Abstract
The effects of feed supplementation with the approved antimicrobial agents bambermycin, penicillin, salinomycin, and bacitracin or a combination of salinomycin plus bacitracin were evaluated for the incidence and distribution of antibiotic resistance in 197 commensal Escherichia coli isolates from broiler chickens over 35 days. All isolates showed some degree of multiple antibiotic resistance. Resistance to tetracycline (68.5%), amoxicillin (61.4%), ceftiofur (51.3%), spectinomycin (47.2%), and sulfonamides (42%) was most frequent. The levels of resistance to streptomycin, chloramphenicol, and gentamicin were 33.5, 35.5, and 25.3%, respectively. The overall resistance levels decreased from day 7 to day 35 (P < 0.001). Comparing treatments, the levels of resistance to ceftiofur, spectinomycin, and gentamicin (except for resistance to bacitracin treatment) were significantly higher in isolates from chickens receiving feed supplemented with salinomycin than from the other feeds (P < 0.001). Using a DNA microarray analysis capable of detecting commonly found antimicrobial resistance genes, we characterized 104 tetracycline-resistant E. coli isolates from 7- to 28-day-old chickens fed different growth promoters. Results showed a decrease in the incidence of isolates harboring tet(B), blaTEM, sulI, and aadA and class 1 integron from days 7 to 35 (P < 0.01). Of the 84 tetracycline-ceftiofur-resistant E. coli isolates, 76 (90.5%) were positive for blaCMY-2. The proportions of isolates positive for sulI, aadA, and integron class 1 were significantly higher in salinomycin-treated chickens than in the control or other treatment groups (P < 0.05). These data demonstrate that multiantibiotic-resistant E. coli isolates can be found in broiler chickens regardless of the antimicrobial growth promoters used. However, the phenotype and the distribution of resistance determinants in E. coli can be modulated by feed supplementation with some of the antimicrobial agents used in broiler chicken production.
Several classes of antibiotics, including glycolipids (bambermycin), polypeptides (bacitracin), ionophores (salinomycin) and β-lactams (penicillin), are used in broiler chicken production for growth promotion and prevention of infectious diseases (10, 43). Salinomycin and bacitracin are widely used in starter, grower, and finisher feeds for broilers. These antibiotics improve feed conversion and body weight gain presumably by altering the composition and activities of microflora (14, 31). This practice may modify the intestinal flora and create a selective pressure in favor of resistant bacteria (1, 43). In response to the emergence of antibiotic resistance, several European countries have restricted or banned the use of antibiotics as growth promoters (3). According to Apajalahti et al. (4), the identity of only about 10% of the chicken gastrointestinal tract bacteria is known. Little research has been conducted to systematically evaluate the potential effects antibiotics may have on the dynamics of the overall gut microflora of chicken. Much work needs to be done to study the distribution of antibiotic resistance genes among commensal bacteria in chickens fed antimicrobial growth promoter agents.
Escherichia coli is a ubiquitous organism in the chicken gastrointestinal tract and is regarded as a major pathogen of worldwide importance in commercially produced poultry (25, 47). It can cause diseases including colibacillosis and air sacculitis in poultry, resulting in significant economic losses (47). In commercial broiler chicken farms, the rations fed to chickens may legally contain up to three antimicrobial agents. However, the overall distribution of antibiotic resistance determinants among commensal bacteria isolated from healthy chickens fed with such feed is largely unknown. Publications on resistance to therapeutically used antibiotics do not provide a complete picture of the situation (27).
Fairchild et al. (21) showed that the oral administration of tetracycline did not induce significant changes in the chicken cecal bacterial community but that Enterococcus spp. and E. coli showed high tetracycline MICs. Escherichia coli isolates were found to harbor the tetracycline resistance gene tet(A) or tet(B), while Enterococcus isolates were positive for tet(M), tet(L), tet(K), and tet(O), with the latter gene conferring tetracycline resistance in Campylobacter jejuni isolates (21). The authors suggested that complex ecological and genetic factors could contribute to the prevalence and transfer of antibiotic resistance genes in the chicken production environment. Multiply drug-resistant E. coli strains isolated from healthy broiler chickens and humans were found to harbor similar genes encoding tetracycline resistance, suggesting the possibility that chickens may be reservoirs for tetracycline resistance genes (36).
Commensal intestinal bacteria including E. coli are commonly used to monitor resistance to therapeutically valuable antibiotics in food animals and in humans (16). In a previous study, we reported that multiple-antibiotic-resistant commensal E. coli strains carrying virulence and resistance genes can be found in samples from commercial broiler chicken farms and provide a reservoir for these genes in chicken production facilities (17). Such bacteria could later find their way into chicken products and other foods as well as manure, soil, and water. The impact of the agricultural use of antimicrobial agents on human and animal health has been the subject of several reports (3, 10, 47). For public health concerns, it is important to know the changes that occur in the intestinal flora of chickens treated with various antimicrobial growth promoters. Knowledge about the diversity and distribution of antimicrobial resistance determinants in bacteria from the chicken gut and the environment will be useful for understanding the ecology of the gut microflora as well as the epidemiology of antibiotic resistance (8). This study investigated genotypic and phenotypic changes in the intestinal E. coli population of broiler chickens fed with different antimicrobial agents as growth promoters.
MATERIALS AND METHODS
Broiler chickens and treatments.
Studies were performed with 900 1-day-old male broiler chickens. The birds were placed in 18 pens (50 birds per pen) that were assigned at random to six treatments: a control group fed without antibiotics and five groups fed rations containing (per kg of feed) 2 mg bambermycin, 2.2 mg procaine penicillin, 60 mg salinomycin, 4.4 mg bacitracin, and a combination of 3.3 mg bacitracin plus 1.1 mg salinomycin. All of these additives are approved for use in poultry production in Canada, but salinomycin and bacitracin and their combination are among the most popular antibiotics used in British Columbia (Canada). The composition of the feed used in this study is presented in Table 1. The starter, grower, and finisher diets were formulated with wheat, barley, and corn as the principal cereals and soybean and canola meals as protein concentrates to meet the National Research Council nutrient requirements for broiler chickens (39). Analyses of dry matter, total proteins, soluble carbohydrates, fatty acids, and some of the most common minerals were performed at the Centre de Recherche en Sciences Animales de Deschambault (CRSAD, Deschambault, QC, Canada) by the usual laboratory analysis methods. Heat was provided by gas-fired brooders; water was offered through nipple drinkers and feed through tube feeders to allow for ad libitum consumption. The clean and disinfected concrete floor was covered with approximately three inches (7.6 cm) of clean softwood shavings, and the bird density was approximately 0.75 square feet (0.07 m2) per bird, which is the industry standard. Ventilation was provided by negative pressure with fans. Performance traits (body weight, weight gain, feed consumption, and feed efficiency) were measured at days 14, 28, and 35. All experimental procedures performed in this study were approved by the Animal Care Committee of the Pacific Agri-Food Research Center and followed principles described by the Canadian Council on Animal Care (11).
TABLE 1.
Composition of the feed used in this study
| Ingredient/nutrient profile | % of inclusion in diet
|
||
|---|---|---|---|
| Starter (days 0-14) | Grower (days 15-28) | Finisher (days 29-35) | |
| Ingredient | |||
| Wheat | 34.96 | 35.03 | 40.79 |
| Soya | 23 | 0 | 0.51 |
| Barley | 10 | 0 | 0 |
| Canola | 9 | 22 | 18 |
| Canola oil | 8.6 | 7 | 7 |
| Corn | 7 | 25 | 25 |
| Corn gluten | 2.3 | 6 | 4 |
| Limestone | 1.6 | 1.3 | 1.2 |
| Dicalcium phosphatea | 1.6 | 1.5 | 1.4 |
| Vitamin-mineral mixtureb | 1 | 1 | 1 |
| Lysine | 0.4 | 0.71 | 0.63 |
| Avizymec | 0.05 | 0.05 | 0.05 |
| Analyzed nutrientd | |||
| Dry matter | 89 | 88.7 | 88.7 |
| Ash | 6.41 | 5.74 | 5.74 |
| Proteins | 24.8 | 21.3 | 21.3 |
| Fat | 10.6 | 10.4 | 10.4 |
| Glucose | 19.3 | 17.3 | 18.6 |
| Fructose | 22.0 | 22.7 | 24.4 |
| Acid detergent fiber | 7.42 | 7.18 | 7.18 |
| Neutral detergent fiber | 13.35 | 13.72 | 13.72 |
| Ca | 1.0 | 1.0 | 1.0 |
| Mg | 0.2 | 0.2 | 0.2 |
| K | 0.89 | 0.55 | 0.55 |
| P | 0.80 | 0.81 | 0.81 |
| Na | 0.22 | 0.26 | 0.26 |
| Fe | 0.04 | 0.04 | 0.04 |
| Zn | 0.02 | 0.02 | 0.02 |
| Mn | 0.02 | 0.02 | 0.02 |
A mixture of mono- and dicalcium phosphate containing 18% calcium and 21% phosphate.
