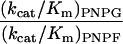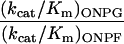Abstract
An efficient β-fucosidase was evolved by DNA shuffling from the Escherichia coli lacZ β-galactosidase. Seven rounds of DNA shuffling and colony screening on chromogenic fucose substrates were performed, using 10,000 colonies per round. Compared with native β-galactosidase, the evolved enzyme purified from cells from the final round showed a 1,000-fold increased substrate specificity for o-nitrophenyl fucopyranoside versus o-nitrophenyl galactopyranoside and a 300-fold increased substrate specificity for p-nitrophenyl fucopyranoside versus p-nitrophenyl galactopyranoside. The evolved cell line showed a 66-fold increase in p-nitrophenyl fucosidase specific activity. The evolved fucosidase has a 10- to 20-fold increased kcat/Km for the fucose substrates compared with the native enzyme. The DNA sequence of the evolved fucosidase gene showed 13 base changes, resulting in six amino acid changes from the native enzyme. This effort shows that the library size that is required to obtain significant enhancements in specificity and activity by reiterative DNA shuffling and screening, even for an enzyme of 109 kDa, is within range of existing high-throughput technology. Reiterative generation of libraries and stepwise accumulation of improvements based on addition of beneficial mutations appears to be a promising alternative to rational design.
Proteins and enzymes with novel functions and properties can be obtained either by searching the largely unknown natural species or by improving upon currently known natural proteins or enzymes. The latter approach may be more suitable for creating properties for which natural evolutionary processes are unlikely to have been selected.
One promising strategy to create such novel properties is by directed molecular evolution. Starting with known natural protein(s), multiple rounds of mutagenesis, functional screening, and amplification can be carried out. When the mutation rate, library size, and selection pressures are properly balanced, the desired phenotype of a protein generally increases with each round (1–8). The advantage of such a process is that it can be used to rapidly evolve any protein, without any knowledge of its structure.
A number of different mutagenesis strategies exist, such as oligonucleotide cassette mutagenesis, point mutagenesis by error-prone PCR or the use of mutator strains, as well as DNA shuffling (1–5, 8). A theoretical approach to choosing a preferred mutagenesis strategy would be to determine the target protein’s fitness landscape (9), which is a plot of fitness (on the y axis) versus sequence space (on the x axis). However, because the sequence space of an average protein of 500 amino acids is 20500, determination of even a fraction of the fitness landscape is a nearly impossible and impractical undertaking. Because there are just a few fundamental ways to search sequence space, it may be informative to compare the performance of these methods for specific model systems.
Natural genes are thought to have evolved by mutation and recombination within a population of diverse, but highly related, sequences. We suggest that a search algorithm similar to that which slowly created the fitness landscape of a natural protein in the first place is likely to also be the preferred method for further searching this natural sequence landscape (5, 10). This approach is supported by our demonstration of the advantage of recombining mutations (over introduction of point mutations alone) for increasing the activity of a natural β-lactamase protein (2). However, recombination may not always be the best search algorithm. For searching the fitness landscapes of nonnatural sequences under unusual conditions, it is conceivable that a different approach may be more optimal.
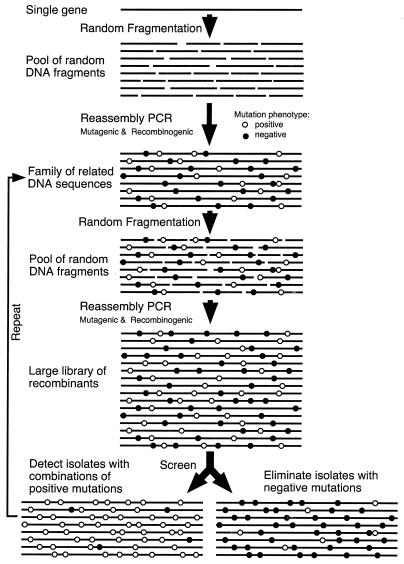
We obtain in vitro recombination of infrequent point mutations by a PCR-based technique called DNA shuffling (1–5). A pool of closely related sequences is fragmented randomly, and these fragments are reassembled into full-length genes via self-priming PCR and extension in a process we call reassembly PCR (4). This process yields crossovers between related sequences due to template switching. Shuffling allows rapid combination of positive-acting mutations and simultaneously flushes out negative-acting mutations from the sequence pool (Fig. 1). When coupled with effective selection and applied reiteratively, such that the output of one cycle is the input for the next cycle, reiterative DNA shuffling has been demonstrated to be an efficient process for directed molecular evolution (1–3).
