Abstract
Phosphoinositides are minor phospholipid constituents of virtually every biological membrane yet they play fundamental roles in controlling membrane-bound signalling events. Phosphoinositides are produced from phosphatidylinositol (PtdIns) by phosphorylation of one or more of three positions (3, 4 and 5) of the inositol headgroup located at the membrane cytoplasmic interface by distinct families of inositol lipid kinases. Intriguingly, many of the kinase reactions are catalysed by more than one form of the kinases even in simple organisms and these enzymes often assume non-redundant functions. A similar diversity is seen with inositide phosphatases, the enzymes that dephosphorylate phosphoinositides with a certain degree of specificity and the impairments of which are often linked to human diseases. This degree of multiplicity at the enzyme level together with the universal roles of these lipids in cell regulation assumes that inositol lipids are spatially and functionally restricted in specific membrane compartments. Studying the compartmentalized roles of these lipids at the cellular level represents a major methodological challenge. Over the last 10 years significant progress has been made in creating reagents that can monitor inositol lipid changes in live cells with fluorescence or confocal microscopy. New methods are also being developed to manipulate these lipids in specific membrane compartments in a regulated fashion. This article recalls some historical aspects of inositide research and describes the new methodological advances highlighting their great potential as well as the problems one can encounter with their use.
Introduction and historical perspectives
Agonist-induced changes in phosphatidylinositol (PtdIns) turnover were first described in the 1950s (Hokin & Hokin, 1953) and the structures of brain phosphoinositides determined in the 1960s (Ballou & Lee, 1964). However, the notion that phosphoinositide turnover is part of the signalling cascade initiated by cell surface receptors of the calcium-mobilizing type was introduced only in 1975 in the seminal review by Robert Michell (Michell, 1975). Early cell fractionation studies have shown that PtdIns is primarily synthesized in the endoplasmic reticulum (ER), while further phosphorylation of PtdIns to PtdIns4P and PtdIns(4,5)P2 occurs in the plasma membrane (PM) as well as in other internal membranes (Michell & Hawthorne, 1965; Michell et al. 1967). The full details of the receptor-mediated changes in PtdIns turnover and its connection to Ca2+ signalling have been clarified by the mid 1980s (Fig. 1A). By then it was understood that activated receptors stimulate the enzyme phospholipase C (PLC) at the PM to hydrolyse PtdIns(4,5)P2 yielding two messenger molecules: the water-soluble Ins(1,4,5)P3 (InsP3) and the membrane-bound diacylglycerol (DAG) (reviewed in Berridge & Irvine (1984). The former binds to InsP3 receptors located in the ER membrane to release stored Ca2+ and regulate Ca2+-dependent enzymes while the latter binds and activates protein kinase C (PKC) enzymes to phosphorylate downstream effectors characteristic of the cell in question. Important further details of this basic signalling scheme have been clarified subsequently: these included the characterization of the G-proteins involved in coupling receptors to PLC (Smrcka & Sternweis, 1993), the identification of various forms and activation mechanisms of the PLC enzymes (Suh et al. 1988), as well as the three forms of InsP3 receptors (Maeda et al. 1991) and the ever increasing number of PKC isoforms (Nishizuka, 1988). However, several observations also indicated that phosphoinositides are also involved in other aspects of cell regulation (Fig. 1B). For example, insulin was shown to activate PI kinases without stimulating PLC (Sale et al. 1986), PLD activation by PtdIns(4,5)P2 was observed without an apparent link to receptor activation (Liscovitch et al. 1994), and regulation of the small GTPase Arf1 and coat assembly by phosphoinositides in the Golgi was described (Randazzo & Kahn, 1994). It was also reported that regulated secretion and fusion of secretory vesicles with the PM also required phosphoinositides (Eberhard et al. 1990).
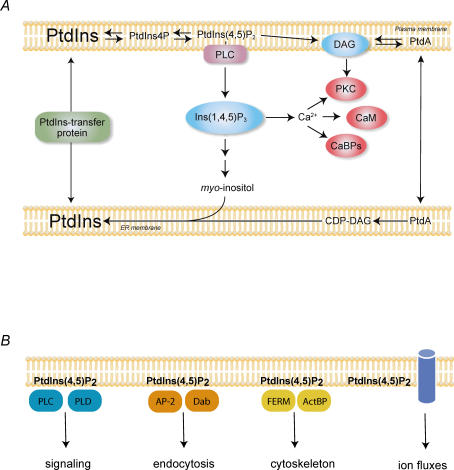
Figure 1. The phosphoinositide signalling cascade and processes at the PM controlled by PtdIns(4,5)P2.
A, the canonical phosphoinositide signalling cascade. PM PtdIns(4,5)P2 is hydrolysed by activated PLC enzymes upon receptor stimulation to yield the two messengers, Ins(1,4,5)P3 and DAG. Ins(1,4,5)P3 mobilizes intracellular Ca2+ via its receptors located in the ER that function as Ca2+ channels. DAG helps recruit PKC enzymes to the membrane and the cytosoloic Ca2+ increase stimulates Ca2+ sensitive enzymes such as calmodulin (CaM) or other Ca2+ binding proteins (CaBPs) to regulate downstream effectors. Ins(1,4,5)P3 is rapidly degraded by sequential dephosphorylations to myo-inositol, which is then reused for PtdIns synthesis in the ER by conjugation with CDP-DAG. PtdIns transfer proteins facilitate the movement of PtdIns between membranes. B, PtdIns(4,5)P2 controls numerous processes at the PM. It activates enzymes such as PLCδ and PLD, it helps recruit clathrin adaptor proteins, such as AP-2 or Dab2, it provides a link between the membrane and the cytoskeleton via interaction with the FERM domain containing or other actin binding proteins (ActBP) and also regulates ion channels and transporters. It is very likely that the affinities of interaction of the lipid with the various proteins show big variations and when the overall level of the lipid drops in the membrane, it will affect processes that are controlled by weaker interactions. In contrast, effectors with tight PtdIns(4,5)P2 binding are less affected by global PtdIns(4,5)P2 changes but perhaps are sensitive to very local changes that are evoked by colocalized kinases or phosphatases.