Amounts supplied per kilogram of diet: vitamin A, 9,000 IU; cholecalciferol, 1,500 IU; vitamin E, 10 IU; vitamin K, 0.5 mg; vitamin B12, 0.007 mg; thiamine, 0.4 mg; riboflavin, 6 mg; folic acid, 1 mg; biotin, 0.15 mg; niacin, 135 mg; pyridoxine, 4 mg; choline chloride, 1,000 mg; dl-methionine, 1,184 mg; ethoxyquine, 125 mg; NaCl, 2 g; manganese sulfate, 60 mg; copper sulfate, 5 mg; selenium (sodium selenium), 0.1 mg; iodine, 0.35 mg; zinc sulfate, 50 mg.
Multienzyme system for wheat-based poultry feed (Halchemix Canada, Inc., Toronto, ON, Canada) containing 5,000 U/g xylanase and 1,600 U/g protease.
The nutrient contents, analyzed on a dry matter basis, were determined at CRESAD.
Sample collection and bacteriological analysis.
Ten chicks (day 0) and two birds per pen were killed by cervical dislocation at each sampling time (7, 14, 21, 28, and 35 days of age). Cecal contents and cloacal samples from the two sacrificed birds were aseptically collected and transferred to peptone buffer in test tubes and sterile “whirl-pack” plastic bags, respectively, for bacteriological culture. The samples were placed on ice and transported to the microbiology laboratory for bacteriological analysis that was carried out on the same day. Sample weights were estimated by subtracting the weight of the container without the sample from the weight with the samples.
Bacteriological analyses were performed with a total of 90 fecal and 90 cecal samples. The generic E. coli population was estimated using E. coli and coliform Petrifilms (3M, St. Paul, MN) as previously described (17, 33). After incubation at 37°C for 24 h, blue-colored, gas-producing colonies were counted as generic E. coli. Results were expressed as CFU per gram of material. At each sampling time, six presumptive E. coli samples from each treatment group (two colonies per pen) were purified on blood agar and confirmed as E. coli by using API20E strips (bioMérieux, St-Laurent, QC, Canada) according to the manufacturer's specifications. Enterococcus populations were determined by spreading 10-fold dilutions of samples on KF streptococcal agar CM0701 (Oxoid, Nepean, ON, Canada) and incubating at 37°C for 48 h (as described by Hayes et al. [29]). Clostridium perfringens cells were enumerated according to the description by Knarreborg et al. (31). Briefly, samples were spread on tryptose sulfite agar (Oxoid) supplemented with cycloserine (SR088E; Oxoid) and incubated under anaerobic conditions for 24 h at 37°C. At the end of the study (day 35), two litter samples were taken from each pen for bacterial analysis as described above.
Determination of antimicrobial susceptibility.
Determination of the antibiotic MICs was performed with all E. coli isolates, using a Sensititre automated system (Trek Diagnostic Systems, Cleveland, OH), according to the Clinical Laboratory Standard Institute's (CLSI [formerly NCCLS]) guidelines with E. coli ATCC 25922 as the control (38). The following antimicrobials were tested on Sensititre Avian plates: amoxicillin, penicillin, ceftiofur, erythromycin, tylosin, clindamycin, spectinomycin, streptomycin, gentamicin, neomycin, oxytetracycline, tetracycline, enrofloxacin, sarafloxacin, novobiocin, sulfadimethoxime, sulfathiazoletrimethoprim-sulfadimethoxazole, and chloramphenicol. In addition, antibiotic resistance profiles were determined for all tetracycline-resistant E. coli isolates using a Sensititre system with National Antimicrobial Resistance Monitoring System (NARMS) plates for gram-negative bacteria. The MIC results were interpreted according to the breakpoints of the CLSI and the 2005 Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS [12]) guidelines.
DNA extraction.
E. coli isolates were grown overnight in 3 ml of beef heart infusion broth (Becton Dickinson, Sparks, MD) at 37°C. Two hundred microliters of this culture was transferred to 1.5-ml centrifuge tubes and centrifuged at 14,000 × g for 2 min. The supernatants were removed, and the bacterial pellets were resuspended in 200 μl of sterile water with vortexing. The suspension was boiled for 10 min and centrifuged as described before, and 150 μl of the supernatant containing DNA was removed for testing. E. coli isolates that were phenotypically resistant to tetracycline and ceftiofur were analyzed by PCR for the presence of the extended-spectrum beta-lactamase-encoding gene blaCMY-2 as previously described (17).
E. coli DNA labeling.
Bacterial DNA was labeled using Bioprime DNA labeling system (Invitrogen Life Technologies, Burlington, ON, Canada). Fifteen microliters of the supernatant containing DNA was added to a final volume of 32.5 μl containing 10 μl of a random primer solution, 0.5 μl of high-concentration DNA polymerase (Klenow fragment, 40 U/μl), 5 μl of a deoxyribonucleoside triphosphate (dNTP) mixture (1.2 mM dATP, 1.2 mM dGTP, 1.2 mM dTTP, and 0.6 mM dCTP in 10 mM Tris [pH 8.0] and 1 mM EDTA), and 2 μl of 1 mM Cy5-dCTP. Labeling reactions were performed in the dark at 37°C for 3.5 h and stopped by the addition of 5 μl Na2EDTA 0.5 M (pH 8.0). The labeled samples were then purified with a PureLink PCR purification kit (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's protocol. The amount of incorporated fluorescent Cy5 dye was then quantified by scanning the DNA sample with a NanoDrop ND-1000 spectrophotometer from 200 to 700 nm. Data were analyzed using a Web-based percent incorporation calculator (http://www.pangloss.com/seidel/Protocols/percent_inc.html).
DNA microarrays.
The antimicrobial resistance determinants in selected tetracycline-resistant isolates were detected using specific probes. The microarray used in this study is based on earlier published work (9) and carries oligonucleotides of 70 bases in length targeting 38 antimicrobial resistance or antimicrobial resistance-related genes. The microarray also carries five positive controls for E. coli derived from the sequences of genes encoding tryptophanase (tnaA), beta-glucuronidase (uidA), lactose permease (lacY), beta-galactosidase (lacZ), and glutamate decarboxylase (gad). Negative controls added to this microarray consist of oligonucleotides derived from the gene sequences for the green fluorescent protein of Aequorea victoria, the lactose permease of Citrobacter freundii, and the chlorophyll synthase from Arabidopsis thaliana.
Hybridization of labeled DNA.
Prehybridization and hybridization were performed as previously described by Hamelin et al. (28), with the following modifications: the habitation was performed using a SlideBooster hybridization workstation (model SB800; Advalytix, Germany), and scanning was performed at a resolution of 5 μm at 95% laser power, using a ScanArray Lite fluorescent microarray analysis system (Perkin-Elmer, Mississauga, ON, Canada). Acquisition of fluorescent spots and quantification of fluorescent spot intensities were performed as described by Hamelin et al. (28).
Statistical analysis.
Data were analyzed according to a randomized complete block design using the GLM procedure of SAS software (33, 45), with the individual pens as experimental units (three pens per treatment group). The association test of Cochran-Mantel-Haenszel and logistic analysis (proportional odds model) were used to determine the relationship among feed supplementation, phenotype, and genotype by using the FREQ procedure of SAS Institute (45). Associations between resistance genes and class 1 integrons were determined using Pearson's chi-square exact test (35). The P value of 0.05 was used to declare significance.
RESULTS
Broiler performance.
The effects of diet supplementation with bambermycin, penicillin, salinomycin, bacitracin, and salinomycin plus bacitracin on body weight, feed intake, feed efficiency, and mortality are presented in Table 2. No significant differences were noted between the treatment groups for body weight and feed intake (P > 0.05). Although bambermycin and penicillin increased the body weight from days 15 to 28, these increases were not statistically significant (P = 0.09). From days 0 to 14 and from days 15 to 28, penicillin improved feed efficiency (with reduced feed intake required to achieve a kilogram of gain). This improvement in feed efficiency with penicillin was significant on a cumulative response from days 0 to 35 (P < 0.05). No significant differences were observed for the cumulative mortality rate (P > 0.05), with 12% and 5% mortality recorded for the salinomycin-treated and the untreated control group, respectively, but less than 5% in the group receiving salinomycin-plus-bacitracin treatment.
TABLE 2.
