Abstract
The Ku70 protein, a product of the XRCC6 gene, is a component of the nonhomologous end-joining (NHEJ) pathway of DNA repair, which protects cells from the effects of radiation-induced DNA damage. Although the spatial expression of Ku70 during vertebrate embryogenesis has not been described, DNA repair proteins are generally considered to be “housekeeping” genes, which are required for radioprotection in all cells. Here, we report the cloning and characterization of the zebrafish Ku70 ortholog. In situ hybridization and RT-PCR analyses demonstrate that Ku70 mRNA is maternally provided and expressed uniformly among embryonic blastomeres. Later during embryogenesis, zygotically transcribed Ku70 mRNA specifically accumulates in neural tissue, including the retina and proliferative regions of the developing brain. In the absence of genotoxic stress, morpholino-mediated knockdown of Ku70 expression does not affect zebrafish embryogenesis. However, exposure of Ku70 morpholino-injected embryos to low doses of ionizing radiation leads to marked cell death throughout the developing brain, spinal cord, and tail. These results suggest that Ku70 protein plays a crucial role in protecting the developing nervous system from radiation-induced DNA damage during embryogenesis.
Keywords: Ku protein, Ku70, XRCC6, embryo, ionizing radiation, nonhomologous end-joining, DNA repair, TUNEL, apoptosis, zebrafish
The physical interaction of ionizing radiation with duplex DNA results in double-strand breaks (DSBs), which are one of the most potent types of DNA lesion. In proliferating cells, the cytotoxic effects of DSBs can be acute or latently manifest as chromosomal translocations and other genomic aberrations [1]. Natural sources of DSBs include ionizing radiation and, to a lesser extent, byproducts of normal metabolic processes, such as reactive oxygen species. To mitigate the biological effects of DSBs, organisms have developed protective strategies that sense and rapidly repair this type of DNA damage. Following DSB induction, chromosomal integrity is typically reestablished either by the nonhomologous end-joining (NHEJ) or homologous recombination pathways of DNA repair (reviewed in [2, 3]).
The NHEJ pathway of DSB repair requires the function of at least five genes: XRCC5, which encodes the Ku80 protein [4-6], XRCC6, which encodes the Ku70 protein [7-9], LIG4 [10-13], XRCC4 [14], and PRKDC [15-18]. Although each of these NHEJ components are required for DNA repair activity in vitro and are necessary for radioprotection in vivo (e.g., postnatal or adult stages), the expression and function of many of these genes during embryogenesis are largely unknown. In part, this is due to the a priori assumption that many, if not most, of the DNA repair proteins are expressed ubiquitously. However, in contrast to this expectation, we have shown that expression of Ku80 mRNA varies greatly among tissues during embryogenesis, with expression highest in developing neural tissues [19].
To determine whether the expression of other NHEJ DNA repair components are similarly tissue-specific during embryogenesis, we cloned and characterized the zebrafish ortholog of mammalian Ku70 protein. Ku70 and Ku80 proteins form a stable molecular complex that binds to broken DNA ends and recruits additional NHEJ proteins. However, Ku70 also has unique subunit-specific functions, notably the inhibition of Bax-mediated apoptosis [20-22]. The existence of subunit-specific functions raises the possibility that the pattern of mRNA expression, or the phenotype associated with attenuation of function, might differ for the two Ku subunits.
All experimental protocols using zebrafish (Danio rerio) were reviewed and approved by the MCG Institutional Animal Care and Use Committee. Breeding and staging of zebrafish embryos was performed according to standard protocols [23] and all experiments were carried out with Tuebingen Wild-Type or brass embryos. Each experiment was repeated using at least 2 different embryo clutches, where each clutch contained 200 or more embryos and a minimum of 20 embryos per condition.
To clone the zebrafish Ku70 cDNA we used the sequence of an existing zebrafish Ku70 EST (Accession number: BC053270) to generate Ku70-specific PCR primers, Ku70u (5′-CTCTAGCTAGCGCAAAAGAGAA) and Ku70d (5′-TTACAAATTTCAAGCATTTATTGAATC). Embryo RNA was prepared using the Trizol method (Invitrogen, Carlsbad, California) and first strand cDNA using Omniscript RT (Qiagen, Valencia, CA). Full-length Ku70 cDNA was obtained by PCR and cloned using the pGEM-Teasy Vector System (Promega Corp., Madison, WI). The identities of Ku70 cDNA clones were confirmed by sequencing of both DNA strands.
