Abstract
This article reviews genetically encoded Ca2+ indicators (GECIs), with a focus on the use of these novel molecules in the context of understanding complex cell signalling in mammals, in vivo. The review focuses on the advantages and limitations of specific GECI design strategies and the results of experiments in which these molecules have been expressed in transgenic mice, concentrating particularly on recent experiments from our laboratory in which physiological signalling could be monitored in vivo. Finally, newer strategies for effective genetic specification of GECIs are briefly reviewed.
Physiological processes involve dynamic interactions between cells of different lineages that are mediated by changes in the concentration or structure of cellular molecules that occur on time scales from milliseconds to days. In practice, molecular scale information about such processes is often limited to average measurements of the concentration or biochemical state of a set of molecules obtained from many millions of cells at a single point in time, or the non-quantitative determination of the presence of specific molecules in fixed or frozen tissues. As an alternative, in vitro preparations of cells isolated from the tissue of interest can be studied by a number of techniques that readily provide real-time information at the molecular level. However, almost without exception these preparations inadequately represent the more complex physiological processes of interest. By analogy, imagine the performance of a great symphony orchestra reduced to periodic samples of the sound produced at a few time points, or to the performance of a single orchestra member playing in isolation from all surrounding cues. For the past decade progressive advances have been made in the design of proteins that serve as optical indicators of cellular behaviour at the molecular level and that can be genetically specified; these sensors can serve as the microphones that allow us to listen to individual components of the cellular symphony in live performance. Moreover, parallel advances in genetic technologies have enabled precise targeting of the sensors to key lineages and subcellular locations, or, to extend the analogy, allowing the placement of the microphone on each individual instrument in turn, or on several distinct instruments simultaneously.
Despite the obvious promise of engineering molecular sensors for use in vivo to understand complex physiology and pathophysiology, genetically encoded sensors have had limited application in mammals in vivo largely due to significant technical challenges associated with this application. Thus, while genetically encoded sensors of Ca2+, voltage and small molecules have been used effectively in lower organisms (Bacskai et al. 1993; Kerr et al. 2000; Ng et al. 2002; Higashijima et al. 2003; Fiala & Spall, 2003; Wang et al. 2003) and in vitro mammalian cells (Adams et al. 1991; Romoser et al. 1997; Miyawaki et al. 1997; Siegel & Isacoff, 1997; Honda et al. 2001; Zaccolo & Pozzan, 2002), their use in mammalian cells in vivo, the experimental context in which they provide the most significant advantage, has been surprisingly restricted. The failure to exploit the potential of protein-based signalling molecules is a consequence of the more demanding design criteria required for their effective use in mammalian cells in vivo: sensor signal strength, stability, transition kinetics, and interactions between the sensor and cellular molecules have proven to be major obstacles. However, substantial progress has been made in the modification or redesign of several optical sensors (Griesbeck et al. 2001; Truong et al. 2001; Guerrero et al. 2002; Karasawa et al. 2004; Tallini et al. 2006a; Mank et al. 2006), justifying the significant investment required to develop mammalian preparations in which a wide range of complex biological questions can be investigated. Here I will review the development and use of one family of these sensors in which arguably the most progress has been made, the genetically encoded Ca2+ indicators or GECIs (the reader is referred to a number of excellent reviews of optical biosensors for an appreciation of the range and design strategies that have been developed: Zhang et al. 2002; Tsien, 2003; Rudolf et al. 2003; Miyawaki, 2003; Griesbeck, 2004; Gross & Piwnica-Worms, 2005; Giepmans et al. 2006). I will focus specifically on the use of GECIs in vivo, including parameters and advances in transgenic methods that will almost certainly further enhance their promise, as progressive improvements have begun to enable the use of GECIs to measure Ca2+ in cells of living mammals. This use is likely to greatly augment our understanding of complex signalling between cellular lineages in vivo and profoundly influence physiology over the next decades.
FRET-based genetically encoded Ca2+ indicators (frGECIs)
Development of frGECIs
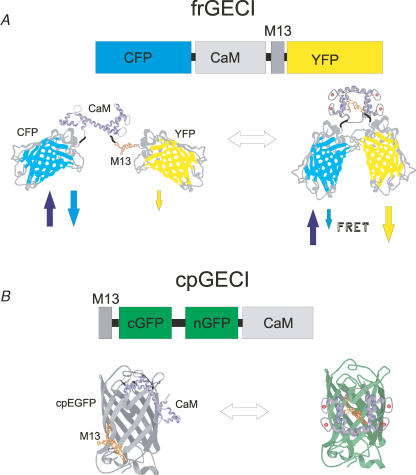
The first detector strategy developed for genetically encoded Ca2+ indicators was based on the transfer of energy between donor and acceptor fluorophores. Forster resonance energy transfer, or FRET, is a convenient way to exploit relatively subtle alterations in molecular structure that occur when a sensor binds its analyte. This approach was pioneered by the development of ‘cameleons’ almost a decade ago by Atsushi Miyawaki, Roger Tsien, and colleagues, in which the energy transfer occurs between members of green fluorescent protein (GFP)-related fluorescent proteins (Miyawaki et al. 1997). FRET was made proportional to the free Ca2+ concentration surrounding these molecules through the insertion of calmodulin and the M13 peptide from myosin light chain kinase between the fluorescent donor and acceptor proteins; the Ca2+-dependent globular condensation of calmodulin around the M13 peptide sequence (Ikura et al. 1992) moves the donor and acceptor closer, resulting in a Ca2+-dependent increase in FRET (Fig. 1). This simple concept has been progressively modified in several ways including a decrease in pH sensitivity of the photon acceptor in the yellow cameleons (Miyawaki et al. 1999; Griesbeck et al. 2001) and a shift to longer wavelength donor and acceptor pairs in the red cameleons (Mizuno et al. 2001), as well as the augmentation of the restricted dynamic range of cameleons through optimization of the donor/acceptor conformation in the Ca2+-bound state by changing the Ca2+ detector molecule from calmodulin to troponin C (Heim & Griesbeck, 2004) or optimizing the donor/acceptor interactions through redesign or optimization of the calmodulin–binding peptide interaction (Truong et al. 2001; Nagai et al. 2004; Nguyen & Daugherty, 2005; Palmer et al. 2006).
Figure 1. GECI design strategies and functional models.
A, FRET-based genetically encoded Ca2+ indicators (frGECIs) exploit the conformational change associated with binding of Ca2+ to a Ca2+-binding molecule. The yellow cameleon model using calmodulin (CaM) and the 13-residue peptide-binding domain of myosin light chain kinase (M13) is shown above. Below, condensation of Ca2+/CaM/M13 reduces the separation between the donor, cyan fluorescent protein (CFP), and the acceptor, yellow fluorescent protein (YFP). Red balls represent Ca2+ ions in EF hands; arrows represent excitation (up arrow) and emission. B, circularly permutated indicators (cpGECIs) attach the Ca2+-binding protein and the target peptide to opposing ends of one of the β sheets forming the cage around the chromophore. Binding of Ca2+ results in an interaction between CaM and M13, rearranging the cage and enabling fluorescence (represented by green hue). cGFP and nGFP refer to the original C and N-terminal sequences.
