393 cluster.
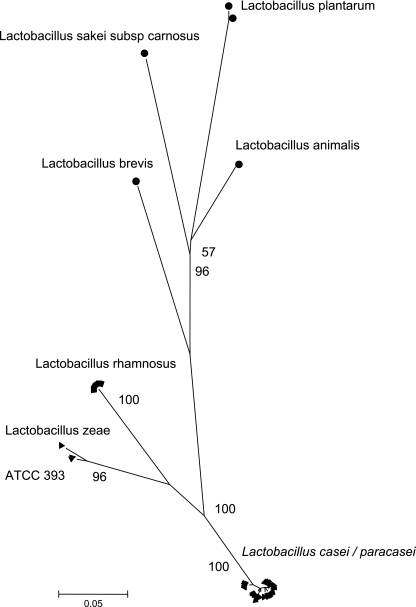
In order to ensure that the study strains belonged to L. casei, we gathered reference and type strains of Lactobacillus plantarum, Lactobacillus brevis, Lactobacillus casei, Lactobacillus zeae, Lactobacillus animalis, Lactobacillus sakei subsp. carnosus, and Lactobacillus rhamnosus. The gene rplB was amplified in all study strains and all of these species, whereas the seven other genes could not be amplified in some of these species. Therefore, the rplB gene sequence was chosen to compare the study strains with the reference/type strains of the other species. Excluding insertion and deletion events (in which two adjacent codons are implicated), a total of 184 (50.3%) polymorphic sites were found based on the alignment of 366 bp. Phylogenetic analysis of the sequence data showed that the 52 study strains had an rplB gene sequence that was very similar to that of L. casei reference strain CIP107868 (= ATCC 334), proposed as the neotype strain for L. casei (10, 13), and clearly distinct from rplB sequences obtained for other species, including the compact cluster formed by two L. zeae strains (including the type strain of this species) and the current taxonomic type strain of L. casei, CIP103137T (= ATCC 393T) (Fig. 1). These results, which support the exceptional position of the current type strain of L. casei, are concordant with those based on sequence analysis of the gene recA (20), gene tuf (6, 56) and ribotyping (52). Concordance between these markers renders it very unlikely that the atypical phylogenetic positioning of strain CIP103137T in single gene-based phylogenies is due to horizontal gene transfer of any of these three protein-coding genes. The gene rplB hence appears as a reliable phylogenetic marker for strains of the L. casei group and could be used in conjunction with tuf and recA for multilocus sequence analysis-based species delineation (24) in this group (6, 20, 56). Strain BL23, which is considered to be a plasmid-cured derivative of the type strain ATCC 393T, clustered within the L. casei rplB cluster. It thus appears distantly related from its supposedly ancestral strain, an observation that confirms previous reports that BL23 is not directly related to ATCC 393T (1).
FIG. 1.
rplB gene-based phylogenetic analysis using the neighbor-joining method and based on a Jukes-Cantor distance matrix. Bootstrap values obtained after 1,000 replicates are given at the nodes. The 52 L. casei (L. paracasei) study strains clearly grouped into a compact cluster together with L. casei strain CIP107868 (= ATCC 334). This cluster was clearly distinct from L. rhamnosus and from a cluster with L. zeae and CIP103137T (= ATCC 393T), as well as from all other included reference or type strains.
We also noted that based on the rplB sequence, all study strains initially referred to as L. paracasei were not separated from L. casei strains (Fig. 1), which supports previous findings. The maximal nucleotide divergence observed between two study strains was 1.4%. We conclude that our study strains can all be considered to belong to a single genomic species, which we refer to as L. casei.
Nucleotide variation.
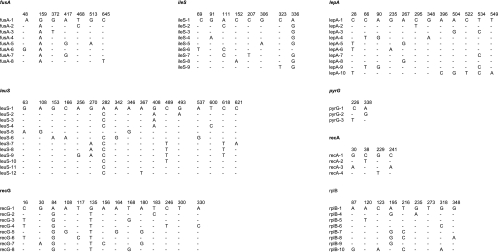
The sequences of the 8 loci were determined for the 52 study strains. Consensus sequence templates ranged in length from 315 bp (recA) to 663 bp (fusA). The proportion of variable sites ranged from 0.58% (pyrG) to 4.09% (recG). Polymorphic sites are given on Fig. 2. Nonsynonymous substitutions per nonsynonymous site were relatively rare compared to synonymous changes per synonymous site (Table 4), indicating selection against amino acid changes and excluding strong positive selection on the observed allelic diversity, as is typically observed for housekeeping genes. The GC percentage observed in all alleles of the eight genes ranged between 47 and 50, thus being close to the GC percent value (46.6) of the complete genome of strain ATCC 334 (35).