Performance of broiler chickens fed diets containing antimicrobialsa
| Parameter | Period (days) | Value for control | Value for treatment with:
|
SEM | P valueb | ||||
|---|---|---|---|---|---|---|---|---|---|
| BBM | PEN | SAL | BAC | SAL + BAC | |||||
| Body wt (g) | Initial | 40.69 | 40.76 | 40.64 | 40.71 | 40.67 | 40.40 | 0.232 | 0.91 |
| 0-14 | 457.31 | 459.89 | 462.87 | 441.32 | 451.67 | 458.77 | 7.224 | 0.38 | |
| 15-28 | 1,252.02 | 1,265.29 | 1,267.10 | 1,209.64 | 1,224.63 | 1,255.29 | 14.605 | 0.09 | |
| 29-35 | 1,815.09 | 1,811.42 | 1,805.12 | 1,783.69 | 1,776.75 | 1,825.11 | 27.805 | 0.80 | |
| Feed intake (g) | 0-14 | 365.90 | 373.21 | 363.39 | 361.82 | 360.72 | 368.93 | 6.272 | 0.72 |
| 15-28 | 896.23 | 906.58 | 868.70 | 861.47 | 870.33 | 901.47 | 15.969 | 0.27 | |
| 29-35 | 1,288.22 | 1,280.62 | 1,223.70 | 1,263.36 | 1,264.54 | 1,329.18 | 23.142 | 0.12 | |
| 0-35 | 3,215.03 | 3,237.87 | 3,084.20 | 3,118.83 | 3,125.57 | 3,239.98 | 47.916 | 0.14 | |
| Feed efficiency | 0-14 | 1.21 | 1.22 | 1.17 | 1.24 | 1.21 | 1.20 | 0.014 | 0.09 |
| (g of feed/g body wt gain) | 15-28 | 1.78 | 1.77 | 1.69 | 1.76 | 1.76 | 1.77 | 0.018 | 0.06 |
| 29-35 | 2.29 | 2.35 | 2.28 | 2.17 | 2.30 | 2.34 | 0.060 | 0.37 | |
| 0-35 | 1.78 | 1.79 | 1.72 | 1.75 | 1.77 | 1.78 | 0.015 | 0.04* | |
| Total mortality (%) | 0-35 | 5.48 | 12.64 | 7.12 | 12.77 | 7.65 | 4.47 | 2.682 | 0.26 |
Values indicate performance parameters of broiler chickens fed diets containing bambermycin (BBM), penicillin (PEN), salinomycin (SAL), bacitracin (BAC), and a salinomycin-bacitracin combination (SAL + BAC) at concentrations specified in Materials and Methods.
P values were obtained by analysis of variance. *, values are statistically different (P < 0.05).
Bacteriological analyses.
Cloacal, cecal, and litter samples were collected as described in Materials and Methods and analyzed in order to determine the concentrations of generic E. coli, C. perfringens, and Enterococcus (Table 3). The concentrations of each bacterial group were higher in the cecal samples than in the cloacal samples (P > 0.05). At day 35, counts of C. perfringens cells in cecal and cloacal samples were significantly higher than those obtained at day 7 (P < 0.05), although counts of this bacterium remained lower than those of E. coli or Enterococcus. The numbers of these two bacteria (E. coli and Enterococcus) were highest on day 7 and slowly declined thereafter. At day 35, analysis of litter samples showed higher E. coli and Enterococcus concentrations compared to the cloacal samples. There was no significant effect of any treatment on bacterial counts in any sample (cloacal, cecal, or litter; P > 0.05). One hundred ninety-seven presumptive E. coli isolates were obtained (32 from the salinomycin-plus-bacitracin treatment group and 33 from each of the five other treatment groups). Antibiotic susceptibility was determined for these 197 isolates.
TABLE 3.
C. perfringens, Enterococcus, and E. coli counts obtained from cecal, cloacal, and litter samplesa
| Bacterium | Day | Sample | Mean CFU/g of sample in treatment
|
SEM | P valueb | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | BBM | PEN | SAL | BAC | SAL + BAC | |||||
| C. perfringens | 7 | CE | 2.15 | 2.22 | 2.93 | 0.62 | 1.86 | 1.76 | 0.530 | 0.39 |
| CL | 1.30 | 0.57 | 1.23 | 1.16 | 1.20 | 1.36 | ||||
| 14 | CE | 2.27 | 0.82 | 1.86 | 2.57 | 1.55 | 1.71 | 0.462 | 0.286 | |
| CL | 1.22 | 0.92 | 0.00 | 0.83 | 0.88 | 0.54 | ||||
| 21 | CE | 2.45 | 2.54 | 2.54 | 2.54 | 2.00 | 2.54 | 0.306 | 0.127 | |
| CL | 1.43 | 2.34 | 1.15 | 2.29 | 2.55 | 2.67 | ||||
| 28 | CE | 3.23 | 2.53 | 2.76 | 3.28 | 3.11 | 3.04 | 0.512 | 0.701 | |
| CL | 2.54 | 2.30 | 1.56 | 1.38 | 2.44 | 2.28 | ||||
| 35 | CE | 3.95 | 3.55 | 3.66 | 3.29 | 3.76 | 3.28 | 0.341 | 0.831 | |
| CL | 2.65 | 2.54 | 2.54 | 2.78 | 2.95 | 2.67 | ||||
| Enterococcus spp. | 7 | CE | 9.41 | 9.55 | 8.06 | 7.63 | 7.51 | 8.40 | 0.782 | 0.7047 |
| CL | 5.73 | 5.73 | 5.73 | 5.73 | 5.73 | 5.73 | 0.958 | |||
| 14 | CE | 6.95 | 4.90 | 4.92 | 6.26 | 5.93 | 6.55 | 0.683 | 0.217 | |
| CL | 4.76 | 5.78 | 4.45 | 6.62 | 5.39 | 6.04 | 0.837 | |||
| 21 | CE | 6.50 | 6.52 | 5.66 | 5.68 | 5.81 | 5.23 | 0.512 | 0.173 | |
| CL | 5.87 | 5.40 | 5.49 | 5.20 | 5.50 | 4.20 | 0.627 | |||
| 28 | CE | 6.09 | 5.28 | 5.73 | 4.73 | 5.16 | 5.22 | 0.472 | 0.072 | |
| CL | 4.47 | 5.68 | 3.17 | 3.41 | 4.59 | 4.20 | 0.578 | |||
| 35 | CE | 5.23 | 5.76 | 5.60 | 5.46 | 5.74 | 5.03 | 0.400 | 0.115 | |
| CL | 6.61 | 7.06 | 4.87 | 5.99 | 6.93 | 6.48 | 0.490 | |||
| Litter | 7.86 | 7.45 | 7.60 | 7.28 | 7.80 | 7.47 | 0.28 | 0.671 | ||
| E. coli | 7 | CE | 10.50 | 9.90 | 9.62 | 10.09 | 9.86 | 10.39 | 0.357 | 0.533 |
| CL | 7.22 | 7.32 | 6.98 | 0.26 | 7.28 | 7.60 | 0.437 | |||
| 14 | CE | 8.41 | 8.00 | 8.80 | 8.77 | 7.98 | 8.70 | 0.425 | 0.106 | |
| CL | 6.71 | 5.73 | 6.16 | 6.64 | 5.73 | 7.17 | 0.520 | |||
| 21 | CE | 7.68 | 7.55 | 9.69 | 8.52 | 7.52 | 7.88 | 0.372 | 0.041* | |
| CL | 7.01 | 6.58 | 6.63 | 6.63 | 6.63 | 6.09 | 0.456 | |||
| CE | 7.89 | 8.71 | 9.27 | 7.92 | 9.74 | 8.06 | 0.643 | 0.52 | ||
| CL | 5.93 | 5.24 | 5.81 | 5.71 | 5.95 | 5.73 | 0.788 | |||
| 35 | CE | 9.57 | 8.67 | 9.43 | 8.29 | 9.17 | 9.10 | 0.542 | 0.493 | |
| CL | 6.75 | 6.31 | 6.80 | 6.53 | 7.38 | 6.47 | 0.664 | |||
| Litter | 8.47 | 8.87 | 8.72 | 8.43 | 8.57 | 8.44 | 0.14 | 0.197 | ||
Values indicate log10 bacterial counts of C. perfringens, Enterococcus, and E. coli cells obtained from cecal (CE), cloacal (CL), and litter (analyzed on day 35 only) samples of broiler chickens fed diets containing bambermycin (BBM), penicillin (PEN), salinomycin (SAL), bacitracin (BAC), and a salinomycin-bacitracin combination (SAL + BAC) at concentrations specified in Materials and Methods.
P values were obtained by analysis of variance. *, values are statistically different (P < 0.05).
Antibiotic susceptibility.