Properties of frGECIs
Despite these modifications, however, cameleons have shown limited utility in transgenic invertebrates, and there are no successful reports of their use in mammals. The failure to exploit the obvious promise of these molecules relates primarily to the limited in vivo signal strength of the frGECIs. In this regard, even the improved molecules have shown a compressed dynamic range when expressed in vivo, often indicating a marked difference in biophysical characteristics when compared to those of the recombinant protein assayed in simple solutions. The most rigorous examination of this phenomenon was by Reiff et al. who expressed several second generation frGECIs in transgenic flies using equivalent promoter strategies (Reiff et al. 2005). These studies demonstrated that the fold change fluorescence ratio (ΔR/Ro), where R is the ratio of fluorescence at the acceptor and donor emission wavelengths, and ΔR is the change in ratio over the background Ro, for a maximum physiological stimulus was between 5.8 and 11.6% (TN-L15, Heim & Griesbeck, 2004, and YC2.0, Miyawaki et al. 1999, respectively) of the change observed in vitro. This marked decrease in performance in non-transformed cells has been observed for many genetic indicators and almost certainly relates to the more complex interactions with other biological molecules that attend transgenic expression in the more complex intracellular milieu of differentiated cells. As calmodulin is known to preassociate with some target molecules (Jurado et al. 1999; Erickson et al. 2001), undergoes covalent modification (Benaim & Villalobo, 2002), and recognizes multiple cellular targets in its Ca2+-bound form, interactions within the complex cytosolic milieu are likely. Moreover, competition by endogenous calmodulin at the level of the calmodulin target peptide, which would markedly decrease probe signal strength, is also expected. While these considerations are also relevant for non-FRET GECIs that incorporate calmodulin as a sensor (see below), the dependence of the FRET signal on a precise orientation between the donor and acceptor fluorophores may confer an enhanced vulnerability to interactions of this sort. A similar compression of dynamic range has been observed with the expression of the yellow cameleon YC2.1 in pancreatic islets using adenovirus (Takahashi et al. 2002) and in neuronal organ cultures using a gene gun (Ikeda et al. 2003), methods that would be expected to overexpress the indicator. Interestingly, this general complication appears to hold for a non-calmodulin frGECI based on a truncated chicken troponin C (TNC-L15, Heim & Griesbeck, 2004), which displayed the lowest relative dynamic range in flies relative to its in vitro performance (Reiff et al. 2005). This decrement cannot be explained by endogenous calmodulin competition, but may relate to troponin C interactions with troponin I that were not observed in HEK293 cells or primary neurons (Heim & Griesbeck, 2004).
A second critical feature of GECIs is the fluorescence on-rate and off-rate following a step change in Ca2+ concentration. While small EF hand-based organic Ca2+ indicators display transition kinetics in the microsecond to low millisecond range (Eberhard & Erne, 1989, 1991), GECIs undergo complex intramolecular transitions resulting in on-rates and off-rates 2–3 orders of magnitude slower. Unfortunately, kinetic determinations of the fluorescence transition kinetics have been performed for only a few GECIs and comparisons of response kinetics, which depend on the rate of rise of Ca2+ in the system under study, endogenous buffer concentration, the volume of the cell, expression of the indicator, and the temperature at which the measurements are performed, must be approached with caution. The fluorescence transition rates are aggregate rate constants that include the processes of Ca2+ binding/unbinding, intramolecular reorganization and fluorescence development or decay. Heim & Griesbeck reported dissociation rates between 560 (TN-humTnC) and 870 ms (YC2.3) for several frGECIs (Heim & Griesbeck, 2004), placing clear constraints on the kinds of cellular signalling that can be effectively interrogated. Time constants for the rise and decay of Ca2+ in electrically stimulated Drosophila presynaptic boutons using several GECIs expressed under equivalent conditions indicated that fluorescence transition kinetics may be slower for the FRET-based probes (Reiff et al. 2005), but also that neither FRET- nor modified GFP-based GECIs accurately reproduced the Ca2+ decay kinetics or was capable of measuring the rise in Ca2+ associated with a single action potential.
Finally, several GECIs have displayed thermal instability, resulting in a marked degradation of fluorescence at mammalian body temperature. The relatively high background and intrinsic optical filtering properties of intact tissues necessitates molecules with high brightness. While this issue tends to be less of a consideration for frGECIs in which the fluorophores are effectively fusion molecules without extensive rearrangement, subtle temperature-dependent structural shifts may markedly alter fluorescent properties, resulting in poor visualization or signalling properties in in vivo experiments. As many of the initial assays of GECIs and other genetic indicators are performed at room temperature, a careful evaluation of brightness and signalling properties at 37°C should be performed before undertaking expensive and time-consuming transgenic expression.
Mammalian expression of frGECIs
To date there have been few studies in which frGECIs have been stably inserted into the mouse genome and these have yielded markedly disappointing results. Miyawaki's group first reported the expression of a frGECI in a transgenic mouse, expressing a plasma membrane-targeted yellow cameleon, YC3.6pm, under control of the ubiquitous β-actin promoter/CMV enhancer construct (pCAAGS) (Nagai et al. 2004). YC3.6 was redesigned to incorporate a circularly permutated yellow fluorescent protein (cpYFP) as the FRET acceptor, thereby markedly increasing the dynamic range to roughly 6-fold in HeLa cells. The indicator was localized to the plasma membrane by incorporating the polyisoprenylated C-terminal CAAX motif of ras proteins (Hancock et al. 1989), separated by a 16-residue linker. Despite achieving excellent targeted expression in the brain, tetanic stimulation of brain slices from YC3.6 transgenic mice produced less than a 3% increase in ΔR/R0, providing a dramatic example of the dynamic range compression of frGECIs discussed above. The related yellow cameleon YC3.0 has also expressed as a transgene under the control of pCAAGS (Tsai et al. 2003), but functional responses could not be recorded from these mice, which have been used for fluorescence visualization and lineage tracking of transplanted cells (Nyqvist et al. 2005). These results reflect the challenges and limitations associated with transgenic expression of the current frGECI molecules. By contrast, frGECIs have been successfully used for in vivo recording following direct transfection or viral infection of tissues. Particular success has been achieved in skeletal muscle, in which cytoplasmic, mitochondrial, and ER signals have been recorded in vivo (Rudolf et al. 2004, 2006). These studies have demonstrated a higher effective dynamic range than has been achieved following stable genome insertion (e.g. a 40% increase in FRET during muscle stimulation), perhaps due to the high level of transient expression from episomal DNA following transfection.