Figure 1.
Schematic illustration of the DNA shuffling process used in the present study.
In our previous shuffling studies we used selection and/or large libraries (1, 2). Our primary goal in this work was to determine whether detection by screening of libraries of 10,000 clones, a number that is within range of any high throughput screening procedure, would be sufficient to obtain significant enhancement of a minor activity of β-galactosidase, a highly specific and complex enzyme, and at 109 kDa, one of the largest single-chain proteins in Escherichia coli. If screening would detect significant improvement, we then would establish that improvements are obtainable by evolution with such small libraries.
E. coli β-galactosidase, encoded by lacZ (11), is widely used, and its biological function, catalytic mechanism, and molecular structures are well characterized (11–15). It is a tetramer of identical subunits of 1,023 amino acids (13, 16, 17). The crystal structure of β-galactosidase is solved and shows that each subunit forms five structural domains (14). Each active site resides mainly in one subunit, but part of another subunit also is involved (14). The native enzyme hydrolyzes β-galactosyl linkages, such as the β(1, 4)-linkage in its natural disaccharide substrate, lactose. The native β-galactosidase is known to be highly specific for β-d-galactosyl substrates. A multistep model of the reaction was proposed (18, 19) based on kinetic studies of the native enzyme for o-nitrophenyl β-d-galactopyranoside (ONPG), p-nitrophenyl β-d-galactopyranoside (PNPG), and other substrates and substrate analogs. The native β-galactosidase acts only weakly on β-d-fucosyl moieties (18–20) and does not act on most substrate analogs.
MATERIALS AND METHODS
E. coli β-galactosidase (EC 3.2.1.23) and the galactosyl and fucosyl substrates 5-bromo-4-chloro-3-indolyl β-galactopyranoside (X-Gal), PNPG, ONPG, 5-bromo-4-chloro-3-indolyl β-d-fucopyranoside (X-Fuc), p-nitrophenyl β-d-fucopyranoside (PNPF), and o-nitrophenyl β-d-fucopyranoside (ONPF) were purchased from Sigma. Plasmid pCH110 containing a lacZ gene was from Pharmacia. E. coli strain TB1 was a gift from Charles Roessner of Texas A&M University.
Construction of Plasmid p18lacZ.
A 3.8-kb HindIII and BamHI restriction nuclease fragment from pCH110 containing a lacZ gene (codon 8 fused to a short N-terminal peptide) and the gpt promoter region (21) was subcloned into the HindIII and BamHI sites of vector p18-sfi-kan-sfi vector, a 2.3-kb pUC18 derivative in which the ampicillin gene is replaced by a kanamycin phosphotransferase gene (2). The resulting plasmid, named p18lacZ, was used for DNA shuffling. DNA fragments of 50–200 bp were used and reassembled as described previously (1, 2). The PCR primers for amplification of the reassembled genes were AGCGCCCAATACGCAAACCGCCTCTCCCCGCGCGTTGGCC (forward) and CTATGCGGCATCAGAGCAGATTGTACTGAGAGTGCACCAT (reverse), located on either side of the BamHI and HindIII fragment. The reassembled gene was digested with restriction enzymes HindIII and BamHI and ligated back into the P18-sfi-kan-sfi vector. The ligation mixture was electroporated into E. coli TB1 competent cells and plated out on Luria–Bertani plates (150 mm) with 40 μg/ml kanamycin and 2 mg/plate of the X-Fuc substrate (22). The plates were incubated 12 to 24 hr at 37°C. The resulting kanamycin-resistant transformants were visually screened for the intensity of the blue color. The 20–40 colonies with the most intense blue color were picked from about 10,000 transformants of each round and used for the next round of DNA shuffling. Seven rounds of DNA shuffling and screening were carried out. The best clone from the final screening round, called evolved β-fucosidase, was characterized in detail.
Enzyme Purification.