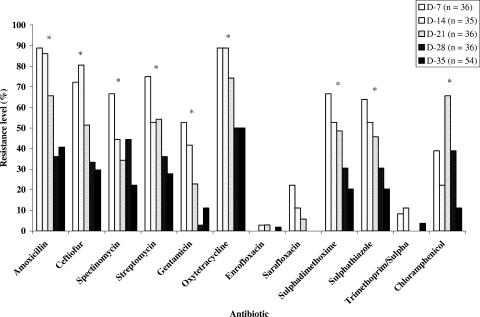
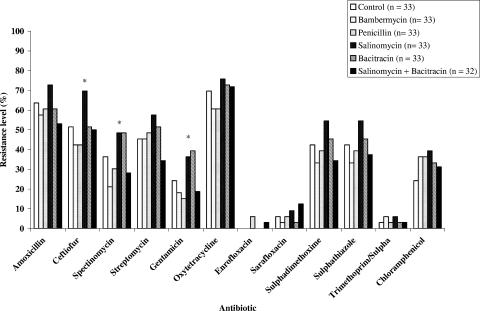
Antibiotic-resistant E. coli isolates were obtained from chickens regardless of the diets they received (Fig. 1). All 197 E. coli isolates tested were multiresistant to penicillin, erythromycin, tylosin, clindamycin, and novobiocin and displayed different resistance levels to the other antibiotics. Low levels of resistance to enrofloxacin, sarafloxacin, and trimethoprim-sulfamethoxazole were noted. Among the 19 antimicrobial agents on the Sensititre Avian plate, resistance levels to tetracycline (68.5%), amoxicillin (61.4%), ceftiofur (51.3%), spectinomycin (47.2%), sulfathiazole (42.1%), and sulfadimethoxime (41.6%) were the most frequent. The levels of resistance to streptomycin, chloramphenicol, and gentamicin were 33.5%, 35.5%, and 25.3%, respectively. Overall resistance levels were highest on day 7 and decreased thereafter (P < 0.001; Fig. 1). Interestingly, resistance to ceftiofur (69.0%) was significantly higher in E. coli isolates from chickens receiving feed supplemented with salinomycin than from the other groups (P < 0.001). Resistance levels to spectinomycin and to gentamicin obtained from salinomycin- and bacitracin-fed chickens were similar and higher than those from the other groups (P < 0.05; Fig. 2).
FIG. 1.
Effect of age on resistance profiles of 197 E. coli isolates from broiler chickens. The percentage of resistance to most antibiotics decreased significantly (P < 0.001) from day 7 to day 35). Asterisks indicate the antibiotics against which the resistance percentages between treatments were statistically different (P < 0.001).
FIG. 2.
Effect of growth promoter diet agents on the frequency of antibiotic resistance levels in 197 E. coli isolates from broiler chickens. The percentage of resistance to most antibiotics decreased significantly (P < 0.001) from day 7 to day 35. Asterisks indicate the antibiotics against which the resistance percentages between treatments were statistically different (P < 0.001).
Distribution of antibiotic resistance phenotypes and their determinants.
Of the 197 E. coli isolates tested, 135 (68.5%) were resistant to tetracycline. One hundred four (77%) of these 135 tetracycline-resistant E. coli isolates were obtained between days 7 and 28 and were further characterized for resistance to antibiotics of importance in human and food animal medicine and for the presence of antibiotic resistance genes (Tables 4 and 5). All were susceptible to ceftriaxone, kanamycin, and amikacin and displayed low resistance levels to quinolones. Higher levels of resistance to β-lactams, aminoglycosines, sulfonamides, and chloramphenicol were noted. In general, a correlation between resistance phenotype and genotype was observed. Antibiotic resistant phenotypes and corresponding resistance genes were found in similar proportions in E. coli isolates from untreated controls and from chickens treated with the test antimicrobial agents.
TABLE 4.
Distribution of antibiotic resistance phenotypes among tetracycline-resistant E. coli isolates from chickens fed antimicrobialsa
| Class | No. (%) of tetracycline-resistant isolates relative to the total no. of isolates (n) tested per treatment
|
||||||
|---|---|---|---|---|---|---|---|
| Control (n = 19) | BBM (n = 14) | PEN (n = 16) | SAL (n = 20) | BAC (n = 19) | SAL + BAC (n = 16) | Total (n = 104)b | |
| β-Lactams | |||||||
| Amoxicillin | 17 (89.5) | 14 (100) | 16 (100) | 20 (100) | 16 (84.2) | 12 (75) | 95 (91.3) |
| Ampicillin | 17 (89.5) | 14 (100) | 15 (93.7) | 19 (95) | 15 (78.5) | 12 (75) | 92 (88.5) |
| Amoxicillin-clavulanic acid | 17 (89.5) | 12 (85.7) | 15 (93.7) | 19 (95) | 15 (78.5) | 12 (75) | 90 (86.5) |
| Cefoxitin | 17 (89.5) | 12 (85.7) | 15 (93.7) | 19 (95) | 14 (73.7) | 12 (75) | 89 (85.6) |
| Ceftiofur | 14 (73.7) | 12 (85.7) | 14 (87.5) | 19 (95) | 13 (68.4) | 12 (75) | 84 (80.8)* |
| Aminoglycosides | |||||||
| Streptomycin | 13 (68.4) | 11 (78.6) | 12 (75) | 15 (75.0) | 15 (78.9) | 7 (43.7) | 73 (70.2) |
| Spectinomycin | 11 (57.9) | 6 (42.8) | 6 (37.5) | 15 (75.0) | 12 (63.1) | 6 (37.5) | 56 (53.8)* |
| Gentamicin | 7 (36.8) | 6 (42.8) | 4 (25.0) | 11 (55.0) | 11 (57.9) | 5 (31.2) | 44 (42.3)* |
| Sulfonamides | |||||||
| Sulfadimethoxime | 12 (63.1) | 10 (71.4) | 10 (62.5) | 17 (85.0) | 13 (68.4) | 7 (43.7) | 69 (66.3) |
| Sulfathiazole | 12 (63.1) | 10 (71.4) | 10 (62.5) | 17 (85.0) | 13 (68.4) | 5 (31.2) | 67 (64.4) |
| Sulfizoxazole | 12 (63.1) | 9 (64.3) | 10 (62.5) | 16 (80.0) | 13 (68.4) | 5 (31.2) | 65 (62.5) |
| Trimethoprim-sulfamethoxazole | 2 (10.5) | 1 (7.1) | 0 (0.0) | 2 (10.0) | 1 (5.3) | 0 (0.0) | 6 (5.8) |
| Phenicols | |||||||
| Chloramphenicol | 10 (52.6) | 9 (64.3) | 9 (56.2) | 13 (65.0) | 12 (63.2) | 5 (31.2) | 58 (55.8) |
| Quinolones | |||||||
| Nalidixic acid | 2 (10.5) | 2 (14.3) | 4 (25.0) | 3 (15) | 2 (10.5) | 2 (12.5) | 15 (14.4) |
| Sarafloxacin | 2 (10.5) | 1 (7.1) | 3 (18.7) | 3 (15) | 2 (10.5) | 2 (12.5) | 13 (12.5) |
| Ciprofloxacin | 0 (0.0) | 1 (7.1) | 3 (18.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (3.8) |
| Enrofloxacin | 0 (0.0) | 0 (0.0) | 2 (12.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.9) |
Distribution of antibiotic resistance phenotypes among tetracycline-resistant E. coli isolates from chickens fed a control diet and diets containing bambermycin (BBM), penicillin (PEN), salinomycin (SAL), bacitracin (BAC), and a salinomycin-bacitracin combination (SAL + BAC) at the concentrations specified in Materials and Methods. All the isolates were resistant to penicillin, erythromycin, tylosin, clindamycin, oxytetracycline, and novobiocin, and all were susceptible to ceftriaxone, kanamycin, and amikacin.
*, values are statistically different (P < 0.05).
TABLE 5.