FIG. 2.
Polymorphic nucleotide sites found among the 52 L. casei study strains at the seven MLST genes and at gene rplB. For each gene, all discovered alleles are compared, and only polymorphic sites are shown. Numbering starts at the beginning of the aligned sequence portion of each gene.
TABLE 4.
Allelic variation in seven housekeeping genesa
| Gene | Size (bp) of analyzed fragment | Coordinate of first template position on coding sequence | No. of alleles | No. of polymorphic sites | No. of synonymous changes | No. of non- synonymous changes | dS | dN | dN/dS | π (population) | π (alleles) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| fusA | 663 | 97 | 8 | 7 | 7 | 0 | 0.00652 | 0 | 0 | 0.00195 | 0.00291 |
| ileS | 360 | 703 | 9 | 8 | 5 | 3 | 0.00941 | 0.00043 | 0.046 | 0.00336 | 0.00617 |
| lepA | 549 | 274 | 10 | 12 | 9 | 3 | 0.00878 | 0.00023 | 0.026 | 0.00343 | 0.00660 |
| leuS | 642 | 547 | 12 | 17 | 11 | 6 | 0.01024 | 0.00062 | 0.061 | 0.00420 | 0.00625 |
| pyrG | 345 | 70 | 3 | 2 | 1 | 1 | 0.00031 | 0.00009 | 0.290 | 0.00022 | 0.00386 |
| recA | 315 | 265 | 4 | 4 | 2 | 2 | 0.00063 | 0.0002 | 0.317 | 0.00049 | 0.00635 |
| recG | 342 | 988 | 9 | 14 | 13 | 1 | 0.0226 | 0.00018 | 0.008 | 0.00763 | 0.0109 |
dS, number of synonymous changes per synonymous site; dN, number of nonsynonymous changes per nonsynonymous site; π, nucleotide diversity.
Considering the 3,582 nucleotides of the 8 gene portions together, there were 73 variable sites (Fig. 2). The nucleotide diversity (average number of nucleotide differences per site between two randomly selected strains) ranged from 0.00022 (pyrG) to 0.0076 (recG). When considering only the distinct alleles, the diversity indices ranged from 0.0038 (pyrG) to 0.0109 (recG). By comparison, allelic diversity values obtained for Mycobacterium prototuberculosis (22) ranged from 0.00552 to 0.020, whereas Escherichia coli alleles (57) show diversity indices ranging from 0.015 (purA) to 0.038 (fumC). Higher values are obtained for most species for which MLST data are available. Therefore, our L. casei strain sample appears to encompass small amounts of nucleotide diversity, although it clearly is more diverse than archetypal homogeneous bacterial groups such as M. tuberculosis or Bacillus anthracis, for which MLST shows little or no strain discrimination.
MLST scheme optimization for future use.
The number of alleles per locus ranged from 3 (pyrG) to 12 (leuS). By combining the 8 gene loci, 31 STs were distinguished. MLST schemes generally include only five to seven gene loci, because adding more genes often does not increase the number of STs that are distinguished. We intended to optimize our MLST scheme by limiting the number of genes (for practical reasons) while retaining the highest number of STs. When rplB was removed from the analysis, the same 31 STs were found based on the 7 remaining genes. In contrast, when either pyrG or recA (the two genes with the lowest number of alleles) was removed, the number of STs decreased to 30. Therefore, we decided to eliminate rplB from our MLST scheme. Another reason to do so was that rplB and fusA are adjacent on the chromosome of strains DN-114 001 (start nucleotides are, respectively, at positions 2673252 and 2678996; see Table 2) and may therefore be horizontally transferred by a single recombination event, which would introduce bias in recombination rate estimations, for example, when using the clonal diversification method (see below).
Strain relationships based on allelic profiles.