Whole-mount in situ hybridization was carried out under standard conditions [24]. Riboprobe templates were prepared by linearizing cDNA plasmids with the appropriate restriction enzyme (sense, BstX1; antisense, SacII) and transcribing with either with T7 (sense) or SP6 (antisense) RNA polymerase in the presence of digoxigenin-labeled nucleotides (Roche Applied Science, Indianapolis, USA). Sense riboprobes were used for control hybridization reactions and resulted in only low level diffuse staining.
Ku70 and control morpholino oligonucleotides (MOs) were designed using cDNA or genomic sequence (Chromosome 12; zebrafish genome assembly Zv6) and purchased from Gene-Tools (Philomath, OR). Their sequences are: atgMO (5′-ATTTCCCCAGTTCGCCATTAAAGTT), atgmmMO (5′-ATTACCCCACTTCGCGATTTAAGAT), sd2MO (5′-AACTTTTTAGGCTCACCTGCATAGT), and sd2mmMO (5′-AACATTTTTACGCTCTCCTGGATACT), with mismatch nucleotides underlined. MOs were diluted to 5 μg/μl in 2× injection buffer [25] and microinjected into the yolk of 1-cell embryos. Titration experiments determined that 4-5 ng of injected MO was not toxic, but sufficient to generate a radiosensitive phenotype. Experiments were repeated 2-3 times using at least two MO doses.
Irradiation (137Cs) was performed using a Gammacell Exactor (MDS Nordion, Ottawa, ON). Dosimetry was performed using thermoluminescent dosimetry devices (Landauer Inc, Glenwood, IL) irradiated simultaneously with embryos. Irradiated embryos were fixed at 24 hpf and assayed by the TUNEL method [19].
An alignment of zebrafish Ku70 amino acid sequence with representative vertebrate orthologs is shown in Fig. 1A. The full-length zebrafish Ku70 cDNA (GenBank accession number: DQ859046) encodes a predicted peptide of 610 amino acids, which is 59% identical and 77% similar to the human Ku70 peptide. Moreover, the domain structure of zebrafish Ku70 is similar to other Ku70 proteins, with an N-terminal von Willebrand Factor type A domain (VWA) domain, a central Ku core region, a Ku70 C-terminal arm domain (Ku_C), and an SAP domain that mediates Ku subunit-specific functions (Fig. 1B). Nucleotide sequence comparison of our cloned cDNA with the original Ku70 EST (GenBank accession number: BC053270) identified a single nucleotide deletion (1169delA) in the EST. This nucleotide (1169A) is present, not only in our cDNA clone, but also in the Ku70 genomic sequence (Chromosome 12). Because the deletion in the EST results in a predicted frameshift and premature termination codon (L410X), it seems likely that the EST sequence is erroneous.
Figure 1.
(A) Predicted amino acid sequence of zebrafish Ku70 aligned with that of human, mouse, and Xenopus. Identical residues in all sequences are marked with an “*”, conserved substitutions with a “:”, and semi-conservative substitutions with a “;”. Color coding of specific domains is according to the schematic in Panel B. (B) Cartoon representation of Ku70 peptide showing the domain structure and organization of zebrafish Ku70. Similar to other Ku70 proteins, zebrafish Ku70 has an N-terminal von Willebrand Factor type A domain (VWA), a central Ku core region, a Ku C-terminal arm, and SAP domain. The SAP domain (named after SAF-A/B, Acinus and PIAS motifs) is found in diverse nuclear proteins. The amino acid similarity between specific domains of zebrafish Ku70 and human Ku70 are shown below the structure, with amino acid identity in parentheses.