Distribution of antibiotic resistance genes among 104 selected tetracycline-resistant E. coli isolatesa
| Antibacterial | Gene | No. (%) of tetracycline-resistant E. coli isolates detected relative to no. tested (n) carrying the indicated gene per treatmentb
|
||||||
|---|---|---|---|---|---|---|---|---|
| C (n = 19) | BBM (n = 14) | PEN (n = 16) | SAL (n = 20) | BAC (n = 19) | SAL + BAC (n = 16) | Total (n = 104)d | ||
| β-Lactam (amoxicillin) | 17 (89.5)c | 14 (100.0) | 16 (100.0) | 20 (100.0) | 16 (84.2) | 12 (75) | 95 (91.3) | |
| blaTEM | 8 (57.14) | 4 (25.00) | 8 (40.0) | 8 (50.0) | 6 (50.5) | 40 (42.1) | ||
| blaSHV | 1 (5.9) | 5 (35.7) | 0 (0.00) | 0 (0.0) | 0 (0.00) | 2 (16.7) | 8 (8.4) | |
| blaTEM + blaSHV | 0 (0.0) | 2 (14.3) | 0 (0.0) | 0 (0.0) | 0 (0.00) | 0 (0.0) | 2 (2.1) | |
| Aminoglycoside (streptomycin) | 13 (68.4) | 11 (78.6) | 12 (75) | 15 (75.0) | 15 (78.9) | 7 (43.7) | 73 (70.2) | |
| ant(3″)-Ia (aadA) | 8 (61.5) | 5 (45.4) | 4 (30.7) | 12 (80.0) | 10 (66.7) | 4 (57.1) | 43 (58.9)* | |
| Tetracyclinec | ||||||||
| tet(A) | 13 (68.4) | 12 (85.7) | 9 (56.2) | 17 (85.0) | 15 (78.9) | 10 (62.5) | 76 (73.1) | |
| tet(B) | 11 (57.9) | 8 (57.1) | 9 (56.2) | 12 (60.0) | 11 (57.9) | 8 (50.0) | 59 (56.7) | |
| tet(A) + tet(B) | 5 (26.3) | 6 (42.8) | 2 (12.5) | 9 (45.0) | 7 (36.8) | 2 (12.5) | 31 (29.8) | |
| tet(A) + tet(C) | 9 (5.3) | 0 (0.0) | 0 (0.0) | 2 (10.0) | 1 (5.3) | 2 (12.5) | 6 (5.8) | |
| tet(A) + tet(B) + tet(C) | 2 (10.5) | 1 (7.1) | 0 (0.0) | 3 (15.0) | 2 (10.5) | 0 (0.0) | 8 (7.7) | |
| Sulfonamide (sulfadimethoxime) | 12 (63.1) | 10 (71.4) | 10 (62.5) | 17 (85.0) | 13 (68.4) | 7 (43.7) | 69 (66.3) | |
| sulI | 7 (58.3) | 5 (50.0) | 2 (20.0) | 12 (70.6) | 10 (76.9) | 4 (57.1) | 40 (58.0)* | |
| sulII | 10 (83.3) | 9 (90.0) | 10 (100.0) | 16 (94.1) | 13 (100.0) | 5 (71.4) | 63 (91.3) | |
| sulI + sulI | 5 (41.7) | 5 (50.0) | 2 (20.0) | 10 (58.8) | 10 (76.9) | 1 (14.3) | 35 (50.7)* | |
| dhfrI + sulII | 1 (8.3) | 0 (0.0) | 1 (10.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (2.9) | |
| Phenicol (chloramphenicol) | 10 (52.6) | 9 (64.3) | 9 (56.2) | 13 (65.0) | 12 (63.2) | 5 (31.2) | 58 (55.8) | |
| floR | 8 (80.0) | 9 (100.0) | 9 (100.0) | 13 (100.0) | 12 (100.0) | 5 (100.0) | 56 (96.6) | |
| Class 1 integron in the 104 tetR genes | ||||||||
| int1(1) + int1(2) + int1(3) | 7 (36.84) | 5 (35.71) | 3 (18.75) | 12 (60.00) | 10 (52.63) | 4 (25.00) | 41 (39.42)* | |
Table shows the distribution of antibiotic resistance genes among 104 selected tetracycline-resistant E. coli isolates from the guts of chickens fed with antibiotic growth promoters.
C, control; BBM, bambermycin; PEN, penicillin; SAL, salinomycin; BAC, bacitracin; SAL + BAC, salinomycin with bacitracin.
None of the isolates were positive to tet(C) alone or to the tet(B)-tet(C) combination.
*, values are statistically different (P < 0.05).
Tetracycline.
Of the 104 tetracycline-resistant isolates, at least one of three tetracycline resistance genes, tet(A), tet(B), or tet(C), was found in each isolate. The tet(A) and tet(B) genes were found in 76 (73.1%) and 59 (56.7%) isolates, respectively. The combinations of tet(A) plus tet(B) and tet(A) plus tet(C) were found in 31 (29.8) and 6 (5.8) isolates, respectively. Eight isolates were positive for all three tetracycline resistance genes tet(A) plus tet(B) plus tet(C). None of the tetracycline-resistant isolates was found to harbor the tet(D), tet(E), or tet(Y) gene. No significant differences were observed for the number of isolates in which the genes were detected (P > 0.05) between the treatment and the control groups.
β-Lactams.
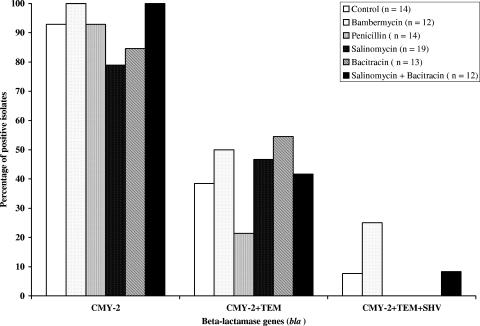
Ninety-five of the 104 tetracycline-resistant E. coli isolates were also resistant to amoxicillin. Of the six resistance genes screened in these 95 isolates, only blaTEM and blaSHV were detected in 42.1% and 8.4% of isolates, respectively. No significant differences were observed for the distribution of these genes (P > 0.05) between treatment and control groups. Both genes were found in two isolates from the bambermycin treatment group. Among the 104 selected tetracycline-resistant E. coli isolates, 84 (80.8%) were resistant to ceftiofur. Among these 84 ceftiofur-resistant E. coli isolates, PCR detection showed that 76 (90.5%) were positive for blaCMY2 (Fig. 3).
FIG. 3.
Prevalence of the bla (CMY-2, TEM, and SHV) genes in 84 tetracycline-ceftiofur-resistant E. coli isolates (14, 12, 14, 19, 13, and 12 for treatment control, bambermycin, penicillin, salinomycin, bacitracin, and salinomycin plus bacitracin, respectively). Detection of the blaCMY-2 gene was performed by PCR as previously described (17).
Aminoglycosides.
Seventy-three of the tetracycline-resistant isolates were phenotypically resistant to streptomycin. Of the six aminoglycoside resistance genes, only ant(3″)-Ia (aadA) was detected in 43 (58.9%) of these 73 isolates. The proportion of isolates carrying this gene was significantly higher from the salinomycin (80%) and bacitracin (66.7.%) treatment groups than from the other groups (P = 0.05). In the isolates from the control group, 61.5% of streptomycin-resistant isolates were also found to harbor the ant(3″)-Ia (aadA) gene. The ant(2")-Ia (aadB) gene encoding the kanamycin, neomycin, and gentamicin resistance phenotypes was not detected, even though 42% of the 104 tetracycline-resistant isolates were also resistant to gentamicin (Table 4).
Sulfonamides.
Among the 69 tetracycline-resistant isolates that were also resistant to sulfadimethoxime, sulI and sulII were found in 40 (58.0%) and 63 (91.3%) isolates, respectively. The proportions of isolates carrying the sulI gene were significantly higher in isolates from the bacitracin (76.9%) and salinomycin (70.6%) treatment groups than from the other groups (P < 0.05). The combination of sulI plus sulII was found in 35 (50.7%) of the 69 sulfadimethoxime-resistant isolates, with a significantly higher proportion found in isolates from the salinomycin (58.8%) and bacitracin (76.9%) treatment groups. Two isolates (one in the control and one in the penicillin treatment groups) were positive for the combination of dhfrI plus sulII.
Phenicols.
Fifty-eight of the 104 tetracycline-resistant isolates were also resistant to chloramphenicol (Tables 4 and 5). Of these 58 isolates, only the floR gene was found in 56 (96.6%) isolates. No significant differences were observed between the groups (P > 0.05) for the presence of this gene. The other phenicol resistance genes, catI, catII, and catIII, were not found in our isolates.
Class 1 integron.
The 104 tetracycline-resistant isolates were screened for the presence of genes related to the class 1 integron in order to investigate the distribution of this resistance-disseminating element. A class 1 integron (qacED1-sulI and integrase gene) was found in 41 (39.4%) of the 104 E. coli isolates. These 41 strains were isolated at different ages (days 7, 14, 21, and 28) from all the experimental groups, and their phenotypes and genotypes are presented in Table 6. Some pens had two or three different isolates over the course of the study. Compared to the isolates of the other groups, significantly higher proportions of class 1 integron were found in isolates from the salinomycin (60.0%) and bacitracin (52.6%) treatment groups (P < 0.05). All 41 isolates were multiresistant to several of the antibiotics tested (Table 6). However, all integron-bearing isolates were susceptible to ceftriaxone, kanamycin, amikacin, and ciprofloxacin. The aminoglycoside [ant(3″)-Ia (aadA)], tetracycline [tet(A), tet(B), or tet(C)], and sulfonamide (sulI or sulII) resistance genes were found in 100% of the integron-positive isolates. The β-lactamase gene (tem) and the phenicol resistance gene floR were found in 78 and 93% of the isolates, respectively (Table 6).
TABLE 6.