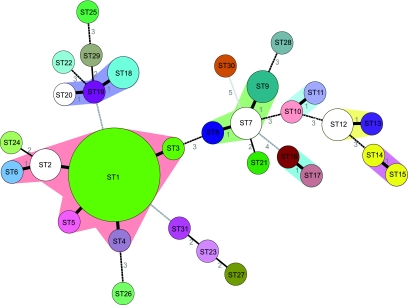
To explore the relationships among the 52 study strains, allelic profile-based phylogenetic analysis was performed using the minimum spanning tree algorithm (Fig. 3), which links profiles so that the sum of the distances (number of distinct alleles between two profiles) is minimized (48). In this representation, strains of the same allelic profile fall in the same circle, the size of which is proportional to the number of strains with that particular profile. Similar to eBURST (17), this approach is less sensitive than nucleotide-based approaches to the disturbing effect of genetic recombination on phylogenetic reconstruction. Figure 3 illustrates that genotype ST1 was dominant in number among our 52 study strains, with 17 (33%) strains in total. Notably, L. casei strain DN-114 001 belonged to ST1. Strain BL23 also belonged to the ST1 cluster, and this affiliation was confirmed based on AFLP (see below), further confirming that it is not a direct derivative of ATCC 393T. Other STs with more than one strain were ST2, ST7, ST9, ST12, and ST18 (Fig. 3; Table 1).
FIG. 3.
Minimum spanning tree analysis of the 52 L. casei strains based on allelic profiles at the seven genes fusA, ileS, lepA, leuS, pyrG, recA, and recG. Each circle corresponds to a sequence type, and the size of the circle is related to the number of strains found with that profile. In order to illustrate the concordance of MLST data with AFLP data, circles were colored based on AFLP types. Each distinct AFLP type corresponds to a distinct color. The color white corresponds to the absence of AFLP data. Colored zones between some groups of circles indicate that these profiles belong to the same clonal complex. Numbers between the circles indicate the number of allelic differences between the profiles. The strength of the link (bold, plain, or discontinuous) is related to the genetic similarity (number of common alleles) between profiles.
Clonal complexes (CCs) (clonal families) can be defined as groups of profiles differing by no more than one gene from at least one other profile of the group (15). Indeed, STs differing by a single gene difference out of seven are very likely to share a common ancestry. Seven CCs were identified (Fig. 3). CC1 was the most common CC, representing 23 study strains. CC1 was composed of ST1 to ST6 and comprised human strains, strains isolated from milk products, and one strain isolated from beer (ST4). Interestingly, ST1 had four single-locus variants (other STs with a single gene difference), suggesting that ST1 is the founder of the group composed of ST1 to -6 (15). The ancestral status of ST1 is consistent with this ST being the most frequent in the population, as a higher frequency increases the likelihood that single locus variants will arise from a given ST (15). ST24 appeared to be closely related to CC1, since it differed from ST2 by only two loci. All other clonal complexes comprised only two or three STs, with a limited number of strains (Fig. 3). The complete genome reference strain ATCC 334 (35) was a single-locus variant of ST25, differing from strain D671 (ST25) by a single nucleotide change in the gene ileS.
Comparison of MLST with AFLP data.
Comparison of MLST data with AFLP data showed very high agreement (see the color pattern in Fig. 3). All strains with different STs had a different AFLP type, with two exceptions. First, strain D659 (ST3), which had the same AFLP type as strains of ST1, differed from ST1 strains at the locus ileS, with two nucleotide changes in this gene between the two STs. Likewise, strains D640 (ST14) and D641 (ST15) differed at the locus recG by five nucleotides, most probably caused by a single recombination event. Therefore, it appears that in both cases, the MLST profile evolved by a single locus change from a common ancestral strain while the AFLP profile remained identical. Concordance between MLST and AFLP was further evident in that most strains of a given ST had the same AFLP type. In particular, this was the case for all strains of ST1, which were all undistinguishable by AFLP. The discriminatory powers found herein for MLST and AFLP were very similar, an observation that is concordant with comparisons of these two methods when used with other bacterial species.
Evidence for homologous recombination in L. casei.
Bacterial species differ widely in their rates of homologous recombination (19). High rates of recombination accelerate the speed of genome diversification, hence affecting the interpretation of genomic differences that are observed among strains. In addition, recombination reduces the linkage between a given genomic background and individual genes, including those possibly involved, e.g., in probiotic characteristics. Because housekeeping genes are unlikely to be positively selected for variation, detection of recombination in these genes would provide an indication that recombination is relatively frequent in the population (19).