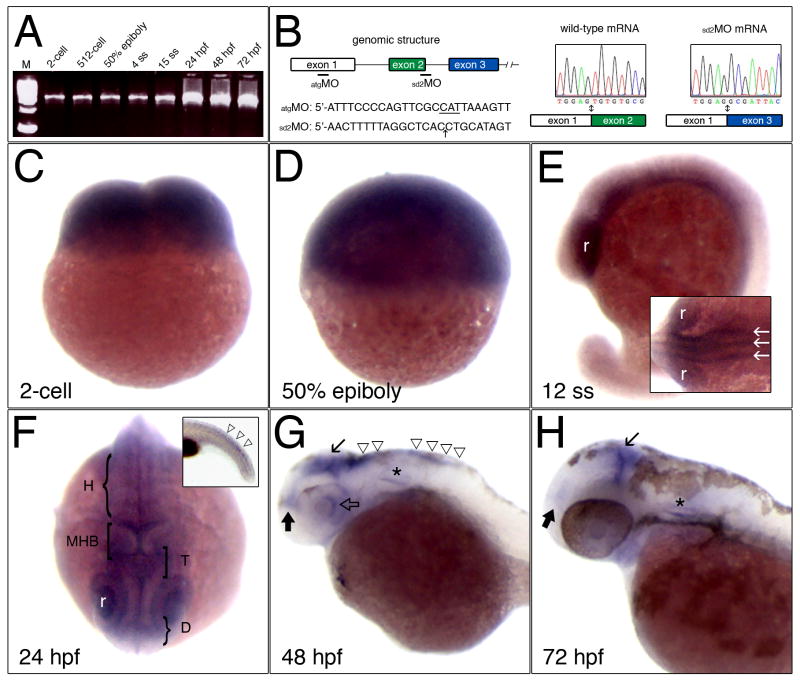
To determine the temporal expression of zebrafish Ku70 mRNA during embryonic development we performed RT-PCR using Ku70-specific primers and RNA prepared from different embryonic stages (Fig. 2A; equal inputs of RNA). Low levels of maternally provided Ku70 mRNA are present at the 2-cell and 512-cell stages of embryogenesis. After the onset of zygotic transcription, levels of Ku70 mRNA remain low at the beginning of gastrulation (50% epiboly) and throughout somite stages. Expression levels of Ku70 mRNA then increase at 24 hpf and remain elevated through at least 72 hpf.
Figure 2.
Expression of Ku70 mRNA during embryogenesis. (A) Temporal expression of Ku70 mRNA by RT-PCR analysis. Stages are as indicated; ss, somite stage; hpf, hours post-fertilization, M, DNA size marker. (B) Genomic structure of exons 1-3 and the locations of morpholino oligonucleotides. Electropherograms to the right show the wild-type exon1/exon2 boundary and the aberrant splicing of exon 1 to exon 3 in sd2MO-injected embryos. (C-H) Expression of zebrafish Ku70 by in situ hybridization. (C) 2-cell stage. (D) 50% epiboly (∼6 hours post-fertilization). (E) 12 somite stage (ss), white arrows indicate three longitudinal proneural domains in the neural keel; r, retina. (F) 24 hpf; H, hindbrain; MHB, midbrain hindbrain boundary; T, tectum; D, diencephalon. Inset shows Ku70 mRNA expression in the developing tail (open triangles). (G) 48 hpf and (H) 72 hpf; Line arrow marks the tectum, solid black arrow marks the diencephalon, and triangles indicate the hindbrain. The otic vesicle is marked by an asterisk. Panels C, D, animal pole is toward top. Panels E, G, inset to F, and H are lateral views with anterior to left and dorsal toward top. Panel F is dorsal view, anterior toward bottom.
We determined the spatial expression of Ku70 mRNA during embryonic development by whole mount in situ hybridization (Fig. 2C-H). At the 2-cell stage, maternally provided Ku70 mRNA is uniformly expressed among blastomeres. Ku70 mRNA continues to accumulate uniformly among blastomeres during gastrulation but then becomes spatially restricted during mid-somitogenesis stages. At the 12 somite stage, Ku70 mRNA is localized to the retina and ventral lateral regions of the head (Fig. 2E). From a dorsal view (inset, Fig. 2E), longitudinal stripes of Ku70 mRNA expression in the central nervous system are consistent with domains of neural fate specification in the neural keel. By 24 hpf, Ku70 expression becomes restricted to regions of the retina, diencephalon, tectum, and hindbrain (Fig. 2F). By increasing the duration of the staining reaction, Ku70 mRNA expression becomes apparent in the developing tail (inset, Fig. 2F). At 48 hpf, Ku70 mRNA is expressed in the posterior portion of the retina and dorsal surfaces of the diencephalon, tectum, and hindbrain (Fig. 2G). By 72 hpf, lower levels of Ku70 mRNA are detected in the diencephalon and tectum (Fig. 2H). The domains of zebrafish Ku70 mRNA expression are similar to those of Ku80 and suggest that, like Ku80, Ku70 is predominantly expressed in proliferative regions of the developing nervous system, such as the ventricular zones of the brain and presumptive ganglion cell layer of the retina [19].
