Abstract
Elemental sulfur (S0) is associated with many geochemically diverse hot springs, yet little is known about the phylogeny, physiology, and ecology of the organisms involved in its cycling. Here we report the isolation, characterization, and ecology of two novel, S0-reducing Crenarchaea from an acid geothermal spring referred to as Dragon Spring. Isolate 18U65 grows optimally at 70 to 72°C and at pH 2.5 to 3.0, while isolate 18D70 grows optimally at 81°C and pH 3.0. Both isolates are chemoorganotrophs, dependent on complex peptide-containing carbon sources, S0, and anaerobic conditions for respiration-dependent growth. Glycerol dialkyl glycerol tetraethers (GDGTs) containing four to six cyclopentyl rings were present in the lipid fraction of isolates 18U65 and 18D70. Physiological characterization suggests that the isolates are adapted to the physicochemical conditions of Dragon Spring and can utilize the natural organic matter in the spring as a carbon and energy source. Quantitative PCR analysis of 16S rRNA genes associated with the S0 flocs recovered from several acid geothermal springs using isolate-specific primers indicates that these two populations together represent 17 to 37% of the floc-associated DNA. The physiological characteristics of isolates 18U65 and 18D70 are consistent with their potential widespread distribution and putative role in the cycling of sulfur in acid geothermal springs throughout the Yellowstone National Park geothermal complex. Based on phenotypic and genetic characterization, the designations Caldisphaera draconis sp. nov. and Acidilobus sulfurireducens sp. nov. are proposed for isolates 18U65 and 18D70, respectively.
Sulfur compounds are common constituents of the gaseous, aqueous, and solid phases of acid geothermal fluid of volcanic origin (26). At the point of surface discharge, gas-phase components typically include sulfur dioxide (SO2), hydrogen sulfide (H2S), and elemental sulfur vapor (29, 35), while the solid phase is typically comprised of flocculent S0 (29, 31). The origin of solid-phase S0 near the point of discharge in acidic geothermal springs can be attributed to biotic oxidation of H2S as well as abiotic H2S oxidation to thiosulfate (S2O32−), which disproportionates under acidic conditions to form sulfite (SO3−) and S0 (38). Abiotic H2S oxidation by metal oxidants (vanadate, ferric iron, cupric copper, etc.) may also contribute to the precipitation and accumulation of S0 in geothermal environments (44); however, these reactions are less important due to the reducing conditions and the absence of these oxidants in source water in many high-temperature acidic geothermal systems such as those found in Norris Geyser Basin, Yellowstone National Park (YNP), WY (29, 31, 38). The S0 formed from H2S oxidation by these mechanisms accumulates in many acid geothermal springs due to its slow reactivity with water below 100°C (37) and thus represents a significant source of electron donor and acceptor for microorganisms in these systems (2, 20).
S0 has been shown to support respiratory metabolisms for a variety of organisms distributed throughout the Bacteria and Archaea (8, 16, 27, 43). Within the Archaea, S0 reduction has been identified in both the Euryarchaeota (10, 13) and Crenarchaeota (22, 23, 41, 51) and is typically coupled with the oxidation of complex organics and/or hydrogen (16). Within the order Desulfurococcales, phylum Crenarchaeota, the dominant metabolism of acidophiles is the fermentation of simple and complex organic compounds (23, 41), with significantly enhanced growth rates in the presence of S0. Many acidic geothermal springs within YNP possess geochemical conditions that should support organisms with metabolisms similar to those of the order Desulfurococcales.
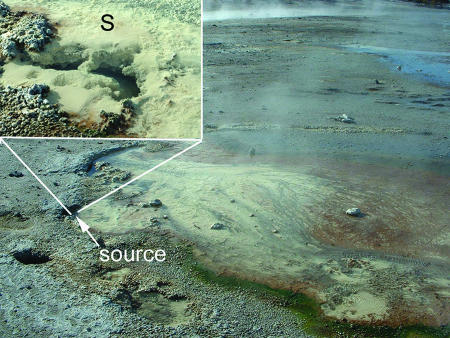
The study summarized herein primarily focused on an acidic geothermal spring referred to as Dragon Spring (24, 29), which is located in Norris Geyser Basin, YNP. Dragon Spring is classified as an acid-sulfate-chloride spring (ASC), so named because of its distinctive chemical signature (6, 29, 49). The source water of Dragon Spring (pH 3.1; 66 to 73°C) contains significant dissolved organic carbon (80 μM) and various inorganic energy sources such as H2S, Fe(II), and As(III) (29). In addition, the entire outflow channel is visually distinguished by copious amounts of a solid phase comprised almost exclusively of S0 (29) (Fig. 1), suggesting that this as an appropriate habitat for biotic S0 oxidation and reduction. While the geochemical conditions at the source of Dragon Spring suggest an environment favorable for biological S0 reduction, phylogenetic analysis of bacterial and archaeal community 16S rRNA genes obtained from S0 precipitates failed to identify phylotypes related to known S0 reducers (unpublished data), thereby precluding efforts to use phylogeny to identify cultivation conditions necessary to culture such organisms, an approach utilized in other studies (42, 46). Alternatively, we analyzed the geochemistry of the spring water to identify the laboratory culture conditions representative of in situ conditions for the enrichment and isolation of microorganisms capable of S0 reduction. Here, we report the isolation, characterization, and ecology of two novel, S0-respiring heterotrophs that cluster both phylogenetically and phenotypically within the “Acidilobus group” (proposed name) of the order Desulfurococcales, phylum Crenarchaeota. The physiological properties which could convey pertinent information on the ecological role of each isolate in ASC geothermal springs were determined. Results suggest that both isolates are adapted to the geochemical conditions present in several ASC springs in the Norris Geyser Basin, YNP.
FIG. 1.
Dragon Spring source with abundant S0 flocs (S in inset).
MATERIALS AND METHODS
Sample collection and enrichment and isolation of spring microorganisms.
Samples of precipitated (flocculent) S0 were collected using sterile syringes and serum bottles from the source of Dragon Spring (44°43′55" N, 110°42′39" W), Beowulf Spring (44°43′53.4" N, 110°42′40.9" W), and Succession Spring (44°43′75.7" N, 110°42′72.7" W), all of which are ASC thermal springs located in the Hundred Springs Plain area of Norris Geyser Basin, YNP, WY. Samples were collected from the point of discharge at the source of Dragon (66°C; pH 3.1), Succession (69°C; pH 3.1), and Beowulf (68°C; pH 3.0) Springs. Upon collection, samples were stored at 65°C under anaerobic conditions during the 90-min transit to Montana State University, Bozeman, MT. Separate samples were placed on dry ice during transit to Montana State University, where they were stored at −20°C until processed for DNA extraction. Peptone-S0 (PS) medium was used to enrich for S0-reducing thermophiles. PS medium consisted of NH4Cl (0.33 g liter−1), KCl (0.33 g liter−1), CaCl2·2H2O (0.33 g liter−1), MgSO4·7H2O (0.33 g liter−1), and KH2PO4 (0.33 g liter−1), peptone (0.05% [wt/vol]), S0 (5 g liter−1), Wolfe's vitamins (1 ml liter−1) (5), and SL-10 trace metals (1 ml liter−1) (48). Following autoclave sterilization of the salts and peptone components in serum bottles, filter-sterilized Wolfe's vitamins, filter-sterilized SL-10 trace metals, and S0 (sterilized by baking at 100°C for 24 h) were added, and the bottles and contents were deoxygenated by purging with sterile nitrogen gas passed over heated (210°C) copper shavings. The serum bottles were sealed with butyl rubber stoppers and heated to 80°C prior to the replacement of the headspace with H2 and addition of sodium ascorbate to a final concentration of 50 μM. The serum bottles were then inoculated with S0 precipitates recovered from the source of Dragon Spring and were incubated at 65 or 70°C. A pure culture of isolate 18U65 was obtained from the 65°C enrichment by dilution to extinction. A pure culture of isolate 18D70 was obtained from the 70°C enrichment by dilution to extinction and increased incubation temperature (80°C). Multiple rounds of dilution to extinction were performed for each isolate to ensure pure culture status. Enrichment isolation progress was monitored using epifluorescence microscopy and denaturing gradient gel electrophoresis (DGGE) (see below).