For purification of the native β-galactosidase and the evolved β-fucosidase, a histidine tag (His6) was fused to the N terminus of both enzymes by PCR with two primers [5′-(P)CATCACCATCACCACCATATCGTCACCTGGGACATGT and 5′-(P)GTATTTTTCGCTCATGTGAA] in a standard PCR. The histidine-tagged native and evolved enzymes were purified from overnight TB1 cell cultures harboring the corresponding plasmid (23). The crude cell extract, in 50 mM phosphate (pH 7.0) with 100 mM NaCl and 0.2 mM of phenylmethylsulfonyl fluoride protease inhibitor was passed through a 20-ml Ni-nitrilotriacetic acid agarose (Qiagen) column. The bound protein was stepwise-eluted with the same buffer containing 5 mM, 10 mM, 25 mM, and 100 mM imidazole. The active fractions from the metal affinity column were desalted and loaded on a DEAE column in 20 mM Tris (pH 7.5), followed by elution with a 0 to 1 M NaCl gradient. The active fractions were concentrated and loaded on a Superose 12 gel filtration column in an FPLC protein purification unit (Pharmacia). SDS/PAGE analysis (data not shown) showed that the native galactosidase and the evolved fucosidase were greater than 90% pure.
Enzyme Kinetics.
β-Galactosidase activity was assayed using the synthetic chromogenic substrates ONPG and PNPG. β-Fucosidase activity was assayed using chromogenic fucosyl substrates ONPF and PNPF. Enzyme assays were performed at 25°C and pH 7.0 in 30 mM N-tris(hydroxymethyl)methylaminoethanesulfonic acid with 1 mM MgCl2 and 150 mM NaCl. The absorbance change at 420 nm was recorded with time, and product formation was quantitated using the absorption extinction coefficient (2.65 mM−1·cm−1 for o-nitrophenol and 6.7 mM−1·cm−1 for p-nitrophenol). For kinetic parameter measurements, the initial velocity Vo (when less than 10% of the substrate was converted into product) was determined with varied substrate concentrations. The values of Vmax and Km were calculated using the simple weighting method of Cornish-Bowden (24). The Vmax values were converted to kcat values, the turnover number per active site, by normalizing for the enzyme concentrations by the molecular mass of the monomer. The Km and kcat values of the wild-type β-galactosidase for ONPF could not be determined directly because of the low activity on this substrate. The kcat/Km value had to be estimated from the enzyme dilution factor required for the native enzyme to generate the same amount of o-nitrophenol product from ONPG after the same period of time (usually several hours) and from the kcat/Km value of the wild-type enzyme on ONPG.
Sequencing of the Evolved lacZ Gene.
The 3.8-kb DNA fragment encoding the evolved β-galactosidase and its flanking regions was sequenced in both forward and reverse directions with 20 primers using an Applied Biosystems 391 DNA sequencer.
RESULTS AND DISCUSSION
Strategy for Evolving β-Galactosidase.
The primary goal of the experiment was to determine if a substantial enhancement in the specificity and/or activity of a large model enzyme could be obtained by reiterative screening of libraries of a size (10,000 clones) that is routinely accessible by high throughput detection assays. No structural information was used in the design of the experiment, but the structure of β-galactosidase is useful for interpretation of results.
Screening for Improved Fucosidase Activity.
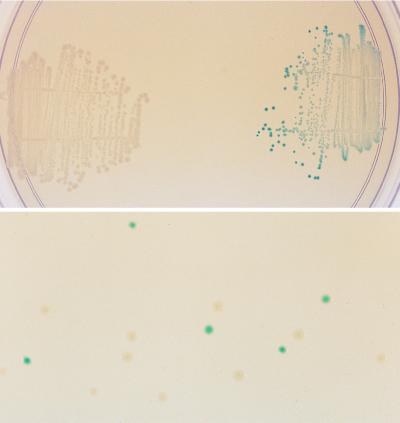
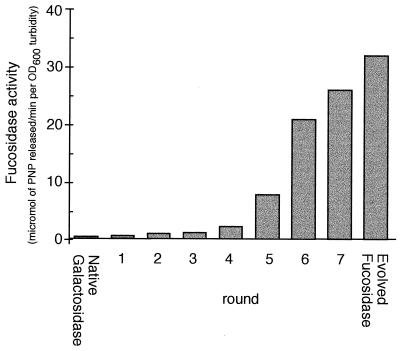
The 3.8-kb DNA fragment of p18LacZ containing the lacZ gene was shuffled as described previously (1–3), and the reassembled genes were digested with restriction enzymes (HindIII and BamHI) and ligated back into the vector p18-sfi-kan-sfi. The initial diversity was introduced into the native lacZ gene by random point mutagenesis, which occurs by shuffling of small fragments (1, 2). We previously showed that shuffling with 10- to 50-bp fragments resulted in a 0.7% rate of point mutation. Here we used fragments of 50 to 200 bp, which results in a much lower rate of point mutation, resulting in inactivation of approximately 20% of the clones. X-Fuc was chosen as the indicator substrate for the plate assay because of the nondiffusable nature of the colored product and the high sensitivity (22). After each round of DNA shuffling, 10,000 kanamycin-resistant transformants, growing on plates supplemented with X-Fuc, were visually screened for enhanced blue color formation. About 2–5% of the transformant colonies in each round showed colonies that were more highly blue-colored than the bulk of the population. The 20–40 bluest colonies (0.2–0.4%) were picked at each round, individually verified to be more active than the pool from the previous round by plate assays, and then used as a pool for the source of DNA to initiate the next round of DNA shuffling. This number of colonies was chosen as a compromise between obtaining too little diversity (<10 colonies) and obtaining suboptimal selection pressure (≫100 colonies), which could limit the rate of improvement. In the seventh round of DNA shuffling some colonies developed a deep blue color after overnight growth (Fig. 2). One mutant from this seventh and final round of shuffling showed a 66-fold increase in fucosidase activity on 1 mM PNPF (Fig. 3).