Homologous recombination introduces conflict among nucleotide sites in sequence data, which can be visually detected using split decomposition analysis and representing the conflicting relationships among sequences by a network rather than a tree (25). The concatenated sequence of the seven MLST genes did not show a network-like structure (see Fig. S1A in the supplemental material), suggesting that overall the seven gene portions are compatible among themselves, which excludes widespread associative recombination among genes. Accordingly, the phylogeny obtained using the neighbor-joining method (see Fig. S1B in the supplemental material) was very similar to the split network.
A method based on likelihood analysis has been proposed to evaluate the long-term consequences of recombination for the congruence of gene genealogies derived from independent genes (16). We found a significant congruence with the concatenated sequence of the eight genes (including rplB) for the gene trees derived from lepA, leuS, recG, rplB, ileS, and fusA (see Fig. S4, last panel, in the supplemental material). In addition, all of these genes but fusA were congruent with each other (see Fig. S4, first six panels, in the supplemental material). For fusA, the lack of congruence with the other genes could be explained by the presence of only one phylogenetically informative (i.e., a polymorphism present in at least two sequences) site (Fig. 2), which does not provide enough phylogenetic signal. For pyrG and recA, there was not a single phylogenetically informative site (Fig. 2), and for these two genes the obtained phylogenies were not statistically different from the random phylogenies (see Fig. S4 in the supplemental material). Overall, the above results indicate that recombination is not frequent enough to disrupt the phylogenetic signal, but they do not exclude low rates of recombination.
When the nucleotide sequences of individual genes were analyzed (see Fig. S2 in the supplemental material), they showed no or few network-like relationships (splits), except for lepA. Visual inspection of the nucleotide polymorphisms confirmed the existence of a number of conflicting partitions among some sites in lepA sequences. For example, site 28 partitions alleles 5, 6, and 10 versus all other alleles, whereas site 267 would group allele 5 with allele 8. Accordingly, lepA-5 appears to be related either to lepA-8 or to the node leading to lepA-6 (with the lepA-6 branch being explained by a singleton polymorphism, A at position 235).
Although recombination can introduce conflicts between sites, other explanations are possible, for example, homoplasy (reversion or parallel mutations in independent lineages). One more direct way to detect intragenic recombination is the observation of clustered distribution of polymorphisms along the sequence length (mosaic structure). In the case of lepA-10, we observed five differences from lepA-1 between positions 396 and 549 but none between positions 29 and 395 (Fig. 2). The existence of intragenic recombination among lepA alleles was also suggested by a positive Sawyer's test (P = 0.009 using uncondensed fragments) and the chi-square intragenic recombination test (P < 0.01). Overall, these results suggest that homologous recombination occurs in L. casei but at a rate that is not sufficiently high to eliminate most of the phylogenetic signal contained in the nucleotide sequences.
Although the above-used methods can detect recombination, they do not provide a direct estimate of the relative contributions of recombination and mutation in the evolution of strains. A way to quantify the recent contribution of recombination to the generation of genotypic diversity is the clonal diversification method (18, 23). In this method, for each pair of profiles that differ by only one gene (out of seven) along the evolutionary tree, the number of nucleotide changes between the two alleles that differ is counted. A single nucleotide difference is considered to be likely caused by mutation, whereas more than one mutation is considered to derive from recombination. Out of 13 allelic changes, 8 could be attributed to recombination, representing 21 nucleotide changes in total (see Table S2 in the supplemental material). Therefore, nucleotides are approximately four times (21/5) more likely to change by recombination than by mutation in this data set. This result is comparable to the relative contributions of recombination and mutation in E. coli: when the 527 STs available on the E. coli MLST web site (http://web.mpiib-berlin.mpg.de/mlst/) were considered, allelic changes observed within the 81 clonal complexes were found to be caused by recombination 0.85 times as frequently as by mutation, with nucleotides being 5.18 times more likely to change by recombination than mutation. The species E. coli can be considered weakly clonal, with statistical congruence among gene phylogenies but with evidence for localized recombination and gene mosaicism (16, 23, 57). As for E. coli, the apparently paradoxical observation for L. casei of both recombination and phylogenetic congruence among genes can be reconciled by the low rate of recombination or by ecological structuration of the natural populations (16, 23).
Characterization of VNTR loci.
CC1 represented a high proportion of strains in our collection. Therefore, we were interested in identifying genetic markers that would allow discrimination among members of this complex and in particular for members of ST1. MLVA has been shown to be a powerful method for subtyping very closely related strains that are not distinguished by MLST (31). This method is based on tandem repeat copy number differences between strains at well-defined loci. These differences result in locus size variation, which can be detected efficiently by PCR amplification using locus-specific primers targeting the regions that flank the tandem repeats.