DNA extraction and DGGE.
Enrichment progress was monitored using DGGE analysis of PCR-amplified 16S rRNA gene fragments recovered from enrichment cultures. CS2 was added prior to DNA extraction to remove S0 and to improve the recovery of DNA. Biomass for DNA extraction was concentrated by centrifugation (14,000 × g for 10 min at 4°C) and was resuspended in a minimal volume of sterile PS medium (lacking S0). CS2 was added to a final concentration of 10% (vol/vol), and following phase separation, the top aqueous phase containing cells was collected and concentrated by centrifugation (14,000 × g for 10 min at 4°C). Preliminary experiments indicate that CS2 treatment did not significantly affect the integrity or recovery (79% ± 9.9% recovery) of cells as determined by epifluorescence microscopy (data not shown). The cell pellet was subjected to DNA extraction using a FastDNA Spin for Soil kit (MP Biomedicals, Solon, OH) according to the manufacturer's directions with the following exception: 500 μl of Tris-buffered phenol (pH 8.0) (Sigma, St. Louis, MO) was substituted for 500 μl of the 978 μl of sodium phosphate buffer. Following extraction, the DNA concentration was determined using a High DNA mass ladder (Invitrogen, Carlsbad, CA) for use in PCR. PCR was performed according to previously published protocols (9) using 10 ng of DNA as a template and the primers archaeal 931F (5′-AAGGAATTGGCGGGGGAGCA-3′) or bacterial 1070F (5′-ATGGCTGTCGTCAGCT-3′), in conjunction with universal 1392R (5′-ACGGGCGGTGTGTRC-3′) or 1492R (5′-GGTTACCTTGTTACGACTT-3′); an annealing temperature of 55°C was used for all primer combinations. Each of the reverse primers was conjugated with a 40-bp GC-clamp attached to the 5′ terminus (Integrated DNA Technologies, Coralville, IA). DGGE (60V at 60°C for 18 h) was performed according to previously published protocols (24) in a vertical gradient of 50 to 70% (archaeal amplicons) or 35 to 55% (bacterial amplicons) denaturant.
Characterization of physicochemical properties of isolates.
The cardinal temperatures, pH, and Cl− concentrations were determined for each isolate. The minimum, optimum, and maximum temperatures for growth of each isolate were determined in PS medium adjusted to pH 3.0. Minimum, optimum, and maximum pH for growth of isolates 18U65 and 18D70 were determined in PS medium incubated at 65 and 80°C, respectively. The requirement for and tolerance to Cl− for each isolate was determined in PS medium with the pH and incubation temperature adjusted to the optimum for each isolate. Cl− concentrations were adjusted by additions of NaCl.
Carbon source and electron donors.
Identification of carbon sources that support growth of the isolates when S0 is used as the terminal electron acceptor (TEA) was evaluated using PS medium adjusted to pH 3.0, where the candidate carbon source was substituted for peptone. Glucose, galactose, fructose, ribose, lactose, maltose, xylose, sucrose arabinose, mannose, yeast extract, peptone, Casamino Acids, tryptone, glycerin, cellobiose, cellulose, starch, soya extract, beef extract, gelatin, and glycogen were added to achieve a final concentration of 0.1% (wt/vol). Methane, propane, propylene, ethanol, butanol, propanol, isopropanol, methanol, and acetone were added to a final concentration of 10 mM. Butyrate, propionate, succinate, lactate, pyruvate, acetate, citrate, malate, formate, fumarate, and benzoate were added to a final concentration of 4 mM. Individual amino acids and a mixture of 20 essential individual amino acids mixed at equal molar concentrations were added to a final concentration of 0.02% (wt/vol). Carbon sources were tested for supporting growth in medium containing a 1:1 ratio of N2 gas:aqueous phase, with the pH and incubation temperature adjusted to the optimum for each isolate.
An extract of lodgepole pine needles was also tested as a carbon source for the isolates since needle litter is a potential source of organic carbon in situ (unpublished observations). Extract was prepared by incubating 10 g of fresh lodgepole pine needles in a 150-ml serum bottle containing 100 ml of pH 3.0 peptone-free PS medium at 81°C for 24 h, after which the aqueous phase was recovered, filter sterilized, and frozen. One milliliter of thawed extract was added to 100 ml of peptone-free PS medium as the sole carbon and energy source for the isolates.
Growth of isolates 18U65 and 18D70 on the different carbon sources was determined at 65°C and 81°C, respectively, in cultures that initially contained 100% N2 in the headspace. After a carbon source was determined to support growth of the isolates when S0 was used as the TEA, fermentative growth with that carbon source was assessed by deleting S0 from the medium (beef extract, glycogen, and gelatin were not evaluated).
H2 was tested as a potential electron donor by comparing growth rates and final cell densities in PS medium containing 100% H2 in the headspace at the time of inoculation to values observed when the headspace contained 100% N2. In addition, H2 was also used as an electron donor in tests for chemolithoautotrophic growth with CO2 as a sole C source in serum bottles containing peptone-free PS medium and a headspace gas mixture of 20%:80% CO2-H2 (headspace:aqueous phase ratio of 1:1).
Alternative TEAs.
The ability of the isolates to utilize TEAs other than S0 for growth was determined in S0-free PS medium. Sodium sulfate, sodium sulfite, sodium thiosulfate, potassium nitrate, and potassium nitrite were each added at a concentration of 10 mM; Fe(III) citrate and FeCl3 were each added at a concentration of 500 μM; sodium arsenate was added at a concentration of 100 μM; and cysteine was added at a concentration of 0.02% (wt/vol).
Evaluation of growth.
Growth of each isolate on different carbon sources and with different electron donors and electron acceptors was quantified both in terms of generation time and H2S production. Generation time was determined by collecting a sample of suspended cells at different times following inoculation, adding SYBR Gold (Invitrogen) diluted 1/4,000 (vol/vol), collecting the cells on black 0.22-μm-pore-size polycarbonate filters (Millipore, Billerica, MA), and enumerating the cells by direct epifluorescence microscopy using an Olympus Β-max microscope with a WIBA filter cube combination (Olympus, Center Valley, PA).
The total sulfide produced by cultures respiring S0 was determined using the methylene blue reduction method (14). Triplicate measurements of dissolved sulfide for each culture were averaged for three replicate cultures to calculate the average sulfide concentration and the standard deviation among replicates. Total sulfide production was calculated using standard gas-phase equilibrium calculations (see the supplemental material).
Influence of O2 on growth of isolates.
The influence of oxygen concentration on the growth of isolates was determined by replacing different volumes of headspace N2 with air in serum bottles containing PS medium with the pH and incubation temperature adjusted to the optimum for each isolate. The O2 concentration in the aqueous phase was calculated using standard gas-phase equilibrium calculations (see the supplemental material).