Antibiotic resistance phenotypes and genotypes of 41 class 1 integron-positive E. coli isolates from broiler chickensa
| Strain | Treatmentb | Pen | Age (days) | Phenotype (resistance pattern)c | Genotype
|
|||
|---|---|---|---|---|---|---|---|---|
| bla | tet | sul | floR | |||||
| 2440-CE-2 | Control | 2 | 7 | Amo-AmoCla-Amp-Cefx-Spec-Stre-Gent-Sulx-Sulz | A, C | sulI | − | |
| 2447-CE-10 | Control | 10 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2448-CE-17 | Control | 17 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B, C | sulI, sulII | + |
| 2451-CL-17 | Control | 17 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B, C | sulI, sulII | + |
| 2453-CE-10 | Control | 10 | 21 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2454-CE-17 | Control | 17 | 21 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2458-CE2 | Control | 2 | 28 | Amo-AmoCla-Amp-Cefx-Spec-Stre-Gent-Sulx-Sulz | A | sulI | − | |
| 2465-CE-5 | BBM | 5 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2466-CE-18 | BBM | 18 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem, shv | A, B | sulI, sulII | + |
| 2467-CL-1 | BBM | 1 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem, shv | A, B | sulI, sulII | + |
| 2468-CL-5 | BBM | 5 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2478-CE-18 | BBM | 18 | 21 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B, C | sulI, sulII | + |
| 2488-CE-6 | PEN | 6 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2489-CE-9 | PEN | 9 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2492-CL-9 | PEN | 9 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A | sulII | + |
| 2511-CE-3 | SAL | 3 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sara-Nali-Sulx-Sulz-Trm/Sul | A | sulI, sulII | − | |
| 2513-CE-14 | SAL | 14 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2514-CL-3 | SAL | 3 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Sara-Nali-Sulx-Sulz-Trm/Sul | A | sulI, sulII | − | |
| 2515-CL-4 | SAL | 4 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2516-CL-14 | SAL | 14 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2519-CE-14 | SAL | 14 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Clho | A, B | sulI, sulII | + | |
| 2520-CL-3 | SAL | 3 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sara-Nali-Sulx-Sulz | A | sulI, sulII | − | |
| 2521-CL-4 | SAL | 4 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B, C | sulI, sulII | + |
| 2522-CL-14 | SAL | 14 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B, C | sulI, sulII | + |
| 2523-CE-3 | SAL | 3 | 21 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B, C | sulI, sulII | + |
| 2525-CE-14 | SAL | 14 | 21 | Amo-AmoCla-Amp-Cefx-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2534-CL-14 | SAL | 14 | 28 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2535-CE-7 | BAC | 7 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sul-Clho | A | sulI, sulII | + | |
| 2536-CE-11 | BAC | 11 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2537-CE-16 | BAC | 16 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B, C | sulI, sulII | + |
| 2538-CL-7 | BAC | 7 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B, C | sulI, sulII | + |
| 2541-CE-7 | BAC | 7 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2542-CE-11 | BAC | 11 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2543-CE-16 | BAC | 16 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2544-CL-7 | BAC | 7 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sara-Nali-Sulx-Sulz-Trm/Sul | A, C | sulI, sulII | − | |
| 2545-CL-11 | BAC | 11 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2550-CL-7 | BAC | 7 | 21 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | A | sulI, sulII | + | |
| 2560-CE-12 | SAL + BAC | 12 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sara-Nali-Clho | tem | A, C | sulI, sulII | + |
| 2563-CL-12 | SAL + BAC | 12 | 7 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sara-Nali-Sulx-Sulz-Clho | tem | A | sulI, sulII | + |
| 2566-CE-12 | SAL + BAC | 12 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
| 2569-CL-12 | SAL + BAC | 12 | 14 | Amo-AmoCla-Amp-Cefx-Cefti-Spec-Stre-Gent-Sulx-Sulz-Clho | tem | A, B | sulI, sulII | + |
Table shows antibiotic resistance phenotypes and genotypes of the 41 class 1 integron-positive E. coli isolates from broiler chickens. All the isolates were resistant to penicillin, erythromycin, tylosin, clindamycin, oxytetracycline, novobiocin, and sulfizoxazol. All were susceptible to ceftriaxone, kanamycin, amikacin, and ciprofloxacin.
BBM, bambermycin; PEN, penicillin; SAL, salinomycin; BAC, bacitracin; SAL + BAC, salinomycin with bacitracin.
Amo, amoxicillin; AmoCla, amoxicillin-clavulanic acid; Amp, ampicillin; Cefx, cefoxitin; Cefti, ceftiofur; Spec, spectinomycin; Stre, streptomycin; Gent, gentamicin; Sara, saraloxacin; Nali, nalidixic acid; Sulx, sulfadimethoxime; Sulz, sulfathiazole;Trm/Sul, trimethoprim-sulfamethoxazole; Clho; chloramphenicol.
Association between genetic resistance determinants.
The significance of the association between resistance determinants was analyzed. The β-lactamase gene blaSHV and the sulfonamide (trimethoprim) resistance gene dhfrI were not associated with any of the resistance genes or class 1 integron. Associations between the β-lactamase (tem), tetracycline (tet), sulfonamide (sulI or sulII), aminoglycoside [ant(3″)-Ia (aadA)], and phenicol resistance (floR) genes and class 1 integron were found.
DISCUSSION
The poultry industry has developed in recent years due to the continuous integration of various disciplines for production such as poultry health, nutrition, breeding, husbandry, and knowledge of poultry products (3). However, poultry production in Canada and in the United States is facing constraints. The consequences of poultry production for environmental, food safety, and animal welfare issues are now part of consumers’ opinions and demands (2, 18). In the present study, the effects of feed supplementation with bambermycin, penicillin, salinomycin, bacitracin, and a combination of salinomycin plus bacitracin on the growth performance and gut microflora of broiler chickens were investigated over a 35-day period. In accordance with findings by Feighner et al. (22), penicillin was found to increase growth performance. No effects were induced by bambermycin, salinomycin, and/or bacitracin. The lack of significant effects of these compounds on growth performance is likely due to the low repetition numbers (three repetitions/treatment group) and the high hygienic and biosecurity practices used before and throughout the experimental protocol. In a similar experiment conducted with virginiamycin, Dumonceaux et al. (19) also found no significant growth promotion effects for broiler chickens.
The growth of normal intestinal bacteria varies with the gut environment, and there is an increasing interest in the commensal components of the gut microfloras associated with food-producing animals (19, 20, 21, 32, 44). Escherichia coli and Enterococcus and Clostridium spp. are normal inhabitants of the gastrointestinal tract of the chicken. In the present study, feed supplementation with antimicrobial agents had no effect on the concentration of these commensal bacterial species in the cecum and cloaca of the broiler chicken. Enterococcus and E. coli viable counts were higher at day 7 than at day 35, in contrast to C. perfringens counts, which increased from days 7 to 35. Our data confirmed that bacterial numbers in the chicken gut change as a function of age (18). However, E. coli counts recovered from the ceca in the present study were higher than those reported in the study by Gabriel et al. (23). The pathogenicity of the bacteria found in our study needs to be investigated to establish their potential health risks for chickens or humans.
The development of antibiotic resistance in E. coli isolates from poultry is a well-known phenomenon (5, 47, 48). All of the E. coli isolates in the present study were susceptible to ceftriaxone, kanamycin, and amikacin; however, all were multiresistant to several antibiotics. A high rate of resistance to β-lactam (amoxicillin and ceftiofur), tetracycline, streptomycin sulfonamides, and chloramphenicol was noted in the present work. In contrast, Smith et al. (44) detected a low prevalence of resistance to amoxicillin in poultry. The chickens in our study did not receive any anticoccidial or antibacterial agents, other than those used in the experimental design. Care was taken to avoid contamination, and clean pens and fresh wood shavings were used. Our results agree with data reported by Smith et al. (44), who showed a high prevalence of resistance to antimicrobials that are not commonly used in broiler chicken production.
We also found that antibiotic resistance levels decreased with increasing bird age. In chickens, the diets and the environments can affect the microbial status of the gastrointestinal tract (4). Litter and other management practices also can change microbial composition of the chicken gut directly by providing a continuous source of bacteria or indirectly by influencing the defense mechanisms of the birds (4). The reason for the decreased antibiotic resistance level in this study is unclear and may be due to the composition of diets fed at different growth phases, from the starter to the finisher (Table 1), or to other unknown parameters resulting in microbial flora turnover. Our data suggest that day-old chicks are colonized with some resistant strains that are replaced by the normal susceptible bacteria as the birds age. The origin of resistance to the other antibiotics is unknown and could be derived from the broiler production environment, including the litter (37), feed, or caretakers (15).