Despite the failure to fully exploit the promise of frGECIs to date, there are several distinct advantages to this approach, which could be exploited if the above discussed limitations can be circumvented. These advantages include an inherently simpler design that does not involve complex rearrangements of the native fluorophore structure (see below), simple ratiometric imaging enabling calibration of the fluorescence signals, and a wider range of indicator spectral characteristics. Moreover, several recent approaches give cause for some optimism regarding perturbing sensor interactions. Palmer et al. have redesigned the calmodulin-binding peptide to develop pairs that do not interact with native calmodulin, display an enhanced dynamic range, and operate over a wide range of [Ca2+]i (Palmer et al. 2006). Similar approaches may eliminate covalent modulation of the sensor calmodulin or its binding to endogenous targets. Improvements have also been made in troponin C-based frGECIs: by incorporating a circularly permutated FRET acceptor (citrine circularly permutated at residue 174) and altering the C-terminal lobe of troponin C to eliminate magnesium sensitivity, Mank et al. developed TN-XL, which has an improved dynamic range and response kinetics (Mank et al. 2006). Hopefully these improved properties will be retained following expression in transgenic mice.
Circular permutation of fluorescent proteins
Development of cpGECIs
An alternative approach to the development of GECIs exploits Ca2+-dependent rearrangements within a single fluorophore: Ca2+ binding to calmodulin within a fusion protein containing the fluorescent protein and calmodulin (with or without a binding peptide) alters the protein conformation resulting in an increase in fluorescence, providing a system in which fluorescence intensity is a function of Ca2+ concentration. The fluorescence of GFP and related fluorescent proteins arises from a post-translational modification of three amino acids within the core of 11 surrounding β sheets, which form a ‘β can’ structure; the three residues autocatalyse to an imidazolidone ring with oxygen atoms forming hydrogen bonds with several of the surrounding β sheet basic residues. In 1998, Roger Tsien and colleagues reported that GFP is surprisingly tolerant to the insertion of relatively large peptides at several locations within the β sheets or β sheet linkers and exploited this discovery by placing calmodulin at an insertion-permissive site (Baird et al. 1999). Compaction of calmodulin following Ca2+ binding markedly increased the fluorescence of the molecule, with a roughly 7-fold increase in fluorescence intensity (FCa/F0Ca), and these molecules were termed ‘camgaroos’, based on the placement of calmodulin (CaM) within a ‘pouch’ (Baird et al. 1999). While the initial YFP-inserted molecule, camgaroo1, suffered from poor thermal stability and weak fluorescence, an improved variant, camgaroo2, was produced by error-prone PCR, identifying a Lys to Met substitution at residue 69 (Griesbeck et al. 2001). The relatively high KD of camgaroo2, as well as the low brightness at resting cytosolic Ca2+ levels (roughly 10% of GFP in resting cells; Pologruto et al. 2004) are disadvantages for cytosolic Ca2+ sensors, but provide distinct advantages in the context of mitochondrial or ER sensors. Tsien and colleagues further showed that GFPs could be circularly permutated, or re-encoded beginning from one or more of the insertion sites and connecting the original C- and N-termini through a flexible linker. For example circularly permutated enhanced green fluorescent protein (cpEGFP) beginning with the N-terminal fragment 145–238 was linked with GGTGGS to EGFP 1–144, resulting in a protein with an emission peak at 512 nm, albeit with decreased 490 nm absorbance relative to 400 nm (Baird et al. 1999).
Two laboratories independently extended this development in 2001 to create a new GECI design in which calmodulin and its target peptide sequence were incorporated within cpEGFP (GCaMP1.3, now termed GCaMP1; Nakai et al. 2001) or cpEYFP (pericams; Nagai et al. 2001). The general functional scheme of circularly permutated GECIs (cpGECIs) is shown in Fig. 1: the 13-residue peptide of myosin light chain kinase that forms the binding domain of Ca2+–calmodulin (M13) is inserted before the C-terminal fragment of EGFP or EYFP at a permissive site as identified by Baird et al. (1999); the N-terminal fragment and calmodulin follow, with each element separated by a linker that has been optimized. The circular permutation of the fluorescent protein thus allows the attachment of calmodulin and its protein-binding domain at either end of the molecule, and the Ca2+-dependent binding of these ends ‘closes’ the cage surrounding the fluorophore. Pericams and GCaMPs utilize the 144–145 break identified by Tsien and colleagues (Baird et al. 1999; Nagai et al. 2001), whereas GCaMPs use the same insertion site, but remove peptides 145–148 (Nakai et al. 2001; Ohkura et al. 2005; Tallini et al. 2006a). While the structures of the Ca2+ bound and free forms of cgGECIs are not known, unbound M13 peptide and/or extended apocalmodulin at the broken β sheet most likely disrupt fluorescence either by (1) altering solvent penetration within the protein core; (2) interference with fluorophore–β sheet interactions; or (3) direct steric interactions with the chromophore. Presumably the inhibitory action is relieved by condensation of Ca2+-bound calmodulin around the M13 peptide.
The first generation of cpGECIs displayed the high signal strength characteristic of the previous insertion molecules (FCa/F0Ca of 6–8) and the KD values (235 and 700 nm for GCaMP1 and Flash-pericam, respectively) were well suited for measurements of cytosolic Ca2+. Flash-pericam, the highest signal pericam, was reported to have lower cooperativity and a higher KD than GCaMP1 (Hill coefficients = 0.7 and 3.3; KD = 0.7 and 0.23 μm, respectively), but subsequent analysis in transfected neurons indicated that pericams have a markedly higher level of cooperativity, and GCaMP1 a higher KD, in cells (Pologruto et al. 2004). As discussed below, the alinear relationship between Ca2+ and fluorescence intensity is a significant factor in the use of cpGECIs for quantitative analyses. The first generation cpGECIs were also significantly less bright than native fluorescent proteins and displayed variable stability above 30°C. Thus the quantum yield of Flash-pericam and GCaMP1 are 0.2 and 0.05, whereas EYFP and EGFP are above 0.6, and the extinction coefficients are also significantly lower than the parent proteins, with GCaMP1 absorption being less than 3% of EGFP. A more promising aspect of the early cpGECIs, however, was the rates of transition between the Ca2+ bound and unbound states. While rate constants were not reported for pericams, stop-flow measurements of GCaMP1 indicated a Ca2+ association time constant (aggregate time constant for all process including Ca2+ binding, protein rearrangement, and fluorescence rise) of less than 10 ms over a range of Ca2+ from 500 nm to 1 μm. The dissociation rate constant was markedly slower (188 ms); however, these measurements, performed at room temperature, indicate in vitro transition kinetics markedly faster than reported for any frGECIs (Reiff et al. 2005). One caution regarding these measurements is the potential difference between transition kinetics measured in vitro and those observed in live cells. As discussed above, interactions between GECIs and endogenous proteins can markedly degrade signal sensor performance; while providing optimal quantification, stop-flow measurements should be conducted in cytosol where possible, to provide the most meaningful estimate of cellular performance.