Metabolic by-products.
The production of the metabolic by-products acetate, lactate, NH4+, and H2 was determined for both isolates during growth at optimal pH and temperatures in PS medium. Following inoculation, cultures were sampled daily for acetate, lactate, and NH4+ for 10 days. Culture samples were filtered through a 0.22-μm-pore-size membrane, and the filtrate was analyzed using a Bioprofile 300A biochemistry analyzer (Nova Biomedical Corporation, Waltham, MA) calibrated with level 6 and 7 certified calibration standards (Nova Biomedical Corporation). Culture headspace H2 concentration was determined during logarithmic growth using a GC8A gas chromatograph (Shimadzu, Columbia, MD) equipped with an 80/100 ProPak N column (Supelco, St. Louis, MO). Total sulfide was determined as described above.
Amino acid analysis.
Amino acid analysis was performed on culture supernatant collected from isolate 18D70 grown in PS medium with the pH and incubation temperature adjusted to the optimum. Amino acids were determined in filtered (0.22-μm pore size) culture supernatant collected at the time of inoculation and following 120 h of growth using an Applied Biosystems 420A derivatizer coupled to an Applied Biosystems 130A separator system. Samples were hydrolyzed in 6 M HCl plus trace phenol in HCl vapors for 1 h and then in a vacuum at 150°C. After hydrolysis, norleucine was added as an internal standard.
Calculation of cell yields.
Cell yields were calculated using 100 fg/μm3 as the conversion factor for carbon content to biovolume (36). Cell yields were computed during log-phase growth using the average cell diameter as determined by transmission electron microscopy (see below). The cell yield for Acidilobus aceticus was calculated from data extrapolated from Fig. 5 of Prokofeva et al. (41).
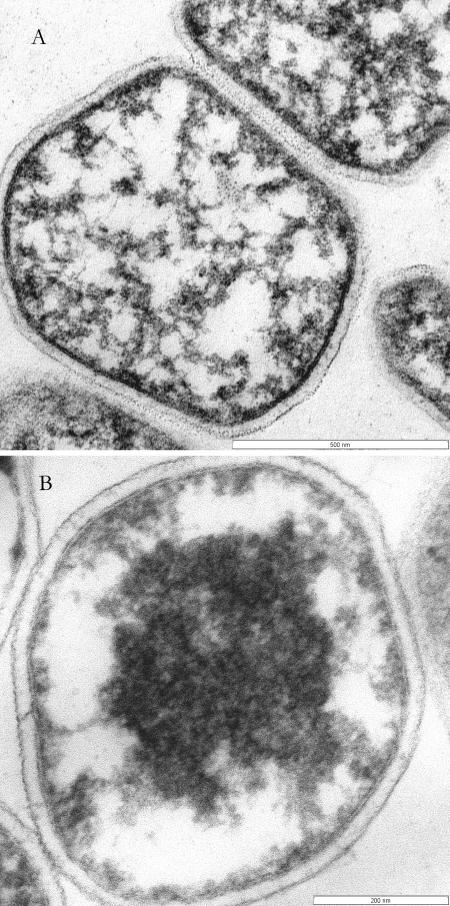
FIG. 5.
Electron micrograph of isolates 18U65 (A) and 18D70 (B). Scale bars, 500 nm (A) and 200 nm (B).
16S rRNA gene analysis.
PCR for determination of the 16S rRNA gene sequence was performed as described above for DGGE analysis using archaeal 21F (5′-TTCCGGTTGATCCYGCCGGA-3′) and universal 1492R (Integrated DNA Technologies) primers. Amplicons were purified, cloned, sequenced, assembled, and analyzed according to previously published protocols (9). Pairwise alignment of 1,309 bp of isolate 18U65, isolate 18D70, and archaeal reference 16S rRNA gene sequences was performed using Clustal W (version 1.83) (45). Pairwise evolutionary distances were computed employing the correction of Jukes and Cantor (25). The neighbor-joining algorithm and bootstrap resampling were used to construct and evaluate the phylogenetic tree, respectively, using programs within the TREECON package (47) with Methanococcus vannielii as the outgroup.
qPCR analysis.
Quantitative PCR (qPCR) was used to determine the relative abundance values of isolate 18U65 and 18D70 DNA associated with S0 floc materials sampled from the sources of Dragon, Succession, and Beowulf Springs. At the laboratory, samples of S0 floc were treated to remove residual S0 and subjected to total DNA extraction as described above for DGGE analysis. DNA was quantified using the nucleic acid binding fluorochrome SYBR Green I (Invitrogen) added to achieve a final concentration of 1/1,000 (vol/vol), an ND-3300 Fluorospectrometer (NanoDrop Technologies, Wilmington, DE), and lambda phage DNA (Promega) as the standard. Strain-specific forward primers for isolate 18U65 and 18D70 were designed from the nearly full-length 16S rRNA gene. To ensure that qPCR amplicons generated in each reaction arose from the template of either isolate 18U65 or 18D70, forward primers were designed to contain 3- to 4-bp sequence mismatches compared to the 16S rRNA gene sequence of A. aceticus and Caldisphaera lagunensis, respectively. The primer pair 18U65-1062F (5′-GCTCTTAGTTGCTATCCC-3′) and 1392R and the pair 18D70-1073F (5′-CTGCGGGCGACCGTG-3′) and 1392R were used to amplify segments of the 16S rRNA gene from isolates 18U65 and 18D70, respectively. For each set of sequence-specific primers, a series of qPCRs was performed over a range of annealing temperatures, primer concentrations, and SYBR Green I concentrations to optimize the qPCR. qPCRs contained the same concentrations of reagents as used above for PCR with the following exceptions: primers were added to a final concentration of 400 nM, and SYBR Green I was added to a final dilution of 1.6× (diluted 1:6,250). A range of DNA template concentrations was used in the qPCRs, and the final reaction mixture volume was then adjusted to 25 μl with nuclease-free H2O (Sigma). qPCRs were performed in 0.5-ml optically pure PCR tubes (Corbett, Sydney, Australia) in a RotorGene3000 quantitative real-time PCR machine (Corbett) according to the following cycling conditions: initial denaturation (95°C for 10 min), followed by 35 cycles of denaturation (95°C for 10 s), annealing (56°C for 15 s), and extension (72°C for 20 s). Standards were prepared from genomic DNA for both isolates, and serial dilution of these standards were used for qPCR, resulting in a standard curve relating DNA template concentration to the qPCR threshold amplification signal generated for that amount of template. Abundance values for each isolate in each spring were generated by comparing qPCR threshold amplification signals from reaction mixtures containing known quantities of spring DNA extract to isolate-specific standard curves. Negative control qPCRs for isolate 18U65-specific primers contained isolate 18D70 genomic DNA as a template, and negative control qPCRs for 18D70-specific primers contained isolate 18U65 genomic DNA as a template. Criteria for selecting a given template dilution for reporting in the present study included the following: (i) the threshold amplification signal must have been generated between PCR cycles 10 to 25, (ii) the degree of similarity of DNA abundance values for a given template dilution must have resembled the DNA abundance values from the other template dilutions, and (iii) the template dilution must have yielded the lowest variability among replicates within a single template dilution. The standard deviation reported for qPCR results reflects the standard deviation of three replicate qPCRs for the selected template concentration.
G+C analysis.
G+C content (moles percent) was determined according to the methods of Gonzalez and Saiz-Jimenez (15) on a RotorGene3000 quantitative real-time PCR machine (Corbett) using Pseudomonas aeruginosa strain PAO1 and Escherichia coli strain K12 as reference strains. The standard deviation reported for G+C analysis reflects the deviation of three replicate determinations.