It has been reported that selection and maintenance of tetracycline-streptomycin-sulfonamide-resistant E. coli may be due to environmental components independent of antibiotic selection (30). Interestingly, we found higher incidences of ceftiofur, spectinomycin, and gentamicin resistance in E. coli isolates from chickens receiving feed supplemented with salinomycin than with other feeds. Higher percentages of gentamicin-resistant isolates were also observed for bacitracin-fed chickens. Coliforms from birds fed salinomycin were found to have more multiresistance patterns with significant numbers resistant to streptomycin, ampicillin, carbenicillin, and cephalothin (24). Our results confirm that multiresistant commensal E. coli strains may be present in conventional broiler chicken production independently of specific antibiotic selection pressure.
Characterization of 104 selected tetracycline-resistant E. coli isolates showed resistance to several antibiotics of human importance. We used a DNA microarray hybridization method to evaluate the presence and distribution of antibiotic resistance determinants among these tetracycline-resistant isolates (9, 28). In E. coli isolates, tetracycline resistance is frequently regulated by several efflux genes on large plasmids that frequently carry other antibiotic and heavy metal resistance genes (13). At least one of three tetracycline resistance genes, tet(A), tet(B), or tet(C), was found in all the 104 tetracycline-resistant isolates. The tet(D), tet(E), and tet(Y) genes were not found in any of the isolates, while the tet(A) and tet(B) genes were detected in 76 and 59 isolates, respectively. Few isolates were positive for tet(C), which was seen only in combination with tet(A) or tet(A) plus tet(B). Fairchild et al. (21) reported the presence of tet(A) and tet(B) but not tet(C) or tet(D) in intestinal E. coli isolates after oral administration of tetracycline to chickens.
β-Lactams are among the most clinically important antibiotics in both human and veterinary medicine, and yet resistance to this class of antibiotics is increasing at an alarming rate (34). Previously, we reported the presence of the extended-spectrum-β-lactamase blaCMY-2 gene in a large percentage of avian E. coli isolates that were resistant to ceftiofur (17). In this study, the blaCMY-2 gene was detected in 90.5% of ceftiofur-resistant isolates, indicating that this gene is widespread in commensal E. coli isolates from chickens. However, blaTEM, and blaSHV were found in 41 (43%) and 9 (10%) of 95 tetracycline-amoxicillin-resistant isolates, respectively. These genes were similarly distributed among the treatment groups. These results indicate that other resistant genes may be implicated in the resistance to this class of antibiotics in our isolates.
Of the 73 aminoglycoside-resistant isolates, 43 were positive for the aminoglycoside nucleotidyltransferase gene ant(3″)-Ia (aadA). The remaining aminoglycoside-resistant isolates, in which none of the other six genes was detected, suggests the presence of different aminoglycoside resistance determinants. Interestingly, we found that there was a higher incidence of ant(3″)-Ia (aadA)-positive E. coli (80%) in the chickens receiving salinomycin than in the other antimicrobial treatment groups. This result suggests that salinomycin may play a role in the selection and maintenance of streptomycin/spectinomycin resistance in broiler chickens.
Chloramphenicol has not been used in chicken production in Canada since 1980 (26). Nevertheless, 56 of the 58 tetracycline-resistant isolates that were also resistant to chloramphenicol bore the floR gene, with a similar distribution among the treatment groups. The chloramphenicol resistance genes floR, cat, and cml were also reported in enterotoxigenic E. coli and nonenterotoxigenic E. coli isolated from swine in Ontario, Canada (46). In avian species, flo was detected in phenicol-resistant E. coli isolates in the United States, where chloramphenicol is likewise not used (47). We did not find the phenicol resistance genes catI, catII, and catIII in any of our isolates.
Historically, sulfonamides played an important role in the development of broiler chicken production systems by allowing birds to be raised in higher densities. However, the development of resistance to this class of antibiotic has reduced its role in poultry production (40). In the present work, the sulI and sulII genes were the sulfonamide resistance genes most frequently found, alone or in combination. More than 70% of the sulfonamide-resistant E. coli isolates from the salinomycin (70.6%) and bacitracin (76.9%) treatment groups were positive for sulI. A high incidence (76.9%) of the sulI-plus-sulII combination was found in the bacitracin treatment group.
The ability of bacteria to acquire and disseminate exogenous genes is a major factor in the development of multiple antibiotic resistance. Integrons are gene expression elements that contribute to the spread of antimicrobial resistance by gene transfer in a variety of enteric bacteria (6, 35, 42). The presence of integrons in enteric bacteria from poultry has been previously reported (17, 37, 41). The 41 integron-positive isolates found in our study were all multiresistant. They all harbored the genes ant(3″)-Ia (aadA), sulI, and/or sulII and tet(A), tet(B), or tet(C). The β-lactam (blaTEM)- and phenicol (floR)-resistant genes were found in 32 (78%) and 35 (85%) isolates, respectively. We did not determine if these genes are physically linked; however, the statistical analysis clearly showed significant associations not only between them but also with the class 1 integron. Associations have been observed between the tet(A), sulI, and aadA genes in porcine E. coli isolates (7). The coexistence of antimicrobial resistance genes in association with integrons may increase the selection and dissemination of multidrug-resistant bacteria (34). We found that isolates from the salinomycin and bacitracin treatment groups showed the highest incidence of the class 1 integron. Phenotypic and genotypic analyses suggested that these two growth promoters may play a role in the development and/or maintenance of antibiotic resistance in broiler chicken production.
Our data confirm that the gastrointestinal tract of broiler chickens can be colonized by multidrug-resistant E. coli bacteria and that the use of growth promoter agents like salinomycin or bacitracin may exercise pressure for selection for such bacteria. The presence of the class 1 integron in E. coli indicates a potential for lateral antibiotic resistance gene transfer between this bacterium and other chicken gut bacteria. These bacteria have the potential to spread in the environment through the litter (37) and subsequently to farm workers and processing plants. Our results also suggest that in the absence of specific antibiotic selection pressure, some specific resistance genes can be maintained due to the association with the genes encoding resistance to other antimicrobials that are currently approved for use in broiler chicken production.
Acknowledgments
This work was financially supported by Agriculture and Agri-Food Canada through the Agriculture Policy Framework GAPs program (Pacific Agri-Food Research Centre—Agassiz contribution number 756).
We thank H. Rempel, L. Struthers (Pacific Agri-Food Research Centre, Agassiz, BC, Canada), and William R. Cox (Canadian Animal Health Management Services Ltd., Chilliwack, BC, Canada) for technical assistance. We acknowledge the technical assistance of S. Methot, K. Hildebrandt, and S. Garcia.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Aarestrup, F. M. 2000. Occurrence, selection and spread of resistance to antimicrobial agents used for growth promotion for food animals in Denmark. APMIS Suppl. 101:1-48. [PubMed] [Google Scholar]
- 2.Altekruse, S. F., M. L. Cohen, and D. L. Swerdlow. 1997. Emerging foodborne diseases. Emerg. Infect. Dis. 3:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 2007. Antimicrobial resistance: implication for food system. Institute of Food Technologists Expert Panel. Comp. Rev. Food Sci. Safety 5:71-137. [Google Scholar]
- 4.Apajalahti, J., A. Kettunen, and H. Graham. 2004. Characteristics of the gastrointestinal microbial communities, with special reference to chicken. World's Poult. Sci. J. 60:223-232. [Google Scholar]
- 5.Asai, T., A. Kojima, K. Harada, K. Ishihara, T. Takahashi, and Y. Tamura. 2005. Correlation between the usage volume of veterinary therapeutic antimicrobials and resistance in Escherichia coli isolated from the feces of food-producing animals in Japan. Jpn. J. Infect. Dis. 58:369-372. [PubMed] [Google Scholar]
- 6.Bass, L., C. A. Liebert, M. D. Lee, A. O. Summers, D. G. White, S. G. Thayer, and J. J. Maurer. 1999. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 43:2925-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boerlin, P., R. Travis, C. L. Gyles, R. Reid-Smith, N. J. Heather Lim, V. Nicholson, S. A. McEwen, R. M. Friendship, and M. Archambault. 2005. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 71:6753-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boerlin, P. 2004. Molecular epidemiology of antimicrobial resistance in veterinary medicine: where do we go? Anim. Health Res. Rev. 5:95-102. [DOI] [PubMed] [Google Scholar]
- 9.Bruant, G., C. Maynard, S. Bekal, I. Gaucher, L. Masson, R. Brousseau, and J. Harel. 2006. Development and validation of an oligonucleotide microarray for the detection of multiple virulence and antimicrobial resistance genes in Escherichia coli. Appl. Environ. Microbiol. 72:3780-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butaye, P., L. A. Devriese, and F. Haesebrouck. 2003. Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on gram-positive bacteria. Clin. Microbiol. Rev. 16:175-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canadian Council on Animal Care. 1993. Guide to the care and use of experimental animals, 2nd ed., vol. 1. CCAC, Ottawa, ON, Canada.