Miyawaki and colleagues also designed a dual excitation pericam by substituting at the 203 position, which had previously been shown to augment YFP fluorescence when excited at 400 nm (Dickson et al. 1997). Optimization of this molecule resulted in the creation of Ratiometric-pericam, in which the 418 nm absorption peak decreases and that at 494 nm increases in the Ca2+-bound state, thereby mitigating a significant disadvantage of the cpGECIs through the use of path length-insensitive ratiometric imaging (Nagai et al. 2001). GCaMP1 was subsequently improved (GCaMP1.6; Ohkura et al. 2005) by the introduction of the stabilizing mutations V163A and S175G (Heim & Tsien, 1996; Siemering et al. 1996), markedly improving the brightness of the molecule and decreasing the pH sensitivity. However, despite these stabilizing mutations the fluorescence of GCaMP1.6 was markedly degraded in cells recorded at 35°C. Designed mutations were also made in the calmodulin EF hands to decrease Ca2+ buffering; the E140K mutation retained sensor signal strength, while slightly lowering the KD. Interestingly, mutation of one of each of the EF hand pairs (E67K/E140K) markedly decreased the Hill coefficient (from 3.8 to 1.4), albeit at the expense of dynamic range (4.9–1.9) (Ohkura et al. 2005). A comparison of GECIs expressed in transgenic flies indicated that GCaMP1.6 reported the largest and most rapid signals reflecting neural activity, although photobleaching was a consistent finding.
Recently, GCaMP1.6 was further modified and the problem of temperature dependence eliminated (Tallini et al. 2006a). The A206K mutation, which prevents chromophore dimerization (Zacharias et al. 2002), was made and a polyHis linker attached to the N-terminus (pRSET) for protein purification. The resultant cDNA was subjected to random mutagenesis by low fidelity PCR amplification and the clones screened for expression in bacteria at 37°C; brightly fluorescent colonies were isolated and sequenced, leading to the identification of two new mutations (D180Y and V93I). The resulting sensor, GCaMP2, is brighter than wild-type GFP, maintains a 5-fold dynamic range, displays the reduced pH sensitivity of GCaMP1.6, and is fully functional at 37°C. Interestingly, this stability results from a serendipitous effect of adding the polyHis linker, as removal of the linker eliminates fluorescence at 37°C. The combination of high brightness and signal strength, thermal stability, rapid kinetics (see below), and favourable KD render GCaMP2 an extremely promising cpGECI for in vivo physiology. However, the complex relationship between fluorescence and Ca2+, as well as non-ratiometric imaging, remain as limitations to the current sensors. Critical attributes of GFP-based (cpGECIs and insertion GECIs) are listed in Table 1, with information provided that should proved useful in selecting sensors for mammalian expression.
Table 1.
GFP-based GECIs
| GECI | Fluorophore | Sensor | F/FEGFP* | KD (in vivo†) | Hill coeff. (in vivo†) | ΔF/F0 (in vivo†) | 37°C | References |
|---|---|---|---|---|---|---|---|---|
| Camgaroo | EYFP(S65G,V68L,Q69K,T203Y) | CaM | n.d. | 7 μm (n.d.) | 1.6 | 7 | No | Baird et al. (1999) |
| Camgaroo-2 | EYFP(Q69M,S72A) and above | CaM/M13 | n.d. | 5.3 (8) | 1.2 (1.4) | 6 (2) | Yes | Griesbeck et al. (2001) |
| Fl-pericam | cp145EYFP (S65G,V68L,Q69K, V163A, S175G,T203H) | CaM/M13 | 0.09 | 0.7 | 0.7 | 8 | No | Nagai et al. (2001) |
| Rat-pericam | cp145EYFP (H148D,T203F) and above | CaM/M13 | 0.05 | 1.7 | 1.1 | 10 | Yes | Nagai et al. (2001) |
| Inv-pericam | cp145EYFP (H148T,T203F) and above | CaM/M13 | 0.49 | 0.2 (0.9) | 1.0 (3.8) | −7 (−0.25) | Yes | Nagai et al. (2001) |
| GCaMP1 | cp145–148EGFP | CaM/M13 | 0.002 | 0.24 (1.7) | 3.3 (3.3) | 4.5 (1.8) | No | Nakai et al. (2001) |
| GCaMP1.6 | cp145–148EGFP (V163A,S175G) | CaM/M13 | 0.08 | 0.15 | 3.8 | 4.9 | No | Ohkura et al. (2005) |
| GCaMP2 | cp145–148EGFP (V93I,D180Y,A206K)pRSET | CaM/M13 | 0.45 | 0.15 | 3.8 | 5 | Yes | Tallini et al. (2006a) |
Brightness relative to EGFP ((ɛGECI×ΦGECI)/(ɛEGFP×ΦEGFP)) in saturating Ca2+ at peak emission wavelength, where ɛ = molar extinction coefficient, Φ = quantum yield, and the values for EGFP (F64L/S65T) are 56 000 cm−1m−1 and 0.7, respectively.
In vivo determinations from Pologruto et al. (2004) are in parentheses for comparison. Fl, Flash; Rat, Ratiometric; Inv, Inverse; n.d., not determined.
Expression of cpGECIs in transgenic mice
GCaMP1 (Ji et al. 2004), camgaroo-2 and Inverse-pericam (Hasan et al. 2004) have been stably expressed in transgenic mice and used to report Ca2+ signalling in complex tissues. The expression of GCaMP1 was driven by a large, smooth muscle-specific promoter and lineage-specific Ca2+ signalling examined in intact smooth muscle tissues and in isolated smooth muscle cells by confocal microscopy at 28°C. Despite the low fluorescence intensity of GCaMP1, robust Ca2+ signals in individual myocytes were readily apparent, allowing the dissection of distinct postsynaptic responses in myocytes evoked by nerve stimulation (Ji et al. 2004). Signal increases (ΔF/F0) of 50–100% were routinely observed in cells and virtually all myocytes within tissue segments responded to nerve stimulation. Photobleaching was observed in these experiments, but appeared to be complex and did not eliminate Ca2+-dependent signalling. That is, there was a rapid, exponential decrease in fluorescence during laser excitation over the course of a few seconds, but this process decreased fluorescence to roughly steady state, from which photobleaching was much less significant; cell signalling was observed from the decreased baseline fluorescence. This complex behaviour of GCaMP1 further complicates potential quantification efforts, however. Hasan et al. used a bi-allelic system consisting of CMV promoter/tet response element/GECI transgenic mice crossed with mice expressing αCaMKII–tTA; the double transgenic mice expressed Inverse-pericam and camgaroo-2 in the brain in an inducible (doxycycline-off) manner (Hasan et al. 2004). GECI transgenic founders were screened based on expression of a reporter gene not under doxycycline control (bidirectional construct), allowing strong-expressing founders to be determined. Strong expression was observed in brain slices from these mice and by multiphoton imaging of the brain in vivo. Synaptic stimulation and multiphoton imaging of brain slices from mice expressing camgaroo-2 and Inverse-pericam produced responses from 30 to 100% (ΔF/F0), but signals from 3 to 8% in widefield studies. Photobleaching was also observed in these and there was some suggestion of a fraction of protein that was fluorescent, but not Ca2+ responsive, for both indicators. In general, most selected lines expressed the indicators weakly and were poorly responsive, or expressed the transgene concentrated in a punctate pattern; these findings may suggest that much of the protein is poorly folded or bound by other molecules, although weak expression could also result from insufficient transgene copy number/promoter activity. This problem was not observed in GCaMP1 transgenic mice, in which the cpGECI was found to be diffusely present in the cytosol of isolated myocytes (Tallini et al. 2006a). Consistent with drawbacks discussed above, no functional responses were observed in transgenic lines expressing the frGECI YC3.12.