GDGT analysis.
Isolates 18U65 and 18D70 were cultivated in PS medium with the pH and incubation temperature adjusted to the optimum for each isolate to determine the glycerol dialkyl glycerol tetraether (GDGT) lipid composition. Cells were harvested in mid-exponential growth phase by centrifugation (10,000 × g for 20 min) and were resuspended in a minimal volume of S0-free PS medium. Cells were treated with CS2 to remove residual S0 as described above for DNA extraction. The S0-free cell pellet was then subjected to lipid analysis as described previously (40, 50).
Electron microscopy.
Logarithmic-phase cultures of isolates 18U65 and 18D70 were fixed in 0.5% glutaraldehyde for 8 h at their respective cultivation temperatures. Fifty milliliters of each fixed cell suspension was centrifuged (5,000 × g for 15 min), and the cell pellet was resuspended in 1 ml of glycine-HCl buffer (0.05 M; pH 3.0), transferred to a 1.0-ml microcentrifuge tube, and centrifuged (13,000 × g) to form an intact cell pellet. The cell pellet was dehydrated by overlaying a series of solutions of ethanol (30%, 50%, 70%, 90%, and 100%) and 100% propylene oxide for 30 min in each solution. The cell pellet was infiltrated using different mixtures of propylene oxide and Spurr's embedding resin (1:1, 1:2, and, finally, pure resin). The pellets were then transferred to vial block molds and overlaid with additional resin, and the resin was polymerized (at 70°C for 8 h). The polymerized resin blocks were removed from the molds and sectioned with an ultramicrotome, and the sections were stained with uranyl acetate and lead citrate before examination with a Zeiss LEO 912 AB electron microscope.
Nucleotide sequence accession numbers.
Nucleotide sequence accession numbers for 16S rRNA gene sequences used as reference for 16S rRNA gene qPCR primer design and phylogenetic analysis are as follows: Desulfurococcus amylolyticus, AF250331; Desulfurococcus fermentans, AY264344; Thermosphaera aggregans, X99556; Sulfophobococcus marinus, X99560; Pyrolobus fumarii, X99555; Pyrodictium occultum, M21087; Hyperthermus butylicus, X99553; Stetteria hydrogenophila, Y07784; Aeropyrum camini, AB109559; “Caldococcus noboribetus” (proposed name) strain NC12, D85038; A. aceticus, AF191225; C. lagunensis, AB087499; Pyrobaculum islandicum, L07511; Caldivirga maquilingensis, AB013926; Thermocladium modesties, AB005296; Stygiolobus azoricus, X90480; Sulfolobus metallicus, D85519; Acidianus brierleyi, X90477; Metallosphaera sedula, X90481; Ignisphaera aggregans strain AQ1, DQ060321; Methanococcus vannielii, M36507; Geogemma pacifica, DQ492259; uncultured Aquificaceae bacterium clone 1, DQ398776; and uncultured Aquificaceae bacterium clone 27, DQ398794. The sequences of the 16S rRNA genes for isolates 18U65 and 18D70 have been deposited in the GenBank, DDBJ, and EMBL databases under accession numbers EF057392 and EF057391, respectively.
RESULTS
Site description.
The aqueous- and solid-phase chemistry of Dragon Spring has been reported previously (29). Spring source water temperature ranged from 66 to 69°C, while the pH remained relatively stable at 2.9 to 3.1 over the course of the study. The spring contains abundant precipitated (flocculant) S0, extending from the source to approximately 4 m down the central transect of the main flow channel (Fig. 1). The aqueous- and solid-phase geochemistry of Succession Spring (69°C; pH 3.1) and Beowulf Spring (68°C; pH 3.0) have been reported previously (21).
Enrichment and isolation.
Incubation (65 or 70°C) of S0 flocs collected from the source of Dragon Spring in PS medium resulted in the production of H2S, indicating the presence of active, S0-respiring populations. DGGE analysis of the 65 and 70°C enrichment cultures indicated the presence of three and eight unique 16S rRNA gene phylotypes, respectively (data not shown). Following dilution to extinction (isolate 18U65) or increased incubation temperature (80°C), DGGE analysis of PCR-amplified 16S rRNA genes generated using multiple primer sets indicated a single 16S rRNA gene phylotype from cultures subjected to these different conditions (data not shown). Epifluorescence microscopic analysis of enrichments yielded a single morphotype in each culture, further indicating the presence of a clonal population of cells. Prior to characterization, cultures of each isolate were maintained at 65 (isolate 18U65) or 80°C (isolate 18D70) through serial transfer (5%, vol/vol) of log-phase cultures and routine (twice weekly) replacement of the headspace gas phase with N2.
Physiological properties.
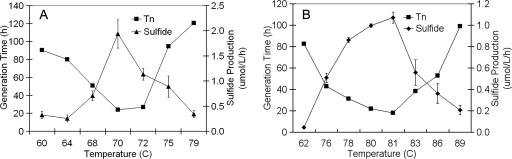
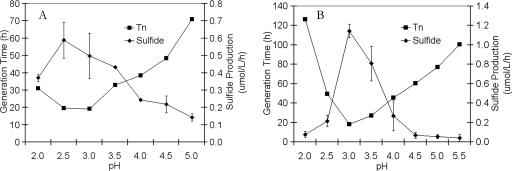
Isolate 18U65 grew over a temperature range of 60 to 79°C with an optimum of 70 to 72°C (Fig. 2) and over a pH range of 2.0 to 5.0 with an optimum of 2.5 to 3.0 (Fig. 3), whereas isolate 18D70 grew over a temperature range of 62 to 89°C with an optimum of 81°C (Fig. 2) and over a pH range of 2.0 to 5.5 with an optimum of 3.0 (Fig. 3). Isolate 18U65 grew over a Cl− concentration range of 0 to 86 mM, with an optimum of 17 to 34 mM, while isolate 18D70 grew over a Cl− concentration range of 0 to 128 mM, with an optimum of 12 mM. The shortest generation times for isolates 18U65 and 18D70 coincided with maximal H2S production (S0-reducing activity) (Fig. 2 and 3). The generation times of isolates 18U65 and 18D70, grown at optimal pH and temperature, were 19 and 17 h, respectively. Sulfide production paralleled culture growth, with maximum culture cell densities for isolates 18U65 and 18D70 being 4.6 × 106 and 2.1 × 107 cells ml−1, respectively.
FIG. 2.
Influence of temperature on generation time (Tn) and total sulfide production for isolates 18U65 (A) and 18D70 (B).
FIG. 3.
Influence of pH on generation time (Tn) and total sulfide production for isolates 18U65 (A) and 18D70 (B).
Both isolates required complex, organic carbon sources for growth. Isolate 18U65 could utilize yeast extract, peptone, tryptone, Casamino Acids, beef extract, glycogen, and gelatin while isolate 18D70 could utilize yeast extract, peptone, glycogen, and gelatin. Both isolates grew on pine needle extract in the absence of peptone, but neither isolate could utilize monomeric or polymeric carbohydrates, fatty acids, alkanes, alkenes, alcohols, ketones, organic acids, individual amino acids, or an equal molar mixture of amino acids. When isolate 18D70 was grown in PS medium, the total free amino acid concentration decreased from 116.4 to 93.9 pmol/μl following 120 h of growth (data not shown), indicating that the isolates could utilize free amino acids when other complex carbon sources were present but not as a sole carbon or energy source. Following peptide hydrolysis, the combined amino acids decreased from 1,017.7 pmol μl−1 at the time of inoculation to 520.3 pmol μl−1, suggesting that peptides are preferred over free amino acids to satisfy a nutritional requirement. The sole TEA utilized by both isolates was S0, although 18U65 could ferment yeast extract and peptone.