Figure 2.
E. coli TB1 cells expressing the native β-galactosidase (white colonies, Upper Left) and the evolved fucosidase of the seventh round (blue colonies, Upper Right) after overnight growth on an Luria–Bertani plus kanamycin plate supplemented with 0.1 mM X-Fuc. (Lower) The results of plating a deliberate mixture of the two types of colonies.
Figure 3.
Whole cell fucosidase activity on PNPF of the pool of colonies selected after each round of DNA shuffling. Rounds 1–7 are pools of colonies. Also shown are the activity of cells expressing the native β-galactosidase and cells expressing the evolved β-fucosidase, both measured as whole cell activity of single clones. The evolved fucosidase is the single-best colony selected after quantitative comparison of the 24 best colonies from the pool of colonies obtained after shuffling round 7. For assay conditions, see Materials and Methods.
Kinetics.
After the final round of selection, (His)6 tags were added to the foreign N terminus of the native β-galactosidase and the evolved β-fucosidase enzymes. Both enzymes were purified, and the kinetic constants of each enzyme on the synthetic chromogenic substrates ONPG, ONPF, PNPG, and PNPF were determined (Table 1). For PNPF, the Km value of the evolved fucosidase is decreased by 20-fold from the Km of wild-type β-galactosidase on the same substrate. The kcat value is decreased about 2-fold. The kcat/Km values thus are increased about 10-fold in the evolved β-fucosidase. The activity of the wild-type enzyme on ONPF was very low and accurate Km and kcat values could not be obtained. By comparing the relative reaction rates of the wild-type enzyme on ONPF and ONPG (at the same enzyme and substrate concentrations), the kcat/Km for ONPF was estimated, assuming that the kcat/Km value is a second order rate constant. The kcat/Km values on ONPF were increased at least 20-fold in the evolved β-fucosidase. These increases in fucosidase activity were accompanied by decreases in galactosidase activity. For the substrates PNPG and ONPG, the kcat/Km is decreased 40-fold and 50-fold, respectively. These kinetic parameter changes suggest that the substrate binding pocket in the evolved β-fucosidase is different from that of the wild-type β-galactosidase.
Table 1.
Kinetic constants for the native and evolved enzymes
| Substrate | Kinetic constant | Native galactosidase | Evolved fucosidase | |
|---|---|---|---|---|
| PNPG | kcat, s−1 | 268 | 30.9 | |
| Km, mM | 0.04 | 0.18 | ||
| kcat/Km, mM−1⋅s−1 | 6,700 | 172 | ||
| PNPF | kcat, s−1 | 209 | 96.6 | |
| Km, mM | 31 | 1.5 | ||
| kcat/Km, mM−1⋅s−1 | 6.7 | 64.4 | ||
| Specificity |
|
1,000 | 2.7 | |
| ONPG | kcat, s−1 | 765 | 14.5 | |
| Km, mM | 0.11 | 0.11 | ||
| kcat/Km, mM−1⋅s−1 | 6,950 | 132 | ||
| ONPF | kcat, s−1 | — | 24.1 | |
| Km, mM | — | 0.55 | ||
| kcat/Km, mM−1⋅s−1 | (2)* | 43.9 | ||
| Specificity |
|
3,200 | 3.0 |
The native galactosidase and the evolved fucosidase were purified, and the enzymes were assayed on four different substrates.