These first transgenic experiments highlighted several challenges for the practical use of GECIs in vivo. First, signal intensity and dynamic range were severely constrained in wide-field measurements, requiring laser illumination and confocal or mutiphoton optical sectioning for useable signals. However, wide-field, low magnification or macro imaging is preferable for some in vivo applications, in particular those with intrinsic motion (e.g. the heart) or where large field sizes are required. Second, while the experiments demonstrated that stable expression could be achieved, the stability and cellular interactions of the GECIs limit the usefulness of some molecules, and effective signals could only be obtained ex vivo. Third, photobleaching and potential alterations in the affinity and cooperativity of GECIs expressed in vivo present very significant challenges to the quantification of fluorescent signals. Finally, these studies did not assess whether the fluorescence transition kinetics of cpGECIs would be sufficient to accurately monitor fast cellular signals in vivo.
Several of these issues have been recently addressed by the expression of the improved cpGECI, GCaMP2 (Tallini et al. 2006a). The significant improvement of GCaMP2 in signal strength and stability led us to direct the expression to the mammalian heart, in which the intrinsic motion, fast transients, and rapid spread impose significant challenges. Transgenic mice incorporating a GCaMP2 construct driven by the heart-specific, mutated α-myosin heavy chain promoter (Sanbe et al. 2003) and tetracycline-inducible enhancer sequences were crossed with cardiac tetracycline transactivator (tTA) transgenic mice and screened for GCaMP2 expression and doxycycline control (dox-off). Two lines were selected with robust expression restricted to myocytes, with no expression in the heart vasculature or other tissues. Interestingly, we observed that unregulated expression of GCaMP2 from birth resulted in significant cardiomegaly in both lines, similar to previous reports for overexpression of GFP (Huang et al. 2000), although the hypertrophy could be avoided by doxycycline suppression of GCaMP2 expression during cardiac development.
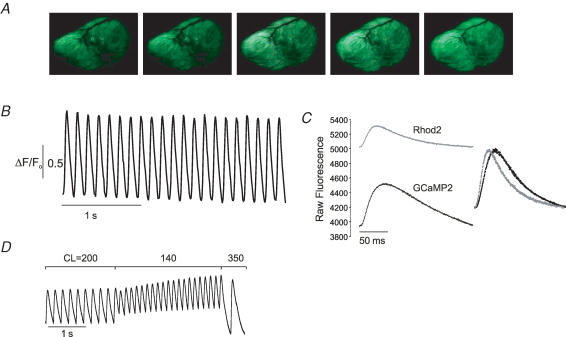
Fluorescence signals were apparent in every region of GCaMP2 transgenic mouse hearts in vivo (open chest, anaesthetized and ventilated mouse) even under the simplest optical preparations. Figure 2A shows a series of images obtained by a consumer grade digital videocamera coupled to a standard fluorescence dissecting microscope; fluorescence (ΔF/F0) increased by approximately 100% from diastole to systole, in vivo. Higher speed cameras and more efficient light collection setups readily allowed imaging in vivo at 256 Hz, sufficient image acquisition rate to acquire highly reproducible Ca2+ transients and to evaluate wave transmission across the myocardial surface, and up to 5 kHz in isolated perfused hearts. Thus transgenic expression of GCaMP2 results in stable and functional GECI expression with sufficient signal strength and dynamic range to record physiological signals in vivo with simple wide-field optics. GCaMP2 was also stably and diffusely expressed in the cytosol of myocytes, without evidence of focal protein accumulation. Photobleaching was also less problematic in these studies, although some rapid fluorescence decay was observed, particularly with intense laser illumination. In practice, however, highly reproducible Ca2+ transients could be recorded from the surface of the heart without signal decay (Fig. 2B).
Figure 2. Cardiac expression of GCaMP2.
A, sequential in vivo images of the left ventricle of an anaesthetized, open-chested transgenic mouse expressing GCaMP2 demonstrate the signal change between end-diastole (left) and peak systole (right). Unprocessed colour images were taken with a consumer grade digital videocamera through a binocular fluorescence dissecting microscope (Leica MZFLIII). B, in vivo fluorescence transients from a 4 pixel area in the left atrium. C, fluorescence transients taken from an isolated, perfused GCaMP2 heart loaded with Rhod2. Note higher ΔF and lower background fluorescence in GCaMP2 signal. Overlay of the normalized traces demonstrates the underestimation of the rate of rise and decay of the Ca2+ signal by GCaMP2 in uncalibrated signals. D, pacing of a GCaMP2 heart in vitro demonstrates the consequence of GCaMP2 transition kinetics for rapid physiological signals. CL = cycle length.
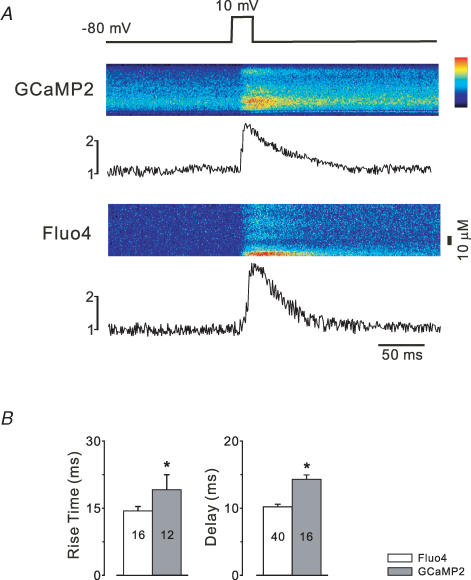
The kinetics of GCaMP2 in cells was rigorously examined in several ways. First, stop-flow transitions were compared between recombinant protein and cardiac cytosol purified from GCaMP2-expressing transgenic mice in vitro. While such experiments inevitably result in substantial dilution of the cytosol, several concentrations of the cytosol were compared with recombinant protein to determine the effect of dilution; this analysis demonstrated that in both jumps from moderate to high Ca2+ and jumps from zero to low Ca2+ transition kinetics (fluorescence on transition) were identical between recombinant protein in physiological solution and the cardiac cytosol (containing transgenic protein), and no differences were observed over an 8-fold dilution range (Tallini et al. 2006a), indicating that protein function is essentially preserved within the cellular milieu. Moreover, the analysis at 37°C indicated that the transition kinetics of GCaMP2 are markedly faster and simpler than reported for GCaMP1 measured at 25°C (Nakai et al. 2001); the fluorescence association and decay time constants are roughly 15 and 75 ms, and are independent of the level of Ca2+. Second, experiments were performed in isolated, perfused GCaMP2-expressing transgenic hearts that were loaded with Rhod2; GCaMP2 and Rhod2 fluorescence transients were measured simultaneously from the same heart region (approximately 40 μm2) during spontaneous and triggered action potentials using spatially registered photodiode arrays (Baker et al. 2000). As shown in Fig. 2C, GCaMP2 and Rhod2 transients were of comparable signal quality and repeatability, with the ΔF/F0 of GCaMP2 signals being higher than Rhod2; the lower KD and high cooperativity of GCaMP2 lead to a distinct rounded appearance of the peak of the Ca2+ transient relative to Rhod2, as GCaMP2 nears saturation at peak systole. Comparison of the transient kinetics revealed a slight underestimation of the rate of rise, and a substantial overestimation of the decay time constant (Tallini et al. 2006a), as would be predicted from the magnitude of the fluorescence transition kinetics (Rhod2 fluorescence transition kinetics are submillisecond). The delay imposed by the fluorescence decay time constant effectively limited the maximum rate in which transients could be recorded, as at cycle lengths below 300 ms the transient did not fully decay before the next upstroke; however, immediately upon slowing the stimulation rate the fluorescence signal returned to the previous baseline (Fig. 2D), indicating a remarkable stability that was consistent over many hours of recording. As a final investigation of the kinetic behaviour of GCaMP2 in cells, we compared Ca2+ transients in single GCaMP2-expressing myocytes with transients in fluo4-loaded cells from GCaMP− littermates, as shown in Fig. 3. These experiments also documented a slight slowing of the Ca2+ upstroke and a more prominent delay in the signal decay, consistent with the transition kinetics measured in vitro.