After in silico identification of tandem repeats in the genome of strain DN-114 001 (see Materials and Methods) and PCR amplification testing on a small strain set, we selected nine loci that gave the best results. The number of repeats at the 9 selected VNTR loci was determined for 40 strains from the Danone Research collection (see Table S1 in the supplemental material). Despite repeated PCR amplification trials, only two loci (VNTR-10 and VNTR-14) could be amplified for all strains. However, VNTR-14 was monomorphic, and VNTR-10 showed a distinct allele only in two strains. These two loci therefore appear to be highly stable in the population of L. casei, both with respect to copy number and flanking sequences used for PCR priming. At the other loci, the number of PCR-negative strains ranged from 2 (for VNTR-2 and VNTR-3) to 26 (for VNTR-9). Notably, there was an obvious association between PCR failure and MLST data (see Table S1 in the supplemental material). For example, ST14 and ST15, which belong to the same MLST clonal complex, were PCR negative for loci VNTR-4, VNTR-9, and VNTR-12. Failure to amplify VNTR loci in strains with a specific phylogenetic background has been observed for other species (31, 33) and can most likely be attributed to sequence variation at the priming sites or absence of the locus in specific phylogenetic lineages.
The number of alleles ranged from 1 (VNTR-14) to five (VNTR-4). Sequencing of the distinct alleles confirmed that size variation was due to repeat number variation (not shown). Not considering negative amplification as a distinctive characteristic, only 14 unique MLVA types were identified among the 55 strains tested. By comparison, 23 STs were distinguished among these strains. Only one pair of strains of the same ST (ST18) had distinct MLVA profiles, which differed only by a single repeat difference at VNTR-4. All analyzed strains of ST1 had the same MLVA pattern. We conclude that MLVA based on the nine tested loci is less powerful than MLST in discriminating L. casei strains.
Despite this lower degree of variation, the observed concordance of MLVA data with MLST data was very high. In all cases, strains with distinct STs were distinct by MLVA, except for the highly related ST1, ST2, and ST3 (see Fig. S3 in the supplemental material).
Ribotyping, CRISPR locus variation, and SNR variation of CC1 strains.
We attempted to find genetic differences between strains of CC1 by three other methods. First, five ST1 strains (including DN 114-001), one ST2 strain (D6.1), and one ST18 strain (D697) were tested by ribotyping. The banding patterns of these six ST1 and ST2 strains were totally identical, since the same six DNA fragments were observed. Therefore, in contrast to MLST, ribotyping using EcoRI could not discriminate between DN-114 001 and D6.1. By comparison, the ribotype pattern of strain D697 (ST18) showed two clearly distinct bands (data not shown), in agreement with the more distant relationship of this strain based on MLST.
We next explored the possible existence of sequence or spacer content variation at one CRISPR locus that we identified (at positions 2393826 to 2394728) in the genome sequence of strain DN-114 001. The CRISPR locus is widely distributed in prokaryotes (27) and is constituted by an array of short (21 to 47 bp in the currently described CRISPR loci) conserved nucleotide stretches interleaved with nonrepeating spacers of similar size. In Mycobacterium tuberculosis, the CRISPR locus is called the DR locus and is the basis of spoligotyping (28). Although M. tuberculosis is highly homogeneous based on nucleotide sequencing of protein-coding genes, M. tuberculosis strains show extensive spacer content variation at CRISPR (5). Hence, spacer content variation can be used for strain subtyping, although this method has as yet been used only on a limited number of bacterial groups (4, 7, 42, 47). Four strains of CC1 (D657, D658, and D573 of ST1 and D6.1 of ST2) were selected, together with three strains of ST19, ST29, and ST30 used for comparison. PCR amplification of the entire CRISPR locus was successful for the four CC1 strains. However, sequence determination of 1,400 bp from the two extremities of the CRISPR locus did not reveal a single nucleotide difference between the four strains. Therefore, we did not consider this method promising for strain discrimination in CC1. PCR amplification failed with the three other strains tested, and we also noted that our primers do not match with the CRISPR locus in strain ATCC 334.