The kcat/Km value for the native galactosidase on ONPF was estimated to be about 2 mM−1⋅s−1 by measuring the hydrolysis rate relative to that of ONPG.
The native enzyme is highly specific for hydrolyzing galactosyl rather than fucosyl substrates. The kcat/Km values we determined for PNPG and PNPF differ by about 1,000-fold, and for ONPG and ONPF the values differ by more than 3,000-fold (Table 1). The values we determined for the native β-galactosidase on ONPG, PNPG, and PNPF are in between the values reported previously (18, 20). The substrate specificity changed dramatically from the native β-galactosidase to the evolved β-fucosidase. For the evolved β-fucosidase the kcat/Km values for substrates PNPG and PNPF differ 2.7-fold and for substrates ONPG and ONPF the kcat/Km values differ 3-fold. Therefore, the relative substrate specificity for fucosyl substrates, from the native to the evolved enzyme, is increased 1,000-fold for the o-nitrophenyl substrates and 300-fold for the p-nitrophenyl substrates. The substrate specificity change was further supported by inhibition of the enzymatic activity by isopropyl β-d-thiogalactopyranoside, a β-galactosidase substrate analog and a competitive inhibitor of galactosyl substrates. The Ki values increased by one order of magnitude from the wild-type enzyme to the evolved β-fucosidase, from 0.1 mM to 0.9 mM. The changes in Km values for the galactosyl substrates showed the same trend, because they either increased severalfold or stayed the same. These results imply that the substrate binding affinity of the evolved β-fucosidase is substantially increased for fucosyl substrates and decreased for galactosyl substrates, and hence the substrate binding pocket is likely to be significantly modified.
DNA Sequence.
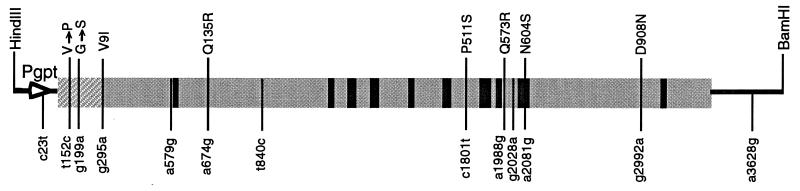
The DNA sequence of the evolved fucosidase gene showed 13 nucleotide substitutions of which 11 were in the coding region. Six of the mutations are predicted to cause amino acid changes in the translated β-galactosidase sequence. Two additional mutations are predicted to cause amino acid changes in the N-terminal fusion peptide (Fig. 4). All 13 nucleotide changes were base transitions between purines and/or between pyrimidines, which usually are more frequent than transversions.
Figure 4.
Nucleotide substitutions in the evolved fucosidase gene. The predicted amino acid changes are shown above the gene by the single-letter denotation, numbered according to the wild-type β-galactosidase sequence (17). Amino acid changes in the N-terminally fused peptide region (hatched area) are indicated by small vertical arrows. Mutations that do not result in amino acid changes are shown below the gene, numbered starting at the HindIII site, as in the parental plasmid pCH110. The gpt promoter is indicated by a thick arrow. The positions of the known active site residues of the wild-type β-galactosidase are indicated by black bars.