Figure 3. Kinetics of GCaMP2 in single heart cells.
A, ventricular myocytes disaggregated from GCaMP2-expressing hearts were step-depolarized in voltage-clamp experiments and compared with myocytes from non-transgenic littermates loaded with Fluo4. Both preparations showed rapid fluorescence responses following step depolarization; however, the GCaMP2 signal decayed at a slower rate. B, the rise time and delay to rise was significantly longer in GECI-expressing cells.
Thus GCaMP2 addresses several of the fundamental limitations that have previously been observed for the use of GECIs in mammals, in vivo. The transgene is homogenously expressed within the cytosol at sufficient concentration to enable practical recording at high bandwidth, and interactions with endogenous proteins do not appear to markedly alter GCaMP2 brightness, dynamic range, Ca2+ affinity, or response kinetics. The sensor also functions effectively as a fusion protein, which has enabled localization of near plasma membrane Ca2+ signals (Lee et al. 2006). However, despite more rapid transitions than any previously described GECI, it is apparent that this class of indicators has limitations in the context of accurately characterizing the most rapid physiological Ca2+ signals.
Bacterial artificial chromosome transgenesis
The foregoing discussion has highlighted several molecules that are likely to be useful in the dissection of complex, multicellular signalling events. To fully exploit these and emerging sensors, however, precise lineage-specific expression will be increasingly important. Recent advances in bacterial artificial chromosome (BAC) transgenesis have enabled such control to a degree worth briefly highlighting in this review. Previously, specification of transgene expression was limited largely to artificial promoter constructs, which variably, but often poorly, approximate endogenous expression patterns, or to knock-ins within a genetic locus that result in the inactivation of one gene allele and are generally limited to the insertion of one or two copies of the transgene cDNA. BAC transgenesis utilizes genomic DNA elements that are approximately 100–200 kb in length and can be chosen to include the entire gene locus as well as extensive 5′ and 3′ sequences. BACs can be modified by homologous recombination in E. coli for efficient insertion of cDNAs, expression of which is controlled by the extensive regulatory elements within these large DNA fragments (Copeland et al. 2001; Heintz, 2001). The engineered BACs can then be injected directly into a one-cell stage embryo and lines screened for transgene insertion. While examples of cis regulatory elements more distant than included in BACs exist, in our experience these DNA constructs provide an outstanding level of transgene control, enabling precise lineage targeting (Tallini et al. 2006b). By determining a gene whose expression pattern demonstrates the desired specificity, one can utilize an appropriate BAC construct to quickly produce a transgene construct with unprecedented transcriptional control without a complex dissection of the underlying regulatory elements. This task is made simple by increasingly powerful BAC engineering systems (Lee et al. 2001; Gong et al. 2002; Warming et al. 2005) and the availability of end-sequenced BAC libraries that cover the entire mouse genome (e.g. http://www.ncbi.nlm.nih.gov/genome/clone/clonefinder/CloneFinder.html). Figure 4 depicts an example of BAC transgenesis in which high expression and specificity of a fluorophore was achieved in lineages that are not accessible by standard transgenesis (Tallini et al. 2006b). This approach is being extensively used to target specific lineages for expression of GCaMP2.
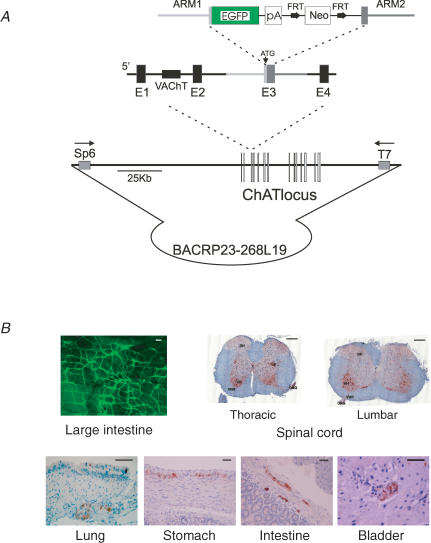
Figure 4. Targeting cell lineages using BAC transgenesis.
A, targeting strategy used to homologously transfer the EGFP coding sequence into the initiation codon of the choline acetyltransferase (ChAT) locus contained within a bacterial artificial chromosome. Note that the vesicular choline acetyltransferase (VAChT) gene is contained within the first intron of ChAT. B, expression of EGFP in BAC transgenic mice demonstrates high expression within the central and peripheral nervous systems, with precise marking of cholinergic enteric neurons. Scale bars are 100 μm (large intestine), 250 μm (spinal cord), and 50 μm (other). Adapted from Tallini et al. (2006b).
Summary
While significant progress has been made in the design and mammalian expression of GECIs, clearly substantial challenges remain. On the side of progress, GECIs have been progressively improved, resulting in the stable expression of GECIs with excellent signal characteristics in mammals sufficient to detect cell–cell signalling in vivo and to provide useful insights into complex physiology. The major remaining challenges are the speed of GECI signalling and the practical quantification of GECI fluorescence. At present, GECIs are substantially slower than synthetic 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA)-based Ca2+ sensors, a disadvantage that is unlikely to be resolved in molecules based on fret or circular permutation. Transition rates of the order of 10–100 ms limit the usefulness of cpGECIs (the fastest GECIs) for the detection of the most rapid signals in muscle and nerve, but are more than adequate for the accurate measurement of Ca2+ signals in most physiological contexts. Quantification of single-wavelength cpGECI signals is further complicated by the highly non-linear relationship between Ca2+ and fluorescence observed for most GECIs. This issue is likely to prove to be more amenable to rational design and random mutagenesis, however, by the introduction of mutations that confer fluorescence at the lower absorption peak of GFP variants (Nagai et al. 2001) and by mutations in calmodulin EF hands that markedly reduce cooperativity, albeit at the expense of dynamic range (Ohkura et al. 2005).