- 12.Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS). 2005. Annual report (2004). Health Canada, Ottawa, ON, Canada.
- 13.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collier, C. T., J. D. van der Klis, B. Deplancke, D. B. Anderson, and H. R. Gaskins. 2003. Effects of tylosin on bacterial mucolysis, Clostridium perfringens colonization, and intestinal barrier function in a chick model of necrotic enteritis. Antimicrob. Agents Chemother. 47:3311-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.da Costa, P. M., M. Oliveira, A. Bica, P. Vaz-Pires, and F. Bernardo. 2007. Antimicrobial resistance in Enterococcus spp. and Escherichia coli isolated from poultry feed and feed ingredients. Vet. Microbiol. 120:122-131. [DOI] [PubMed] [Google Scholar]
- 16.DeVincent, S. J., and C. Viola. 2005. Antimicrobial use in animals in the United States: developments in policy and practice, p. 528-536. In D. G. White, M. N. Alekshun, and P. F. McDermott (ed.), Frontiers in antimicrobial resistance. ASM Press, Washington, DC.
- 17.Diarrassouba, F., M. S. Diarra, S. Bach, P. Delaquis, J. Pritchard, E. Topp, and B. J. Skura. 2007. Antibiotic resistance and virulence genes in commensal Escherichia coli and Salmonella isolated from commercial broiler chicken farms. J. Food Prot. 70:1316-1327. [DOI] [PubMed] [Google Scholar]
- 18.Dibner, J. J., and J. D. Richards. 2005. Antibiotic growth promoter in agriculture: history and mode of action. Poult. Sci. 84:634-643. [DOI] [PubMed] [Google Scholar]
- 19.Dumonceaux, T. J., J. E. Hill, S. M. Hemmingsen, and A. G. Van Kesse. 2006. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl. Environ. Microbiol. 72:2815-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durso, L. M., D. Smith, and R. W. Hutkins. 2004. Measurements of fitness and competition in commensal Escherichia coli and E. coli O157:H7 strains. Appl. Environ. Microbiol. 70:6466-6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fairchild, A. S., J. L. Smith, U. Idris, J. Lu, S. Sanchez, L. B. Purvis, C. Hofacre, and M. D. Lee 2005. Effects of orally administered tetracycline on the intestinal community structure of chickens and on tet determinant carriage by commensal bacteria and Campylobacter jejuni. Appl. Environ. Microbiol. 71:5865-5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feighner, S. D., and M. P. Dashkevicz. 1987. Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl. Environ. Microbiol. 53:331-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel, I., S. Mallet, and P. Sibille. 2005. Digestive microflora of bird: factors of variation and consequences on bird. INRA Prod. Anim. 18:309-322. [Google Scholar]
- 24.George, B. A., A. M. Ford, D. J. Fagerberg, and C. L. Quarles. 1982. Influence of salinomycin on antimicrobial resistance of coliforms and streptococci from broiler chickens. Poult. Sci. 61:1842-1852. [DOI] [PubMed] [Google Scholar]
- 25.Geornaras, I., J. W. Hastings, and A. von Holy. 2001. Genotypic analysis of Escherichia coli strains from poultry carcasses and their susceptibilities to antimicrobial agents. Appl. Environ. Microbiol. 67:1940-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmore, A. 1986. Chloramphenicol and the politics of health. Can. Med. Assoc. J. 134:423. [PMC free article] [PubMed] [Google Scholar]
- 27.Hakanen, A., A. Siitonen, P. Kotilainen, and P. Huovinen. 1999. Increasing fluoroquinolone resistance in Salmonella serotypes in Finland during 1995-1997. J. Antimicrob. Chemother. 43:145-148. [DOI] [PubMed] [Google Scholar]
- 28.Hamelin, K., G. Bruant, A. El-Shaarawi, S. Hill, T. A. Edge, S. Bekal, M. J. Fairbrother, J. Harel, C. Maynard, L. Masson, and R. Brousseau. 2006. A virulence and antimicrobial resistance DNA microarray detects a high frequency of virulence genes in Escherichia coli isolates from Great Lakes recreational waters. Appl. Environ. Microbiol. 72:4200-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes, J. R., L. L. English, P. J. Carter, T Proescholdt, K. Y. Lee, D. D. Wagner, and D. D. White. 2003. Prevalence and antimicrobial resistance of Enterococcus species isolated from retail meats. Appl. Environ. Microbiol. 69:7153-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khachatryan, A. R., T. E. Besser, D. D. Hancock, and D. R. Call. 2006. Use of a nonmedicated dietary supplement correlates with increased prevalence of streptomycin-sulfa-tetracycline-resistant Escherichia coli on a dairy farm. Appl. Environ. Microbiol. 72:4583-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knarreborg, A., M. A. Simon, R. M. Engberg, B. B. Jensen, and G. W. Tannock. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl. Environ. Microbiol. 68:5918-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langhout, D. J., J. B. Schutte, P. Van Leeuwen, J. Wiebenga, and S. Tamminga. 1999. Effect of dietary high- and low-methylated citrus pectin on the activity of the ileal microflora and morphology of the small intestinal wall of broiler chicks. Br. Poult. Sci. 40:340-347. [DOI] [PubMed] [Google Scholar]
- 33.Lefebvre, B., M. S. Diarra, K. Giguère, G. Roy, S. Michaud, and F. Malouin. 2006. Antibiotic-resistance and hypermutability of Escherichia coli O157 from feedlot cattle treated with growth-promoter agents. J. Food Prot. 68:2411-2419. [DOI] [PubMed] [Google Scholar]
- 34.Li, X.-Z., M. Mehrotra, S. Ghimire, and L. Adewoye. 2007. β-Lactam resistance and β-lactamases in bacteria of animal origin. Vet. Microbiol. 121:197-214. [DOI] [PubMed] [Google Scholar]
- 35.Maynard, C., S. Bekal, F. Sanschagrin, R. C. Levesque, R. Brousseau, L. Masson, S. Larivière, and J. Harel. 2004. Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J. Clin. Microbiol. 42:5444-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miles, T. D., W. McLaughlin, and P. D. Brown. 2006. Antimicrobial resistance of Escherichia coli isolates from broiler chickens and humans. BMC Vet. Res. 2:7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nandi, S., J. J. Maurer, C. Hofacre, and A. O. Summers. 2004. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. USA 101:7118-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution of antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 39.National Research Council. 1994. Nutrient requirements of poultry, 9th ed. National Academy Press, Washington, DC.
- 40.Navia, M. A. 2000. A chicken in every pot, thanks to sulfonamide drugs. Science 288:2132-2133. [DOI] [PubMed] [Google Scholar]
- 41.Nógrády, N., J. Pászti, H. Pikó, and B. Nagy. 2006. Class 1 integrons and their conjugal transfer with and without virulence-associated genes in extra-intestinal and intestinal Escherichia coli of poultry. Avian Pathol. 8:349-356. [DOI] [PubMed] [Google Scholar]
- 42.Sáenz, Y., L. Briñas, E. Domínguez, J. Ruiz, M. Zarazaga, J. Vila, and C. Torres. 2004. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 48:3996-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singer, R. S., and C. L. Hofacre. 2006. Potential impacts of antibiotic use in poultry production. Avian Dis. 50:161-172. [DOI] [PubMed] [Google Scholar]
- 44.Smith, J. L., D. J. V. Drum, Y. Dai, J. M. Kim, S. Sanchez, J. J. Maurer, C. L. Hofacre, and M. D. Lee. 2007. Impact of antimicrobial usage on antimicrobial resistance in commensal Escherichia coli strains colonizing broiler chickens. Appl. Environ. Microbiol. 73:1404-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Statistical Analysis System. 2000. Release 8.02. SAS Institute, Inc., Cary, NC.
- 46.Travis, R. M., C. L. Gyles, R. Reid-Smith, C. Poppe, S. A. McEwen, R. Friendship, N. Janecko, and P. Boerlin. 2006. Chloramphenicol and kanamycin resistance among porcine Escherichia coli in Ontario. J. Antimicrob. Chemother. 58:173-177. [DOI] [PubMed] [Google Scholar]
- 47.White, D. G. 2005. Antimicrobial resistance in pathogenic Escherichia coli from animal, p. 145-166. In F. M. Aarestrup (ed.), Antimicrobial resistance in bacteria of animal origin. ASM Press, Washington, DC.
- 48.Zhao, S., J. J. Maurer, S. Hubert, J. F. De Villena, P. F. McDermott, J. Meng, S. Ayers, L. English, and D. G. White. 2005. Antimicrobial susceptibility and molecular characterization of avian pathogenic Escherichia coli isolates. Vet. Microbiol. 107:215-224. [DOI] [PubMed] [Google Scholar]