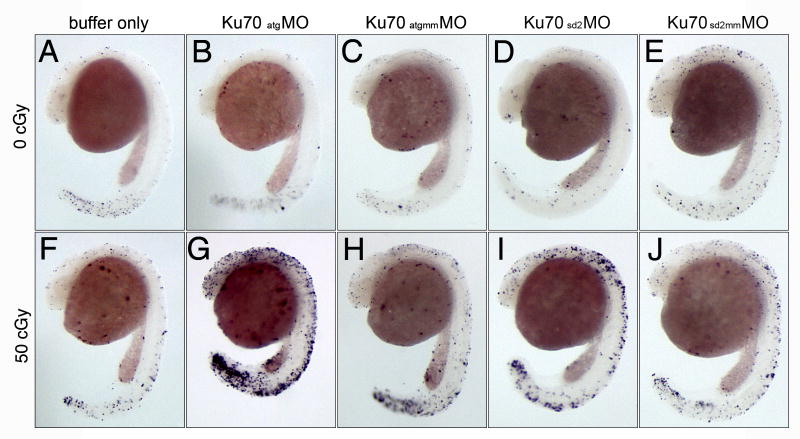
To determine whether expression of Ku70 is required for neural development, we designed an antisense MO [26] complementary to the Ku70 translation initiation site (atgMO; Fig. 2B) and a control MO containing a 5-base mismatch (atgmmMO). Microinjection of Ku70 or control morpholinos at the 1-cell stage did not affect normal embryonic development, which was similar to embryos microinjected with buffer only. To examine more subtle effects on embryogenesis, we performed TUNEL staining to detect cell death (Fig. 3). Neither the Ku70 nor the control mismatch MO increased levels of cell death in embryos assayed at 24 hpf (Fig. 3A-C).
Figure 3.
Expression of Ku70 is required for radioprotection. Whole mount TUNEL assay of zebrafish embryos at 24 hpf. 1-cell stage embryos were microinjected with buffer alone (A, F), Ku70 atgMO (B, G), Ku70 atgmmMO (C, H), Ku70 sd2MO (D, I), or Ku70 sd2mmMO (E, J). At 6 hpf, half of the embryos from each injection group were irradiated with 50 centiGray (cGy) of ionizing radiation (panels F-J) and the remaining embryos served as non-irradiated controls (panels A-E). All embryos were then allowed to develop to 24 hpf, when they were fixed and processed for the TUNEL assay. All panels are lateral view, head is to the top and tail is toward bottom.
In contrast to non-irradiated embryos, irradiation of embryos microinjected with atgMO resulted in a profound effect on gross morphology and cell death. Fig. 3G shows a representative embryo that was microinjected at the 1-cell stage with atgMO, exposed to a low dose of ionizing radiation at 6 hpf, and analyzed by TUNEL staining at 24 hpf. Elevated levels of TUNEL positive cells are seen in the central nervous system, retina, and tail. This hypersensitivity to radiation was not seen in embryos that were microinjected with buffer alone or the control MO (Fig 3F, H).
The Ku70 atgMO is predicted to affect mRNA translation, but an antibody specific to the zebrafish Ku70 protein is not available, so it is not possible to assess the level of knockdown by monitoring changes in Ku70 mRNA or protein. We, therefore, tested a second Ku70 MO that was complementary to the splice donor sequence of intron 2 (sd2MO; Fig. 2B). To confirm that Ku70 mRNA splicing is altered with the sd2MO, we cloned Ku70 cDNAs from buffer-, sd2MO-, and sd2mmMO-injected embryos. By using PCR primers that amplify the complete Ku70 cDNA, we can detect both wild-type and abnormally spliced Ku70 mRNAs (data not shown). Ku70 cDNAs derived from buffer-injected embryos (2/2 clones) and sd2mmMO injected embryos (2/2 clones) only contained wild-type sequence (Fig. 2B). In contrast, all Ku70 cDNAs isolated from sd2MO injected embryos (6/6 clones) had a complete deletion of exon 2, which juxtaposes exon 3 downstream of exon 1 (Fig. 2B) and results in a frameshift and a premature termination codon. The 30 amino acid truncated product is predicted to lack regions of Ku70 that are essential for DNA binding and repair activity [27-29].