In the next several years it is likely that continued improvements in frGECIs and cpGECIs will be made. Importantly, however, many physiological applications do not require high speed detectors or quantified signals. For these applications existing molecules and mice, the characteristics of which are summarized in Tables 1 and 2, comprise useful tools for the understanding of complex biology. Moreover, the availability of novel spectrally shifted GFP mutants (Shaner et al. 2004) and Anthozoa fluorescent proteins (Verkhusha & Lukyanov, 2004) provides additional opportunities for the application of this general strategy to molecules that are likely to have unique properties. One can confidently look forward, within the next few years, to the precise placement of molecular microphones in multiple lineages, enabling the detection of specific components of the cellular symphony.
Table 2.
Transgenic mice expressing GECIs
| GECI | Promoter | Lineage | Expression | Signals in vitro | Signals in vivo | References |
|---|---|---|---|---|---|---|
| GCaMP1 | smMHC | Smooth muscle | Good | 50–100% | No | Ji et al. (2004) |
| CG2 | CMV-CaMKII/tet | Neurons | Good, some punctate | 2–100% | 3% | Hasan et al. (2004) |
| Inv-pericam | CMV-CaMKII/tet | Neurons | Good, some punctate | −(2–30)% | −8% | Hasan et al. (2004) |
| YC3.12 | CMV-CaMKII/tet | Neurons | Good, some punctate | no | No | Hasan et al. (2004) |
| YC3.6 | pCAGGS | Ubiquitous | Good | < 2%ΔF/F0 | No | Nagai et al. (2004) |
| GCaMP2 | αMHC-ΔαMHC/tet | Cardiac | High | 50–100% | 50–100% | Tallini et al. (2006a) |
Acknowledgments
I gratefully acknowledge the many contributions of Junichi Nakai, Yvonne Tallini, Guy Salama, Bo Shui, Jane Lee, Kai Su Greene, Mordy Blaustein, and Ke-Yu Deng to work described here. This study was supported by NIH grants HL45239, DK065992, and DK58795.
References
- Adams SR, Harootunian AT, Buechler YJ, Taylor SS, Tsien RY. Fluorescence ratio imaging of cyclic AMP in single cells. Nature. 1991;349:694–697. doi: 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- Bacskai BJ, Hochner B, Mahaut-Smith M, Adams SR, Kaang BK, Kandel ER, Tsien RY. Spatially resolved dynamics of cAMP and protein kinase A subunits in Aplysia sensory neurons. Science. 1993;260:222–226. doi: 10.1126/science.7682336. [DOI] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LC, London B, Choi BR, Koren G, Salama G. Enhanced dispersion of repolarization and refractoriness in transgenic mouse hearts promotes reentrant ventricular tachycardia. Circ Res. 2000;86:396–407. doi: 10.1161/01.res.86.4.396. [DOI] [PubMed] [Google Scholar]
- Benaim G, Villalobo A. Phosphorylation of calmodulin. Functional implications. Eur J Biochem. 2002;269:3619–3631. doi: 10.1046/j.1432-1033.2002.03038.x. [DOI] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- Dickson RM, Cubitt AB, Tsien RY, Moerner WE. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- Eberhard M, Erne P. Kinetics of calcium binding to fluo-3 determined by stopped-flow fluorescence. Biochem Biophys Res Commun. 1989;163:309–314. doi: 10.1016/0006-291x(89)92136-0. [DOI] [PubMed] [Google Scholar]
- Eberhard M, Erne P. Calcium binding to fluorescent calcium indicators: calcium green, calcium orange and calcium crimson. Biochem Biophys Res Commun. 1991;180:209–215. doi: 10.1016/s0006-291x(05)81278-1. [DOI] [PubMed] [Google Scholar]
- Erickson MG, Alseikhan BA, Peterson BZ, Yue DT. Preassociation of calmodulin with voltage-gated Ca2+ channels revealed by FRET in single living cells. Neuron. 2001;31:973–985. doi: 10.1016/s0896-6273(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Fiala A, Spall T. In vivo calcium imaging of brain activity in Drosophila by transgenic cameleon expression. Sci STKE. 2003;2003:PL6. doi: 10.1126/stke.2003.174.pl6. [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Gong SC, Yang XW, Li CJ, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6K gamma origin of replication. Genome Res. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck O. Fluorescent proteins as sensors for cellular functions. Curr Opin Neurobiol. 2004;14:636–641. doi: 10.1016/j.conb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- Gross S, Piwnica-Worms D. Spying on cancer: molecular imaging in vivo with genetically encoded reporters. Cancer Cell. 2005;7:5–15. doi: 10.1016/j.ccr.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Guerrero G, Siegel MS, Roska B, Loots E, Isacoff EY. Tuning FlaSh: redesign of the dynamics, voltage range, and color of the genetically encoded optical sensor of membrane potential. Biophys J. 2002;83:3607–3618. doi: 10.1016/S0006-3495(02)75361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- Hasan MT, Friedrich RW, Euler T, Larkum ME, Giese G, Both M, Duebel J, Waters J, Bujard H, Griesbeck O, Tsien RY, Nagai T, Miyawaki A, Denk W. Functional fluorescent Ca2+ indicator proteins in transgenic mice under TET control. PLoS Biol. 2004;2:763–775. doi: 10.1371/journal.pbio.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim N, Griesbeck O. Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J Biol Chem. 2004;279:14280–14286. doi: 10.1074/jbc.M312751200. [DOI] [PubMed] [Google Scholar]
- Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Heintz N. BAC to the future: The use of bac transgenic mice for neuroscience research. Nat Rev Neurosci. 2001;2:861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Masino MA, Mandel G, Fetcho JR. Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. J Neurophysiol. 2003;90:3986–3997. doi: 10.1152/jn.00576.2003. [DOI] [PubMed] [Google Scholar]
- Honda A, Adams SR, Sawyer CL, Lev-Ram V, Tsien RY, Dostmann WR. Spatiotemporal dynamics of guanosine 3′,5′-cyclic monophosphate revealed by a genetically encoded, fluorescent indicator. Proc Natl Acad Sci U S A. 2001;98:2437–2442. doi: 10.1073/pnas.051631298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WY, Aramburu J, Douglas PS, Izumo S. Transgenic expression of green fluorescence protein can cause dilated cardiomyopathy. Nat Med. 2000;6:482–483. doi: 10.1038/74914. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Sugiyama T, Wallace CS, Gompf HS, Yoshioka T, Miyawaki A, Allen CN. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron. 2003;38:253–263. doi: 10.1016/s0896-6273(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Ikura M, Clore GM, Gronenborn AM, Zhu G, Klee CB, Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- Ji G, Feldman M, Deng KY, Greene KS, Wilson J, Lee J, Johnston R, Rishniw M, Tallini Y, Zhang J, Wier WG, Blaustein MP, Xin HB, Nakai J, Kotlikoff MI. Ca2+-sensing transgenic mice: postsynaptic signaling in smooth muscle. J Biol Chem. 2004;279:21461–21468. doi: 10.1074/jbc.M401084200. [DOI] [PubMed] [Google Scholar]