SNRs are short sequence stretches of the same nucleotide base at defined genome positions. Some of these mononucleotide stretches were shown to be highly polymorphic, including among strains with the same VNTR type (21, 51, 55). We selected 4 SNR loci with >9 repetitions of the same base (either T or A) in the DN-114 001 genome sequence. Two of these loci were successfully amplified by PCR and sequenced for the seven strains that were tested for CRISPR variation. SNR variation was found at both loci, in the form of a single nucleotide insertion observed in CC1 strains compared to the three non-CC1 strains. In addition, just upstream of each SNR locus, a single nucleotide polymorphism (T/G and A/G) was observed in one non-CC1 strain (ST19 and ST29, respectively). Unfortunately, no variation was found among the CC1 strains tested.
Conclusions.
In conclusion, we developed an MLST scheme for L. casei (L. paracasei) strains and estimated, using 52 strains, some basic population biology parameters of L. casei, including diversity indices and the impact of homologous recombination on the diversification of clones. The present L. casei MLST method is intended to become a common language for strain characterization with L. casei. Our study strains were mostly from food sources, industrial or traditional, creating the possibility that some of these strains were selected several times independently for their appreciated characteristics. Analysis of wider, well-documented strain collections with global strain sampling will precise the population structure of L. casei and could potentially bring interesting information on the history of dairy products and on the genotype-phenotype relationships of strains. To this purpose, we developed an MLST web site for L. casei, publicly available at http://www.pasteur.fr/mlst. The discriminatory power of MLST using the seven proposed genes was very similar to that of AFLP, a more complex, less reproducible, and less portable method. MLST should prove useful for strain collection management or traceability purposes.
We intended, but failed, to develop a complementary typing method that would allow subtyping of strains belonging to the major ST encountered in our strain collection, ST1. The subtyping of strains belonging to ST1 was important to us because they show differences in phenotype (Danone Research, unpublished). Subtyping of this major ST was not achieved using VNTR markers, a result that was surprising given the repeated finding of a large amount of VNTR variation among strains with the same ST in several bacterial species, such as Escherichia coli O157:H7 (32) or Salmonella enterica serotype Typhimurium (33). Similarly, Bacillus anthracis is homogeneous by MLST but shows MLVA variation (29). However, in the case of Enterococcus faecium, a species that is phylogenetically more closely related to lactobacilli based on 16S rRNA gene sequences, MLVA variation was not higher than MLST variation (53), similar to our present finding. These results may indicate an atypical stability of VNTR loci in this group of bacteria.
The other simple methods that were explored for ST1 subtyping (ribotyping, SNR markers, and CRISPR) did not show promise for this purpose. Efforts for further subtyping strategies could be warranted, possibly by undertaking more global mutation discovery approaches, such as whole-genome shotgun sequencing, microarray hybridization-based comparative genome sequencing, or mutation screening in high numbers of selected genes (43).
Supplementary Material
Acknowledgments
We thank Yolande Arnoux for help in nucleic acid extraction. Natalia Bomchil, Stephanie Cools-Portier, and Jean-Michel Faurie are thanked for assistance during the project.
We are grateful to Chantal Bizet and the Collection de l'Institut Pasteur for providing reference and type strains.
Footnotes
Published ahead of print on 17 August 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acedo-Felix, E., and G. Perez-Martinez. 2003. Significant differences between Lactobacillus casei subsp. casei ATCC 393T and a commonly used plasmid-cured derivative revealed by a polyphasic study. Int. J. Syst. Evol. Microbiol. 53:67-75. [DOI] [PubMed] [Google Scholar]
- 2.Achtman, M. 2002. A phylogenetic perspective on molecular epidemiology, p. 485-509. In M. Sussman (ed.), Molecular medical microbiology, vol. 1. Academic Press, London, United Kingdom. [Google Scholar]
- 3.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., B. Quinquis, A. Sorokin, and S. D. Ehrlich. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551-2561. [DOI] [PubMed] [Google Scholar]
- 5.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Guttierez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, M. L. Ho, C. Martin, I. Mokrousov, O. Narvskaia, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. G. Shemyakin, U. B. Singh, A. Somoskovi, R. A. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. M. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavagnat, F., M. Haueter, J. Jimeno, and M. G. Casey. 2002. Comparison of partial tuf gene sequences for the identification of lactobacilli. FEMS Microbiol. Lett. 217:177-183. [DOI] [PubMed] [Google Scholar]
- 7.DeBoy, R. T., E. F. Mongodin, J. B. Emerson, and K. E. Nelson. 2006. Chromosome evolution in the Thermotogales: large-scale inversions and strain diversification of CRISPR sequences. J. Bacteriol. 188:2364-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Las Rivas, B., A. Marcobal, and R. Munoz. 2004. Allelic diversity and population structure in Oenococcus oeni as determined from sequence analysis of housekeeping genes. Appl. Environ. Microbiol. 70:7210-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Las Rivas, B., A. Marcobal, and R. Munoz. 2006. Development of a multilocus sequence typing method for analysis of Lactobacillus plantarum strains. Microbiology 152:85-93. [DOI] [PubMed] [Google Scholar]
- 10.Dellaglio, F., G. E. Felis, and S. Torriani. 2002. The status of the species Lactobacillus casei (Orla-Jensen 1916) Hansen and Lessel 1971 and Lactobacillus paracasei Collins et al. 1989. Request for an opinion. Int. J. Syst. Evol. Microbiol. 52:285-287. [DOI] [PubMed] [Google Scholar]
- 11.Dellaglio, F., and G. E. Felis. 2005. Taxonomy of lactobacilli and bifidobacteria, p. 25-50. In G. W. Tannock (ed.), Probiotics and prebiotics: scientific aspects. Caister Academic Press, Norwich, United Kingdom.