Similar to the Ku70 atgMO, an increase in TUNEL-positive cells was seen in irradiated embryos injected with the Ku70 splice MO, but not the corresponding mismatched MO (Fig. 3I, J). Comparison of different MO-injected embryos (Fig. 3G, I), reveals that the phenotype of sd2MO-injected embryos was consistently less severe (e.g., larger eyes, more brain tissue, and normal body axis) than atgMO-injected embryos, including lower levels of TUNEL staining. Although we did not recover any correctly spliced Ku70 cDNA clones from sd2MO-injected embryos, we cannot rule out the possibility that persistence of a small amount of correctly spliced Ku70 mRNA is sufficient to modify the radiosensitive phenotype in vivo.
In summary, we have described here the expression and embryonic function of Ku70, the product of the XRCC6 gene. Although DNA repair proteins are often considered to be housekeeping enzymes, zygotically expressed Ku70 mRNA accumulates in a tissue-specific pattern during organogenesis and later stages, similar to what was seen previously for Ku80 [19]. Specifically, spatial expression of zebrafish Ku70 mRNA becomes restricted to the developing central nervous system, which would not have been predicted by enzymatic function alone, and suggests a role for this DNA repair protein in formation of neural tissue. Consistent with Ku70 and Ku80 expression [19], LIG4 (another component of the NHEJ pathway) is required for nervous system development in mice, even in the absence of radiation [30-33]. The tissue-specific accumulation of Ku70 mRNA in zebrafish suggests a role for Ku70 in protecting populations of rapidly proliferating cells, like those in the nervous system [34]. Indeed, knockdown of Ku70 expression sensitizes embryos to a 50 cGy dose of ionizing radiation by increasing cell death in the brain and spinal cord. The consequences of radiation exposure during brain development are well-documented [35] and further study of NHEJ protein function during embryogenesis may provide new insights to the protective mechanisms and consequences of low dose radiation exposure, particularly in sensitive populations such as children and pregnant women [36, 37].
Acknowledgments
We thank Dr. B. Yuan and the MCG Transgenic Zebrafish Core Facility for embryo production. Support for this work was provided by grant awards to WSD (DE-FG02-03ER63649 from the U.S. Department of Energy Low Dose Radiation Research Program) and DJK (R01 DC006140 from the National Institutes of Health).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ward JF. Radiation mutagenesis: the initial DNA lesions responsible. Radiat Res. 1995;142(3):362–368. [PubMed] [Google Scholar]
- 2.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005;4(6):639–48. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Wyman C, Ristic D, Kanaar R. Homologous recombination-mediated double-strand break repair. DNA Repair (Amst) 2004;3(89):827–33. doi: 10.1016/j.dnarep.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 4.Getts RC, Stamato TD. Absence of a Ku-like DNA end binding activity in the xrs double-strand DNA repair-deficient mutant. J Biol Chem. 1994;269(23):15981–15984. [PubMed] [Google Scholar]
- 5.Smider V, Rathmell WK, Lieber MR, Chu G. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science. 1994;266(5183):288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 6.Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265(5177):1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 7.Gu Y, Jin S, Gao Y, Weaver DT, Alt FW. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci U S A. 1997;94(15):8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, Davidson L, Cheng HL, Sekiguchi JM, Frank K, Stanhope-Baker P, Schlissel MS, Roth DB, Alt FW. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7(5):653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 9.Ouyang H, Nussenzweig A, Kurimasa A, Soares VC, Li X, Cordon-Cardo C, Li W, Cheong N, Nussenzweig M, Iliakis G, Chen DJ, Li GC. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination In vivo. J Exp Med. 1997;186(6):921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 11.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol. 1997;7(8):588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 12.Teo SH, Jackson SP. Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. Embo J. 1997;16(15):4788–95. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schar P, Herrmann G, Daly G, Lindahl T. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 1997;11(15):1912–24. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, Stamato TD, Taccioli GE, Alt FW. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83(7):1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 15.Blunt T, Finnie NJ, Taccioli GE, Smith GC, Demengeot J, Gottlieb TM, Mizuta R, Varghese AJ, Alt FW, Jeggo PA, Jackson SP. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 16.Kirchgessner CU, Patil CK, Evans JW, Cuomo CA, Fried LM, Carter T, Oettinger MA, Brown JM. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 17.Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day RS, Barron GM, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 18.Peterson SR, Kurimasa A, Oshimura M, Dynan WS, Bradbury EM, Chen DJ. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc Natl Acad Sci U S A. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bladen CL, Lam WK, Dynan WS, Kozlowski DJ. DNA damage response and Ku80 function in the vertebrate embryo. Nucleic Acids Res. 2005;33(9):3002–10. doi: 10.1093/nar/gki613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawada M, Sun W, Hayes P, Leskov K, Boothman DA, Matsuyama S. Ku70 suppresses the apoptotic translocation of Bax to mitochondria. Nat Cell Biol. 2003;5(4):320–9. doi: 10.1038/ncb950. [DOI] [PubMed] [Google Scholar]
- 21.Qin Q, Patil K, Sharma SC. The role of Bax-inhibiting peptide in retinal ganglion cell apoptosis after optic nerve transection. Neurosci Lett. 2004;372(12):17–21. doi: 10.1016/j.neulet.2004.08.075. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian C, Opipari AW, Jr, Bian X, Castle VP, Kwok RP. Ku70 acetylation mediates neuroblastoma cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2005;102(13):4842–7. doi: 10.1073/pnas.0408351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio) 3rd. Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- 24.Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the Zebrafish Snail1 Gene and Its Expression in Wild-Type, Spadetail and No Tail Mutant Embryos. Development. 1993;119(4):1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 25.Kozlowski DJ, Murakami T, Ho RK, Weinberg ES. Regional cell movement and tissue patterning in the zebrafish embryo revealed by fate mapping with caged fluorescein. Biochem Cell Biol. 1997;75(5):551–562. [PubMed] [Google Scholar]
- 26.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26(2):216–20. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Dong X, Kyungjae M, Hendrickson EA, Reeves WH. Identification of Two Domains of the p70 Ku Protein Mediating Dimerization with p80 and DNA Binding. The Journal of Biological Chemistry. 1998;273:842–848. doi: 10.1074/jbc.273.2.842. [DOI] [PubMed] [Google Scholar]
- 28.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412(6847):607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 29.Jin S, Weaver DT. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO Journal. 1997;16:6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol. 1998;8(25):1395–8. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 31.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396(6707):173–7. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 32.Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, Bronson RT, Malynn BA, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin SH, Greenberg ME, Alt FW. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95(7):891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 33.Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Ferguson DO, Zhu C, Manis JP, Horner J, DePinho RA, Alt FW. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell. 2000;5(6):993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 34.Wullimann MF, Knipp S. Proliferation pattern changes in the zebrafish brain from embryonic through early postembryonic stages. Anat Embryol (Berl) 2000;202(5):385–400. doi: 10.1007/s004290000115. [DOI] [PubMed] [Google Scholar]
- 35.Streffer C, Shore R, Konermann G, Meadows A, Uma Devi P, Preston J, Holm LE, Stather J, Mabuchi K, Withers HR. Biological effects after prenatal irradiation (embryo and fetus). A report of the International Commission on Radiological Protection. Ann ICRP. 2003;33(12):5–206. [PubMed] [Google Scholar]
- 36.Brooks AL. Developing a scientific basis for radiation risk estimates: goal of the DOE Low Dose Research Program. Health Phys. 2003;85(1):85–93. doi: 10.1097/00004032-200307000-00016. [DOI] [PubMed] [Google Scholar]
- 37.Coleman CN, Stone HB, Moulder JE, Pellmar TC. Medicine. Modulation of radiation injury. Science. 2004;304(5671):693–4. doi: 10.1126/science.1095956. [DOI] [PubMed] [Google Scholar]