- Jurado LA, Chockalingam PS, Jarrett HW. Apocalmodulin. Physiol Rev. 1999;79:661–682. doi: 10.1152/physrev.1999.79.3.661. [DOI] [PubMed] [Google Scholar]
- Karasawa S, Araki T, Nagai T, Mizuno H, Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr R, Lev-Ram V, Baird G, Vincent P, Tsien RY, Schafer WR. Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron. 2000;26:583–594. doi: 10.1016/s0896-6273(00)81196-4. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu DG, de Velasco JM, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Lee MY, Song H, Nakai J, Ohkura M, Kotlikoff MI, Kinsey SP, Golovina VA, Blaustein MP. Local subplasma membrane Ca2+ signals detected by a tethered Ca2+ sensor. Proc Natl Acad Sci U S A. 2006;103:13232–13237. doi: 10.1073/pnas.0605757103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank M, Reiff DF, Heim N, Friedrich MW, Borst A, Griesbeck O. A FRET-based calcium biosensor with fast signal kinetics and high fluorescence change. Biophys J. 2006;90:1790–1796. doi: 10.1529/biophysj.105.073536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A. Fluorescence imaging of physiological activity in complex systems using GFP-based probes. Curr Opin Neurobiol. 2003;13:591–596. doi: 10.1016/j.conb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Griesbeck O, Heim R, Tsien RY. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc Natl Acad Sci U S A. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Sawano A, Eli P, Hama H, Miyawaki A. Red fluorescent protein from Discosoma as a fusion tag and a partner for fluorescence resonance energy transfer. Biochemistry. 2001;40:2502–2510. doi: 10.1021/bi002263b. [DOI] [PubMed] [Google Scholar]
- Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- Ng M, Roorda RD, Lima SQ, Zemelman BV, Morcillo P, Miesenbock G. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 2002;36:463–474. doi: 10.1016/s0896-6273(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- Nyqvist D, Mattsson G, Kohler M, Lev-Ram V, Andersson A, Carlsson PO, Nordin A, Berggren PO, Jansson L. Pancreatic islet function in a transgenic mouse expressing fluorescent protein. J Endocrinol. 2005;186:333–341. doi: 10.1677/joe.1.06204. [DOI] [PubMed] [Google Scholar]
- Ohkura M, Matsuzaki M, Kasai H, Imoto K, Nakai J. Genetically encoded bright Ca2+ probe applicable for dynamic Ca2+ imaging of dendritic spines. Anal Chem. 2005;77:5861–5869. doi: 10.1021/ac0506837. [DOI] [PubMed] [Google Scholar]
- Palmer AE, Giacomello M, Kortemme T, Hires SA, Lev-Ram V, Baker D, Tsien RY. Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem Biol. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Pologruto TA, Yasuda R, Svoboda K. Monitoring neural activity and [Ca2+] with genetically encoded Ca2+ indicators. J Neurosci. 2004;24:9572–9579. doi: 10.1523/JNEUROSCI.2854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiff DF, Ihring A, Guerrero G, Isacoff EY, Joesch M, Nakai J, Borst A. In vivo performance of genetically encoded indicators of neural activity in flies. J Neurosci. 2005;25:4766–4778. doi: 10.1523/JNEUROSCI.4900-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romoser VA, Hinkle PM, Persechini A. Detection in living cells of Ca2+-dependent changes in the fluorescence emission of an indicator composed of two green fluorescent protein variants linked by a calmodulin-binding sequence. A new class of fluorescent indicators. J Biol Chem. 1997;272:13270–13274. doi: 10.1074/jbc.272.20.13270. [DOI] [PubMed] [Google Scholar]
- Rudolf R, Magalhaes PJ, Pozzan T. Direct in vivo monitoring of sarcoplasmic reticulum Ca2+ and cytosolic cAMP dynamics in mouse skeletal muscle. J Cell Biol. 2006;173:187–193. doi: 10.1083/jcb.200601160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf R, Mongillo M, Magalhaes PJ, Pozzan T. In vivo monitoring of Ca2+ uptake into mitochondria of mouse skeletal muscle during contraction. J Cell Biol. 2004;166:527–536. doi: 10.1083/jcb.200403102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf R, Mongillo M, Rizzuto R, Pozzan T. Looking forward to seeing calcium. Nat Rev Mol Cell Biol. 2003;4:579–586. doi: 10.1038/nrm1153. [DOI] [PubMed] [Google Scholar]
- Sanbe A, Gulick J, Hanks MC, Liang Q, Osinska H, Robbins J. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ Res. 2003;92:609–616. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–741. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- Siemering KR, Golbik R, Sever R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Nemoto T, Kimura R, Tachikawa A, Miwa A, Okado H, Miyashita Y, Iino M, Kadowaki T, Kasai H. Two-photon excitation imaging of pancreatic islets with various fluorescent probes. Diabetes. 2002;51(Suppl. 1):S25–S28. doi: 10.2337/diabetes.51.2007.s25. [DOI] [PubMed] [Google Scholar]
- Tallini Y, Ohkura M, Choi BR, Ji G, Imoto K, Doran R, Lee J, Plan P, Wilson J, Xin HB, Sanbe A, Gulick J, Mathai J, Robbins J, Salama G, Nakai J, Kotlikoff MI. Imaging cellular signals in the heart in vivo: cardiac expression of the high signal Ca2+ indicator GCaMP2. Proc Natl Acad Sci U S A. 2006a;103:4753–4758. doi: 10.1073/pnas.0509378103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallini YN, Shu B, Greene KS, Deng KY, Doran R, Fisher PJ, Zipfel W, Kotlikoff MI. BAC transgenic mice express enhanced green fluorescent protein in central and peripheral cholinergic neurons. Physiol Genomics. 2006b doi: 10.1152/physiolgenomics.00092.2006. in press. [DOI] [PubMed] [Google Scholar]
- Truong K, Sawano A, Mizuno H, Hama H, Tong KI, Mal TK, Miyawaki A, Ikura M. FRET-based in vivo Ca2+ imaging by a new calmodulin-GFP fusion molecule. Nat Struct Biol. 2001;8:1069–1073. doi: 10.1038/nsb728. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Friedman B, Ifarraguerri AI, Thompson BD, Lev-Ram V, Schaffer CB, Xiong Q, Tsien RY, Squier JA, Kleinfeld D. All-optical histology using ultrashort laser pulses. Neuron. 2003;39:27–41. doi: 10.1016/s0896-6273(03)00370-2. [DOI] [PubMed] [Google Scholar]
- Tsien RY. Breeding molecules to spy on cells. Harvey Lect. 2003;99:77–93. [PubMed] [Google Scholar]
- Verkhusha VV, Lukyanov KA. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat Biotechnol. 2004;22:289–296. doi: 10.1038/nbt943. [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccolo M, Pozzan T. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science. 2002;295:1711–1715. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]