One major advantage of in vitro evolution of enzymes over the structural modeling approach is that only minimal information is required for improving the desired phenotype. At each round of our experiment, only colonies with increased fucosidase activity were pooled and used for the next round of DNA shuffling and screening. Although both positive-acting mutations and neutral mutations may accumulate in the evolved fucosidase lacZ gene in each round, we expect that neutral mutations generally do not survive multiple rounds of shuffling and screening due to a back-crossing effect exerted by the consensus sequence (1, 2), combined with the lack of a selective advantage of the neutral mutations. Therefore, only mutations that somewhat contribute to the improved fucosidase activity are likely to accumulate in the evolved fucosidase. While we have not determined the effect of the separate mutations by site-specific mutational studies, we can predict what roles some of the mutations may play based on the three-dimensional structure of the parental β-galactosidase (ref. 14; Fig. 5) and the sophisticated kinetic models based on previous mutations and kinetic analysis of purified proteins (18, 20, 25, 26). Among the six amino acid changes in the β-galactosidase sequence, none appear directly involved in the inter-subunit contact. Three mutations (Pro511Ser, Gln573Arg, and Asn604Ser) are located in domain 3 (residues 334–627) of the wild-type E. coli β-galactosidase (14). Domain 3 in the native protein contains most of the amino acids that form the substrate binding pocket (ref. 14; Fig. 5). Asn604 is one of the amino acids forming this substrate binding pocket in the protein (14), and this residue is conserved in several other known β-galactosidase sequences (25, 27–29), except the evolved galactosidase gene (ebgA) of E. coli (30). In our evolved fucosidase, Asn604 is replaced by Ser. This mutation presumably affects the enzyme’s substrate specificity. All the other mutations found in the evolved β-fucosidase enzyme do not directly affect the active site and substrate binding pocket residues, and therefore they may have no effect or may only subtly change the conformation of the active site and substrate specificity. Gln573, substituted by Arg in the evolved fucosidase, is in close proximity to the substrate binding pocket (Fig. 5). The mutation Pro511Ser is also close to the active site and substrate binding pocket (Fig. 5). These two mutations are likely to affect the enzyme’s active site. Asp908Asn is also close to the active site and may also affect the activity. Additional important catalytic residues of the active site, such as Glu461, Met502, Tyr503, and Glu537 (23, 26, 31), however, are unchanged in the evolved β-fucosidase, implying that the catalytic mechanism of the evolved enzyme remained the same. Therefore the evolved β-fucosidase seems to have only adjusted to fit the fucosyl substrate or its transition state better than the wild-type β-galactosidase does. In addition, one of the nucleotide mutations outside the structural gene (c23t) is very close to the gpt promoter region and could affect transcription (Fig. 4). This mutation, along with the two amino acid mutations in the N-terminal fusion peptide (Fig. 4), may influence the expression level of the protein. Indeed, we found that the evolved β-fucosidase enzyme was expressed at least 2- to 3-fold higher than the wild-type enzyme (data not shown). The mutations Val9Ile and Gln135Arg are far away from the active site and near the surface of the protein (Fig. 5), and may not have any significant effect on the enzymatic activity. The analysis of mutations obtained by molecular evolution of proteins provides a new tool for studying structure–function relationships. However, the real utility of DNA shuffling is the ability to rapidly improve enzyme functions without the need to delineate the myriads of complex molecular mechanisms.
Figure 5.
Ribbon representation of the E. coli β-galactosidase subunit structure (14). The CNO atoms of the six amino acid mutations that conferred the fucosidase activity are shown with stick representation. Two mutations in the active site (Asp604 and Gln572) are shown in red. Two mutations in close proximity of the active site (Pro511 and Asp908) are shown in magenta. Two mutations far away from the active site and on the protein surface (Val9 and Gln135) are shown in green. The rest of the substrate binding and active site residues are shown in yellow.
There are several possible applications for the evolved β-fucosidase. One is as a novel reporter for β-d-fucosyl substrates, in addition to the widely used lacZ gene reporter. The advantage of using the novel enzyme is the low endogenous background of β-fucosidase activity because, unlike α-fucosidases, β-fucosidases are uncommon in nature. This well expressed fucosidase also could be used for the production of fucosyl adducts or for disaccharide synthesis by transglycosylation or reversal of the hydrolysis reactions, because analogous applications already have been demonstrated for the wild-type β-galactosidase (32, 33). Some of these applications may require further evolution of the fucosidase for the specific reaction. The present data suggests that it is reasonable to attempt to obtain such improvements by DNA shuffling and screening of libraries of modest size.
Acknowledgments
We thank A. Crameri for technical assistance and Drs. F. H. Arnold, R. E. Huber, and P. Schatz for useful discussions.