- 12.De Preter, V., T. Vanhoutte, G. Huys, J. Swings, L. De Vuyst, P. Rutgeerts, and K. Verbeke. 2007. Effects of Lactobacillus casei Shirota, Bifidobacterium breve, and oligofructose-enriched inulin on colonic nitrogen-protein metabolism in healthy humans. Am. J. Physiol. Gastrointest. Liver Physiol. 292:G358-G368. [DOI] [PubMed] [Google Scholar]
- 13.Dicks, L. M., E. M. Du Plessis, F. Dellaglio, and E. Lauer. 1996. Reclassification of Lactobacillus casei subsp. casei ATCC 393 and Lactobacillus rhamnosus ATCC 15820 as Lactobacillus zeae nom. rev., designation of ATCC 334 as the neotype of L. casei subsp. casei, and rejection of the name Lactobacillus paracasei. Int. J. Syst. Evol. Microbiol. 46:337-340. [DOI] [PubMed] [Google Scholar]
- 14.Dobson, C. M., B. Chaban, H. Deneer, and B. Ziola. 2004. Lactobacillus casei, Lactobacillus rhamnosus, and Lactobacillus zeae isolates identified by sequence signature and immunoblot phenotype. Can. J. Microbiol. 50:482-488. [DOI] [PubMed] [Google Scholar]
- 15.Feil, E. J. 2004. Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2:483-495. [DOI] [PubMed] [Google Scholar]
- 16.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feil, E. J., M. C. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 19.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 20.Felis, G. E., F. Dellaglio, L. Mizzi, and S. Torriani. 2001. Comparative sequence analysis of a recA gene fragment brings new evidence for a change in the taxonomy of the Lactobacillus casei group. Int. J. Syst. Evol. Microbiol. 51:2113-2117. [DOI] [PubMed] [Google Scholar]
- 21.Gur-Arie, R., C. J. Cohen, Y. Eitan, L. Shelef, E. M. Hallerman, and Y. Kashi. 2000. Simple sequence repeats in Escherichia coli: abundance, distribution, composition, and polymorphism. Genome Res. 10:62-71. [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez, M. C., S. Brisse, R. Brosch, M. Fabre, B. Omais, M. Marmiesse, P. Supply, and V. Vincent. 2005. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathogens 1:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guttman, D. S., and D. E. Dykhuizen. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266:1380-1383. [DOI] [PubMed] [Google Scholar]
- 24.Hanage, W. P., C. Fraser, and B. G. Spratt. 2006. Sequences, sequence clusters and bacterial species. Philos. Trans. R. Soc. Lond. B 361:1917-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 26.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 27.Jansen, R., J. D. Embden, W. Gaastra, and L. M. Schouls. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565-1575. [DOI] [PubMed] [Google Scholar]
- 28.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 31.Lindstedt, B. A. 2005. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 26:2567-2582. [DOI] [PubMed] [Google Scholar]
- 32.Lindstedt, B. A., E. Heir, E. Gjernes, T. Vardund, and G. Kapperud. 2003. DNA fingerprinting of Shiga-toxin producing Escherichia coli O157 based on multiple-locus variable-number tandem-repeats analysis (MLVA). Ann. Clin. Microbiol. Antimicrob. 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindstedt, B. A., T. Vardund, L. Aas, and G. Kapperud. 2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J. Microbiol. Methods 59:163-172. [DOI] [PubMed] [Google Scholar]
- 34.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 37.Mercenier, A., S. Pavan, and B. Pot. 2003. Probiotics as biotherapeutic agents: present knowledge and future prospects. Curr. Pharm. Des. 9:175-191. [DOI] [PubMed] [Google Scholar]