Neither isolate grew under the autotrophic conditions tested in this study. Supplementation of PS medium with H2 or CO2 did not enhance or inhibit S0-respiring activity of either isolate. S0 respiration and growth were inhibited when the concentration of oxygen in the aqueous phase was greater than 62 and 54 nM for isolates 18U65 and 18D70, respectively. Metabolic by-products detected in the cultures of the isolates grown in PS medium include H2S (both isolates) and NH4+ (isolate 18D70 only). Lactate, acetate, and H2 were not detected as metabolic products in the culture supernatant or headspace of either isolate grown in PS medium.
GDGT composition of crenarchaeal isolates.
GDGTs containing four to six cyclopentyl rings were present in isolates 18U65 and 18D70. GDGTs containing four rings (GDGT-4) represented 34% and 4% of the total GDGTs for isolates 18U65 and 18D70, respectively. GDGT-5′, which contains five cyclopentyl rings, represented 41 and 36% of the total GDGTs in isolate 18U65 and 18D70, respectively. GDGT-6, common to both isolates, represented 25 and 55% of the total GDGT in the lipid fraction of isolate 18U65 and 18D70, respectively, while the second six-ringed GDGT (GDGT-6′) was found only in isolate 18D70 (5% of total GDGTs).
Genetic properties of isolates.
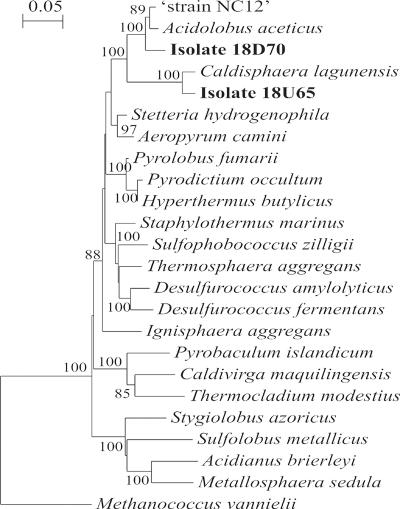
Nearly full-length 16S rRNA gene sequence analysis (corresponding to positions 27 to 1479 of the E. coli 16S rRNA gene) was determined for use in phylogenetic analysis of isolates 18U65 and 18D70. Both isolates clustered within the “Acidilobus group” (23) (phylum Crenarchaeota), with isolate 18U65 clustering within the C. lagunensis lineage (100% bootstrap support) and isolate 18D70 clustering within the A. aceticus lineage (100% bootstrap support) (Fig. 4). Analysis of a 1,430-bp 16S rRNA gene fragment from isolate 18U65 revealed 96% sequence homology with the 16S rRNA gene from C. lagunensis, whereas a 1,432-bp 16S rRNA gene fragment from isolate 18D70 was determined to be 94% similar to the 16S rRNA gene from A. aceticus. The 16S rRNA gene from the uncharacterized crenarchaeotal strain NC12 (“Caldococcus noboribetus” D85038) was 96 and 89% similar to the 16S rRNA gene of isolates 18D70 and 18U65, respectively. The 16S rRNA genes from isolate 18U65 and 18D70 were 88% similar to each other. The G+C content of DNA from isolates 18U65 and 18D70 were found to be 53.9 mol% ± 0.0 mol% and 59.9 mol% ± 0.4 mol%, respectively.
FIG. 4.
Phylogenetic relationships of isolate 18U65 and 18D70 and members of the Crenarchaeota based on comparison of 16S rRNA genes calculated using the neighbor-joining method. Bootstrap values (100 resamplings) are shown at branch points; values greater than 80 are reported. The bar represents 5 nucleotide substitutions per 100 nucleotides. Methanococcus vannielii was used as the outgroup.
Abundance of isolates associated with S0 precipitates.
Primers specific to the 16S rRNA gene of each isolate were used in qPCR to determine the abundance of their DNA relative to that of other members of the S0 precipitate-associated microbial communities of Dragon Spring, where the organisms were isolated, and of two other ASC springs in the Norris Geyser Basin: Beowulf and Succession Springs. The DNA of isolates 18U65 and 18D70 represented 20.2% ± 0.6% and 7.9% ± 0.1% of the DNA associated with Dragon Spring S0 precipitate, respectively; 32.3% ± 1.9% and 5.3% ± 0.6% of the DNA associated with S0 precipitate from Beowulf Spring, respectively; and 12.2% ± 0.8% and 5.6% ± 0.3% of the DNA associated with S0 precipitate from Succession Spring, respectively. There was no indication of a matrix-inhibitory effect in the qPCR assay using S0 precipitate DNA extracts from Dragon, Beowulf, and Succession Springs diluted 2,000-fold or more (data not shown). Treatment of S0 precipitate with CS2 prior to DNA extraction did not alter the results of the qPCR assay (data not shown), suggesting that CS2 treatment did not significantly alter the apparent community composition.
Morphology and ultrastructure.
Cells of isolates 18U65 and 18D70 were examined by epifluorescence and electron microscopy. Both isolates exhibited similar coccoid morphology and ultrastructure (Fig. 5). Cells of isolate 18U65 were 0.8 to 1.0 μm in diameter while cells of isolate 18D70 were 0.4 to 0.6 μm in diameter (Fig. 5). Both isolates were routinely observed singly or in pairs. Electron microscopy of thin sections revealed a cell envelope for both isolates that contained both a cytoplasmic membrane and an outer S-layer (Fig. 5).
Preservation.
Cultures of isolates 18U65 and 18D70 remained viable at room temperature when the headspace was purged with N2 and they were kept in the dark. Isolate 18D70 could be frozen (−80°C) in the presence of 10% (vol/vol) glycerol. Attempts to cryo-preserve cultures of 18U65 have been unsuccessful to date.
DISCUSSION
S0-reducing Crenarchaea have been detected by 16S rRNA gene-based diversity surveys in a variety of geographically distinct, sulfur-rich geothermal springs including those in YNP (31, 33), the Philippines (23), and Japan (3). A traditional laboratory cultivation and enrichment strategy based on aqueous- and solid-phase geochemistry led to the isolation of two novel S0-reducing Crenarchaea from flocs of S0 that had precipitated at the source of Dragon Spring. Comparative sequence analysis of the 16S rRNA gene from the isolates suggested that they each represent novel species within the “Acidilobus group” in the order Desulfurococcales (phylum Crenarchaeota): isolate 18U65 clustering within the C. lagunensis lineage and isolate 18D70 clustering within the A. aceticus lineage. To date, the A. aceticus lineage is comprised of the type strain A. aceticus (41) and the uncharacterized strain NC12 (“Caldococcus noboribetus”) (3), while the C. lagunensis lineage is comprised solely of the type strain (23).
Phenotypic characteristics determined for both isolates 18U65 and 18D70 support the 16S rRNA gene-based phylogenetic assessment placing them in the order Desulfurococcales within the Archaea. The recovery of tetraether-linked GDGTs from the lipid fraction of both isolate 18U65 and 18D70 further supports their placement in the Archaea (11, 28). Within the Archaea, the majority of thermoacidophiles belong to the Crenarchaeota (41), consistent with the clustering of both thermoacidophilic isolates in the crenarchaeal lineage. Within the crenarchaeal order Desulfurococcales, the predominant metabolism is the oxidation of complex organic compounds coupled with the reduction of S0 (7), a phenotype shared by both isolates. In addition, all characterized members of the order Desulfurococcales that are extreme acidophiles (optimum, pH <4.5) cluster solely within the “Acidilobus group” lineage, supporting phylogenetic characterization which placed both acidophilic isolates within this lineage.