ABBREVIATIONS
- ONPG
o-nitrophenyl β-d-galactopyranoside
- ONPF
o-nitrophenyl β-d-fucopyranoside
- PNPG
p-nitrophenyl β-d-galactopyranoside
- PNPF
p-nitrophenyl β-d-fucopyranoside
- X-Fuc
5-bromo-4-chloro-3-indolyl β-d-fucopyranoside
References
- 1.Stemmer W P C. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stemmer W P C. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 3.Crameri A, Whitehorn E A, Tate E, Stemmer W P C. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 4.Stemmer W P C. The Encyclopedia of Molecular Biology. New York: VCH; 1996. pp. 447–457. [Google Scholar]
- 5.Stemmer W P C. Bio/Technology. 1995;13:549–553. [Google Scholar]
- 6.You L, Arnold F H. Protein Eng. 1995;9:77–83. doi: 10.1093/protein/9.1.77. [DOI] [PubMed] [Google Scholar]
- 7.Chen K, Arnold F H. Proc Natl Acad Sci USA. 1993;90:5618–5622. doi: 10.1073/pnas.90.12.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore J C, Arnold F H. Nat Biotechnol. 1996;14:458–467. doi: 10.1038/nbt0496-458. [DOI] [PubMed] [Google Scholar]
- 9.Kauffman S A. The Origins of Order. New York: Oxford Univ. Press; 1993. [Google Scholar]
- 10.Stemmer W P C. Science. 1995;270:1510. doi: 10.1126/science.270.5241.1510. [DOI] [PubMed] [Google Scholar]
- 11.Beckwith J. In: Escherichia coli and Salmonella. Neidhardt F, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1227–1231. [Google Scholar]
- 12.Zabin I, Fowler A V. In: The Operon. Miller J H, Reznikoff W S, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1980. pp. 89–122. [Google Scholar]
- 13.Wallenfels K, Weil R. In: The Enzymes. Boyer P D, editor. New York: Academic; 1972. pp. 618–663. [Google Scholar]
- 14.Jacobson R E, Zhang X-J, DuBose R F, Matthews B W. Nature (London) 1994;369:761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- 15.Huber R E, Gupta M N, Khare S K. Int J Biochem. 1994;26:309–318. doi: 10.1016/0020-711x(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 16.Fowler A, Zabin I. J Biol Chem. 1978;253:5521–5525. [PubMed] [Google Scholar]
- 17.Kalnins A, Otto K, Ruther U, Muller-Hill B. EMBO J. 1983;2:593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth N J, Huber R E. J Biol Chem. 1996;271:14296–14301. doi: 10.1074/jbc.271.24.14296. [DOI] [PubMed] [Google Scholar]
- 19.Huber R E, Gaunt M T. Arch Biochem Biophys. 1983;220:263–271. doi: 10.1016/0003-9861(83)90409-5. [DOI] [PubMed] [Google Scholar]
- 20.Wallenfels K, Lehmann J, Malhotra O P. Biochem Z. 1960;333:209–215. [PubMed] [Google Scholar]
- 21.Hall C V, Jacob P E, Ringold G M, Lee F. J Mol Appl Genet. 1983;2:101–109. [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Cupples C G, Miller J H, Huber R E. J Biol Chem. 1990;265:5512–5518. [PubMed] [Google Scholar]
- 24.Cornish-Bowden A. Principles of Enzyme Kinetics. London: Butterworth; 1976. pp. 168–189. [Google Scholar]
- 25.Buvinger W E, Riley M. J Bact. 1985;163:850–857. doi: 10.1128/jb.163.3.850-857.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gebler J C, Aebersold R, Withers S G. J Biol Chem. 1992;267:11126–11130. [PubMed] [Google Scholar]
- 27.Schroeder C J, Robert C, Lenzen G, McKay L L, Mercenier A. J Gen Microbiol. 1991;137:369–380. doi: 10.1099/00221287-137-2-369. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt B F, Adams R M, Requadt C, Power S, Mainzer S E. J Bacteriol. 1989;171:625–635. doi: 10.1128/jb.171.2.625-635.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hancock K R, Rockman E, Young C A, Pearce L, Maddox I S, Scott D B. J Bacteriol. 1991;173:3084–3095. doi: 10.1128/jb.173.10.3084-3095.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stokes H W, Betts P W, Hall B G. Mol Biol Evol. 1985;2:469–477. doi: 10.1093/oxfordjournals.molbev.a040372. [DOI] [PubMed] [Google Scholar]
- 31.Ring M, Huber R E. Arch Biochem Biophys. 1990;283:342–350. doi: 10.1016/0003-9861(90)90652-f. [DOI] [PubMed] [Google Scholar]
- 32.Hedbys L, Larsson P O, Mosbach K, Svensson S. Biochem Biophys Res Commun. 1984;123:8–15. doi: 10.1016/0006-291x(84)90372-3. [DOI] [PubMed] [Google Scholar]
- 33.Huber R E, Hurlburt K L. Arch Biochem Biophys. 1986;246:411–418. doi: 10.1016/0003-9861(86)90487-x. [DOI] [PubMed] [Google Scholar]