- 38.Ouwehand, A. C., S. Salminen, and E. Isolauri. 2002. Probiotics: an overview of beneficial effects. Antonie Leeuwenhoek 82:279-289. [PubMed] [Google Scholar]
- 39.Pedone, C. A., C. C. Arnaud, E. R. Postaire, C. F. Bouley, and P. Reinert. 2000. Multicentric study of the effect of milk fermented by Lactobacillus casei on the incidence of diarrhoea. Int. J. Clin. Pract. 54:568-571. [PubMed] [Google Scholar]
- 40.Pedone, C. A., A. O. Bernabeu, E. R. Postaire, C. F. Bouley, and P. Reinert. 1999. The effect of supplementation with milk fermented by Lactobacillus casei (strain DN-114 001) on acute diarrhoea in children attending day care centres. Int. J. Clin. Pract. 53:179-184. [PubMed] [Google Scholar]
- 41.Petrovic, T., M. Niksic, and F. Bringel. 2006. Strain typing with ISLpl1 in lactobacilli. FEMS Microbiol. Lett. 255:1-10. [DOI] [PubMed] [Google Scholar]
- 42.Pourcel, C., G. Salvignol, and G. Vergnaud. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653-663. [DOI] [PubMed] [Google Scholar]
- 43.Roumagnac, P., F. X. Weill, C. Dolecek, S. Baker, S. Brisse, N. T. Chinh, T. A. Le, C. J. Acosta, J. Farrar, G. Dougan, and M. Achtman. 2006. Evolutionary history of Salmonella typhi. Science 314:1301-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Santos, S. R., and H. Ochman. 2004. Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ. Microbiol. 6:754-759. [DOI] [PubMed] [Google Scholar]
- 47.Schouls, L. M., S. Reulen, B. Duim, J. A. Wagenaar, R. J. Willems, K. E. Dingle, F. M. Colles, and J. D. Van Embden. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schouls, L. M., H. G. van der Heide, L. Vauterin, P. Vauterin, and F. R. Mooi. 2004. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J. Bacteriol. 186:5496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwenninger, S. M., U. von Ah, B. Niederer, M. Teuber, and L. Meile. 2005. Detection of antifungal properties in Lactobacillus paracasei subsp. paracasei SM20, SM29, and SM63 and molecular typing of the strains. J. Food. Prot. 68:111-119. [DOI] [PubMed] [Google Scholar]
- 50.Severino, P., and S. Brisse. 2005. Ribotyping in clinical microbiology, p. 573-582. In J. M. Walker and R. Rapley (ed.), Medical biomethods handbook. Humana Press Inc., Totowa, NJ.
- 51.Stratilo, C. W., C. T. Lewis, L. Bryden, M. R. Mulvey, and D. Bader. 2006. Single-nucleotide repeat analysis for subtyping Bacillus anthracis isolates. J. Clin. Microbiol. 44:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svec, P., V. Drab, and I. Sedlacek. 2005. Ribotyping of Lactobacillus casei group strains isolated from dairy products. Folia Microbiol. (Praha) 50:223-228. [DOI] [PubMed] [Google Scholar]
- 53.Top, J., L. M. Schouls, M. J. Bonten, and R. J. Willems. 2004. Multiple-locus variable-number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J. Clin. Microbiol. 42:4503-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tynkkynen, S., R. Satokari, M. Saarela, T. Mattila-Sandholm, and M. Saxelin. 1999. Comparison of ribotyping, randomly amplified polymorphic DNA analysis, and pulsed-field gel electrophoresis in typing of Lactobacillus rhamnosus and L. casei strains. Appl. Environ. Microbiol. 65:3908-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ventura, M., C. Canchaya, V. Meylan, T. R. Klaenhammer, and R. Zink. 2003. Analysis, characterization, and loci of the tuf genes in lactobacillus and bifidobacterium species and their direct application for species identification. Appl. Environ. Microbiol. 69:6908-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.