A number of phenotypic traits of isolate 18U65 more closely resemble those of the C. lagunensis lineage than those of the A. aceticus lineage. Cardinal temperatures and pH of isolate 18U65 more closely resemble those of C. lagunensis than those of A. aceticus. The range of carbon sources utilized by and cellular morphology of isolate 18U65 also more closely resemble those of C. lagunensis than those of A. aceticus. While similar to C. lagunensis in many ways, a few phenotypic traits distinguish isolate 18U65 from C. lagunensis. Whereas C. lagunensis can utilize oxygen as a terminal electron acceptor, isolate 18U65 is a strict anaerobe incapable of growth in medium containing nanomolar concentrations of oxygen. Furthermore, C. lagunensis can respire fumarate and sulfate in addition to oxygen (23), while isolate 18U65 can only respire S0.
A number of phenotypic traits of isolate 18D70 more closely resemble those of the A. aceticus lineage than those of the C. lagunensis lineage. Like A. aceticus, but in contrast to C. lagunensis, isolate 18D70 is a strict anaerobe incapable of growth in medium containing nanomolar concentrations of oxygen. Cardinal temperatures and pH of isolate 18D70 more closely resemble those of A. aceticus than those of C. lagunensis. However, isolate 18D70 exhibits phenotypes which distinguish it from A. aceticus. For example, isolate 18D70 is unable to couple the oxidation of starch, soya extract, or beef extract to S0 respiration, all of which support S0 respiration in A. aceticus (41). Furthermore, unlike A. aceticus, isolate 18D70 is unable to support growth through fermentation pathways. Isolate 18D70 also differs from A. aceticus in cell morphology: cells of A. aceticus occur as irregular cocci with a diameter of 1 to 2 μm (41), whereas cells of isolate 18D70 occur as regular cocci with a much smaller diameter of 0.4 to 0.6 μm.
Lipid data are not reported in the characterization of A. aceticus (41), and while both acyclic and cyclic tetraethers are reported in the lipid fraction of C. lagunensis, a detailed description of the structures of these tetraethers is not provided (23). Previous studies have shown that genetically related organisms have similar GDGT profiles (30). While the GDGT profiles of isolates 18U65 and 18D70 are similar, it remains to be determined whether the GDGT composition of isolates 18U65 and 18D70 corroborates genetic and phenotypic properties that support the clustering of these organisms in the C. lagunensis and A. aceticus lineages, respectively.
Lodgepole pines are a predominate form of macrovegetation in the Norris Geyser Basin ecosystem (39), and needle litter is often found in the waters of ASC geothermal springs in this area (personal observation). Thus, needle litter represents a natural source of carbon and energy for heterotrophic consumers inhabiting ASC geothermal environments. Both isolates were capable of coupling S0 reduction with the oxidation of pine needle extract, suggesting a role for these microbes in the mineralization of complex natural organic matter (NOM) in geothermal environments. While NOM utilization has been demonstrated in other Archaeal phyla (17, 18, 32), the results of the current study represent the first time that NOM has been shown to be used as the sole carbon and energy source in the Crenarcheaota.
The results of the laboratory studies reported here indicate that both isolates 18U65 and 18D70 require S0 for respiration. Nine of the 12 recognized genera within the Desulfurococcales are capable of coupling oxidation of organic carbon or hydrogen with S0 reduction. Two genera, Ignicoccus (19) and Staphylothermus (4), can use only S0 as a TEA for respiration. Both Ignicoccus and Staphylothermus are thermophiles that inhabit marine hydrothermal vent ecosystems where S0 precipitates from H2S-containing vent fluids in a process similar to S0 precipitation in ASC springs in the Norris Geyser Basin (24, 29). Habitats such as these with a consistent supply of S0 likely promote the establishment of microbial populations that depend on S0 for respiration, such as Ignicoccus and Staphylothermus and their freshwater counterparts such as the isolates described above.
The greater abundance of isolate 18U65-like phylotypes than isolate 18D70-like phylotypes in the samples analyzed by qPCR may be due in part to their different laboratory-derived optimum temperatures (Topt) for growth. Whereas the Topt (70 to 72°C) of isolate 18U65 was within a few degrees Celsius of the temperature of the spring water where the isolates were recovered (66 to 69°C), the Topt (81°C) of isolate 18D70 was more than 10°C higher than the highest spring water temperature recorded in the spring during the course of this study (66 to 69°C). These results are consistent with those reported for other closely related microbial populations in thermal spring microbial communities (1, 12, 34). In an alkaline hot spring microbial mat, Synechococcus strain A (Topt, 55°C) was detected in an area of the mat exposed to a temperature of 56°C, while Synechococcus strain B (Topt, 50°C) was not. In contrast, Synechococcus strain B was detected in an area of the mat exposed to a temperature of 53°C, whereas Synechococcus strain A was absent (1, 12). Thus, dominance among populations with similar physiologies may be determined by how closely their Topt coincides with the temperature of the environment.
The differences in relative abundance of the two isolates in the S0 precipitate-associated microbial communities in the springs sampled in the present study may also reflect differences in their cell yields when S0 is used as a TEA. The cell yield of the more abundant isolate 18U65 (344 ± 186 pmol of C per nmol of S0) was significantly higher than that of the less abundant isolate 18D70 (39 ± 18 pmol of C per nmol of S0). The greater cell yield of isolate 18U65 than isolate 18D70 could contribute to the greater abundance of the former in the springs sampled in this study if these laboratory-based values apply to the spring environment.
In summary, two novel S0-reducing Crenarchaea were isolated that together represent a significant fraction of the microbial community associated with S0 precipitates in several ASC geothermal springs of Norris Geyser Basin in YNP. Both isolates are capable of utilizing a naturally occurring complex form of carbon as a carbon and energy source and naturally formed S0 as a TEA for respiration-dependent growth. On the basis of phylogenetic and physiological properties, it is proposed that isolates 18U65 and 18D70 represent distinct taxa within the C. lagunensis and A. aceticus lineages, respectively. We propose that isolate 18U65 be assigned to a new type species, Caldisphaera draconis, and that isolate 18D70 be assigned to a new type species, Acidilobus sulfurireducens.
Description of Caldisphaera draconis sp. nov.
Caldisphaera draconis (dra.co′ nis. L. gen. masc. n. draconis of/from dragon, as the organism was isolated from Dragon Springs, Yellowstone). Caldisphaera dracosis: hot spherical cell from Dragon Spring. Growth is anaerobic. Cells are coccoid, 0.8 to 1.0 μm in diameter, and are found singly or in pairs. Cell envelope contains a cytoplasmic membrane and an outer S-layer. Chemoorganotrophic metabolism capable of growth on tryptone, Casamino Acids, peptone, yeast extract, beef extract, glycogen, gelatin, and pine needle extract with S0 as sole TEA. Fermentative growth on peptone and yeast extract. Growth over a pH range of 1.5 to 5.0, a temperature range of 60 to 79°C, and a Cl− range of 0 to 86 mM. Optimal growth occurs at pH 2.5 to 3.0, 70 to 72°C, and 17 to 34 mM Cl−. Generation time under optimal conditions is 19 h, yielding a maximum cell density of 4.6 × 106 cells ml−1. Core lipid fraction contains GDGTs containing four to six cyclopentyl rings. Genomic DNA G+C content is 53.9 mol%. The type strain 18U65 was isolated from Dragon Spring, Norris Geyser Basin, YNP, WY.
Description of Acidilobus sulfurireducens sp. nov.
Acidilobus sulfurireducens (sul.fu.ri.re′ du.cens L. n. sulfur, L. part. adj. reducens leading back, reducing, N.L. part. adj. sulfurireducens reducing sulfur.). Acidicoccus sulfurireducens: acidiphilic coccus that reduces sulfur. Growth is anaerobic. Cells are coccoid, 0.4 to 0.6 μm in diameter, and occur singly or in pairs. The cell envelope contains a cytoplasmic membrane and outer S-layer. Growth occurs over a temperature range of 62 to 89°C, a pH range of 2.0 to 5.5, and a Cl− range of 0 to 128 mM. Optimal growth conditions are 81°C, pH 3.0, and 12 mM Cl−. Generation time under optimal conditions is 17 h, yielding a maximum cell density of 2.1 × 107 cells ml−1. Growth on peptone, yeast extract, glycogen, gelatin, and pine needle extract as carbon and energy source coupled to obligate S0 respiration. Ammonia is produced and excreted into culture medium. Strict anaerobe. Genomic DNA G+C content is 59.9 mol%. Core lipid fraction contains GDGTs containing four to six cyclopentyl rings. The source of isolation was Dragon Spring, Norris Geyser Basin, YNP, WY. The type strain is Acidilobus sulfurireducens strain 18D70.
Supplementary Material
Acknowledgments
We thank Sue Bromfield for assistance with electron microscopy and Kate McInnerney for assistance with genomic G+C mol% content determination. We are also grateful to Hans Trüper for assistance with etymology and Seth D'Imperio for performing gas-phase hydrogen measurements. Special thanks to Tim McDermott for helpful advice during manuscript preparation.
This research was supported by National Science Foundation grant MCB-0132022 to G.G.G. and National Science Foundation grant MCB-0348180 to C.L.Z., with a subcontract award to A.P. E.S.B. was supported by an Inland Northwest Research Alliance Graduate Fellowship. The United States Department of Energy also supported this research through Financial Assistance Award DE-FC09-96SR18546 to the University of Georgia Research Foundation (C.L.Z.).
Footnotes
Published ahead of print on 24 August 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allewalt, J. P., M. M. Bateson, N. P. Revsbech, K. Slack, and D. M. Ward. 2006. Effect of temperature and light on growth of and photosynthesis by Synechococcus isolates typical of those predominating in the Octopus Spring microbial mat community of Yellowstone National Park. Appl. Environ. Microbiol. 72:544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amend, J. P., and E. L. Shock. 2001. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 25:175-243. [DOI] [PubMed] [Google Scholar]
- 3.Aoshima, M., Y. Nishibe, M. Hasegawa, A. Yamagishi, and T. Oshima. 1996. Cloning and sequencing of a gene encoding 16S ribosomal RNA from a novel hyperthermophilic archaebacterium NC12. Gene 180:183-187. [DOI] [PubMed] [Google Scholar]
- 4.Arab, H., H. Volker, and M. Thomm. 2000. Thermococcus aegaeicus sp. nov. and Staphylothermus hellenicus sp. nov., two novel hyperthermophilic archaea isolated from geothermally heated vents off Palaeochori Bay, Milos, Greece. Int. J. Syst. Evol. Microbiol. 50:2101-2108. [DOI] [PubMed] [Google Scholar]
- 5.Atlas, R. M. 1997. Handbook of microbiological media. CRC Press, New York, NY.
- 6.Ball, J. W., R. B. McCleskey, D. K. Nordstrom, J. M. Holloway, and P. L. Verplanck. 2002. Water-chemistry data for selected springs, geysers, and streams in Yellowstone National Park, Wyoming 1999-2000. U.S Geological Survey open file report 02-382. U.S. Geological Survey, Reston, VA.
- 7.Blochl, E., R. Rachel, S. Burggraf, D. Hafenbradl, H. W. Jannasch, and K. O. Stetter. 1997. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of Archaea, extending the upper temperature limit for life to 113 degrees C. Extremophiles 1:14-21. [DOI] [PubMed] [Google Scholar]
- 8.Bonch-Osmolovskaya, E. A. 1994. Bacterial sulfur reduction in hot vents. FEMS Microbiol. Rev. 15:65-77. [Google Scholar]
- 9.Boyd, E. S., D. E. Cummings, and G. G. Geesey. 2007. Mineralogy influences structure and diversity of bacterial communities associated with geological substrata in a pristine aquifer. Microb. Ecol. 54:170-182. [DOI] [PubMed] [Google Scholar]
- 10.Burggraf, S., H. W. Jannasch, B. Nicolaus, and K. O. Stetter. 1990. Archaeoglobus profundus sp. nov., represents a new species within the sulfur-reducing Archaebacteria. Syst. Appl. Microbiol. 13:24-28. [Google Scholar]
- 11.De Rosa, M., and A. Gambacorta. 1988. The lipids of Archaebacteria. Prog. Lipid Res. 27:153-175. [DOI] [PubMed] [Google Scholar]
- 12.Ferris, M., and D. Ward. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63:1375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiala, G., and K. O. Stetter. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145:56-61. [Google Scholar]
- 14.Franson, M. A. H. 1987. Standard methods for the examination of water and wastewater, 17th ed. American Public Health Association, Washington, DC.
- 15.Gonzalez, J. M., and C. Saiz-Jimenez. 2002. A fluorimetric method for the estimation of G+C mol% content in microorganisms by thermal denaturation temperature. Environ. Microbiol. 4:770-773. [DOI] [PubMed] [Google Scholar]
- 16.Hedderich, R., O. Klimmech, A. Kröger, R. Dirmeier, M. Keller, and K. O. Stetter. 1999. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol. Rev. 22:353-381. [Google Scholar]
- 17.Herndl, G. J., T. Reinthaler, E. Teira, H. van Aken, C. Veth, A. Pernthaler, and J. Pernthaler. 2005. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71:2303-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoefs, M. J. L., S. Schouten, J. W. De Leeuw, L. L. King, S. G. Wakeham, and J. S. S. Damste. 1997. Ether lipids of planktonic Archaea in the marine water column. Appl. Environ. Microbiol. 63:3090-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber, H., S. Burggraf, T. Mayer, I. Wyschkony, R. Rachel, and K. Stetter. 2000. Ignicoccus gen. nov., a novel genus of hyperthermophilic, chemolithoautotrophic Archaea, represented by two new species, Ignicoccus islandicus sp. nov. and Ignicoccus pacificus sp. nov. Int. J. Syst. Evol. Microbiol. 50:2093-2100. [DOI] [PubMed] [Google Scholar]
- 20.Inskeep, W. P., G. G. Ackerman, W. P. Taylor, M. Kozubal, S. Korf, and R. E. Macur. 2005. On the energetics of chemolithotrophy in nonequilibrium systems: case studies of geothermal springs in Yellowstone National Park. Geobiology 3:297-317. [Google Scholar]
- 21.Inskeep, W. P., and T. R. McDermott. 2005. Geomicrobiology of acid-sulfate-chloride springs in Yellowstone National Park, p. 143-162. In W. P. Inskeep and T. R. McDermott (ed.), Geothermal biology and geochemistry in Yellowstone National Park, vol. 1. Montana State University, Bozeman. [Google Scholar]
- 22.Itoh, T., K. Suzuki, and T. Nakase. 1998. Thermocladium modestius gen. nov., sp. nov., a new genus of rod-shaped, extremely thermophilic crenarchaeote. Int. J. Syst. Bacteriol. 48:879-887. [DOI] [PubMed] [Google Scholar]
- 23.Itoh, T., K. Suzuki, P. C. Sanchez, and T. Nakase. 2003. Caldisphaera lagunensis gen. nov., sp. nov., a novel thermoacidophilic crenarchaeote isolated from a hot spring at Mt Maquiling, Philippines. Int. J. Syst. Evol. Microbiol. 53:1149-1154. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, C. R., H. W. Langner, J. Donahoe-Christiansen, W. P. Inskeep, and T. R. McDermott. 2001. Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ. Microbiol. 3:532-542. [DOI] [PubMed] [Google Scholar]
- 25.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-123. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 26.Kiyosu, Y., and M. Kurahashi. 1983. Origin of sulfur species in acid sulfate-chloride thermal waters, northeastern Japan. Geochim. Cosmochim. Acta 47:1237-1245. [Google Scholar]
- 27.Kletzin, A., T. Urich, F. Müller, T. M. Bandeiras, and C. M. Gomes. 2004. Dissimilatory oxidation and reduction of elemental sulfur in thermophilic Archaea. J. Bioenerg. Biomembr. 36:77-91. [DOI] [PubMed] [Google Scholar]
- 28.Koga, Y., and H. Morii. 2005. Recent advances in structural research on ether lipids from Archaea including comparative and physiological aspects. Biosci. Biotechnol. Biochem. 69:2019-2034. [DOI] [PubMed] [Google Scholar]
- 29.Langner, H. W., C. R. Jackson, T. R. McDermott, and W. P. Inskeep. 2001. Rapid oxidation of arsenite in a hot spring ecosystem, Yellowstone National Park. Environ. Sci. Technol. 35:3302-3309. [DOI] [PubMed] [Google Scholar]
- 30.Macalady, J. L., M. M. Vestling, D. Baumler, N. Boekelheide, C. W. Kaspar, and J. F. Banfield. 2004. Tetraether-linked membrane monolayers in Ferroplasma spp: a key to survival in acid. Extremophiles 8:411-419. [DOI] [PubMed] [Google Scholar]
- 31.Macur, R. E., H. W. Langner, B. D. Kocar, and W. P. Inskeep. 2004. Linking geochemical processes with microbial community analysis: successional dynamics in an arsenic-rich, acid-sulphate-chloride geothermal spring. Geobiology 2:163-177. [Google Scholar]
- 32.Martone, C. B., O. P. Borla, and J. J. Sanchez. 2005. Fishery by-product as a nutrient source for bacteria and archaea growth media. Bioresour. Technol. 96:383-387. [DOI] [PubMed] [Google Scholar]
- 33.Meyer-Dombard, D. R., E. L. Shock, and J. P. Amend. 2005. Archaeal and bacterial communities in geochemically diverse hot springs of Yellowstone National Park, USA. Geobiology 3:211-227. [Google Scholar]
- 34.Miller, S. R., and R. W. Castenholz. 2000. Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 66:4222-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montegrossi, G., F. Tassi, O. Vaselli, A. Buccianti, and K. Garofalo. 2001. Sulfur species in volcanic gases. Anal. Chem. 73:3709-3715. [DOI] [PubMed] [Google Scholar]
- 36.Nagata, T. 1986. Carbon and nitrogen content of natural planktonic bacteria. Appl. Environ. Microbiol. 52:28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nordstrom, D. K., J. W. Ball, and R. B. McCleskey. 2005. Ground water to surface water: chemistry of thermal outflows in Yellowstone National Park, p. 143-162. In W. P. Inskeep and T. R. McDermott (ed.), Geothermal biology and geochemistry in Yellowstone National Park, vol. 1. Montana State University, Bozeman. [Google Scholar]
- 38.Nordstrom, D. K., J. W. Ball, and R. B. McCleskey. 2004. Oxidation reactions for reduced Fe, As, and S in thermal outflows of Yellowstone National Park: biotic or abiotic?, p. 59-62. In R. B. Wanty and R. R. Seal II (ed.), Water-rock interaction. Taylor and Francis Group, London, United Kingdom.
- 39.Norris, T. B., J. M. Wraith, R. W. Castenholz, and T. R. McDermott. 2002. Soil microbial community structure across a thermal gradient following a geothermal heating event. Appl. Environ. Microbiol. 68:6300-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson, A., Z. Huang, A. E. Ingalls, C. S. Romanek, J. Wiegel, K. H. Freeman, R. H. Smittenberg, and C. L. Zhang. 2004. Nonmarine crenarchaeol in Nevada hot springs. Appl. Environ. Microbiol. 70:5229-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prokofeva, M., M. Miroshnichenko, N. Kostrikina, N. Chernyh, B. Kuznetsov, T. Tourova, and E. Bonch-Osmolovskaya. 2000. Acidilobus aceticus gen. nov., sp. nov., a novel anaerobic thermoacidophilic archaeon from continental hot vents in Kamchatka. Int. J. Syst. Evol. Microbiol. 50:2001-2008. [DOI] [PubMed] [Google Scholar]
- 42.Reysenbach, A.-L., Y. Liu, A. B. Banta, T. J. Beveridge, J. D. Kirshtein, S. Schouten, M. K. Tivey, K. L. Von Damm, and M. A. Voytek. 2006. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature 442:444-447. [DOI] [PubMed] [Google Scholar]
- 43.Schauder, R., and A. Kröger. 1993. Bacterial sulphur respiration. Arch. Microbiol. 159:491-497. [Google Scholar]
- 44.Steudel, R. 1996. Mechanism for the formation of elemental sulfur from aqueous sulfide in chemical and microbiological desulfurization processes. Ind. Eng. Chem. Res. 35:1417-1423. [Google Scholar]
- 45.Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyson, G. W., I. Lo, B. J. Baker, E. E. Allen, P. Hugenholtz, and J. F. Banfield. 2005. Genome-directed isolation of the key nitrogen fixer Leptospirillum ferrodiazotrophum sp. nov. from an acidophilic microbial community. Appl. Environ. Microbiol. 71:6319-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biol. Sci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 48.Widdel, F., G.-W. Kohring, and F. Mayer. 1983. Studies on the dissimilatory sulfate that decomposes fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov., sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-294. [Google Scholar]
- 49.Xu, Y., A. A. Schoonen, D. K. Nordstrom, K. M. Cunningham, and J. W. Ball. 1998. Sulfur geochemistry of hydrothermal waters in Yellowstone National Park: I. The origin of thiosulfate in hot spring waters. Geochim. Cosmochim. Acta 62:3729-3743. [Google Scholar]
- 50.Zhang, C. L., A. Pearson, Y.-L. Li, G. Mills, and J. Wiegel. 2006. Thermophilic temperature optimum for crenarchaeol synthesis and its implication for archaeal evolution. Appl. Environ. Microbiol. 72:4419-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zillig, W., S. Yeats, I. Holz, A. Bock, M. Rettenberger, F. Gropp, and G. Simon. 1986. Desulfurolobus ambivalens, gen. nov., sp. nov., an autotrophic archaebacterium facultatively oxidizing or reducing sulfur. Syst. Appl. Microbiol. 8:197-203. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.