A major development in inositide research was the discovery of the 3-phosphorylation of phosphoinositides and the identification of PI 3-kinases (Whitman et al. 1988). PI 3-kinases (the Class I forms) can phosphorylate PtdIns(4,5)P2 in the PM to generate PtdIns(3,4,5)P3 and their increased activity and association with transforming tyrosine kinase oncogenes has made them very attractive research targets (Cantley, 2002). The Class III PI 3-kinases, on the other hand, phosphorylate PtdIns to PtdIns3P in endosomes and are key regulators of vesicular trafficking (Schu et al. 1993; Volinia et al. 1995). Since 3-phosphorylated inositides are not hydrolysed by phosphoinsitide PLC enzymes, it was understood that these lipids themselves represent a signal in the membrane to initiate cellular responses (Toker & Cantley, 1997). This has turned attention toward proteins that can bind and hence be regulated by phosphoinositides. In the meantime PI 4-kinases have also shown more complexity than originally thought. The highest PI 4-kinase activity known at the time was the tightly membrane-associated enzyme (termed type II PI4K), and it was believed to be responsible for the generation of the PtdIns(4,5)P2 in the PM as exemplified in the red blood cell membrane. Therefore, it came as a surprise when it was found that the generation of the PtdIns(4,5)P2 pool acted upon by agonist-regulated PLCs required another PI 4-kinase that belonged to a new family (termed type III) (Nakanishi et al. 1995). Molecular identification of these latter enzymes (PI4KIIIα and PI4KIIIβ) revealed their similarity to the then recently cloned two yeast PI4Ks, Stt4p and Pik1p, that had been found to serve non-redundant functions (recently reviewed in Balla & Balla, 2006) (Fig. 2). Similar complexity was found with the kinases that phosphorylate PtdIns4P to PtdIns(4,5)P2, termed the PIP kinases. Two such activities were known to exist with unique regulatory properties and the molecular cloning of these proteins revealed two classes of the enzymes, termed type I and type II PIP kinases (Boronenkov & Anderson, 1995; Divecha et al. 1995). Three forms have been identified in both classes, and it was discovered that type II PIP kinases use PtdIns5P as a substrate and, hence are in fact PtdIns(5)P 4-kinases (Rameh et al. 1997). Some of the type I PIP kinases also have additional splice variants that determine their unique subcellular location and regulation (Doughman et al. 2003; Giudici et al. 2004).
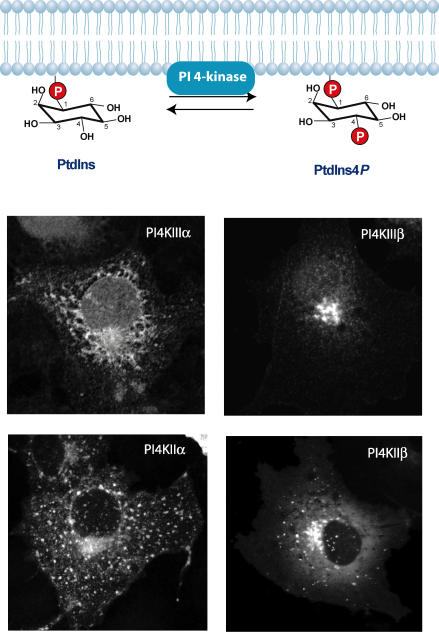
Figure 2. Inositide kinase reactions are often performed by multiple enzymes.
Phosphorylation of PtdIns to PtdIns4P is mediated by four distinct PI 4-kinases (PI4Ks) that localize to unique membrane compartments. All of the enzymes show localization to the Golgi compartment but PI4KIIIα is also found in the ER, while PI4KIIα is also found in endosomes and the PM. PI4IIIβ is exclusively localized to the Golgi while PI4KIIβ shows partial Golgi and some endosomal localization. The localization of the enzymes does not show dramatic changes after stimulation of the cells even when PtdIns4P levels are expected to change in compartments where the enzymes are not enriched (e.g. the PM). This indicates that there is a need to detect their lipid product, PtdIns4P, in addition to following the enzyme distribution to better understand their regulation.
An even larger diversity is found in the enzymes that dephosphorylate phosphoinositides. The importance of these activities from a regulatory standpoint has been initially unappreciated as the focus was on the phosphatases that dephosphorylate InsP3 and its metabolites. The first indication that lipid phosphatases may have an as yet unrecognized significance came when the gene responsible for the OCRL syndrome (oculo–cerebro–renal syndrome of Lowe) was identified as a phosphoinositide 5-phosphatase (Attree et al. 1992). Many groups and forms of inositide phosphatases have since been identified and characterized (Taylor & Dixon, 2003; Astle et al. 2006). Some of these enzymes act both on soluble InsPs and inositol lipids, while others use only lipids as substrates. Some of them show high specificity for a specific position and only act on substrates of specific phosphorylation patterns, while others are more promiscuous in their substrate preference. The unique feature of these proteins is that their impaired functions are often found in human diseases. PTEN (phosphatase and tensin homologue mutated in multiple advanced cancers 1), for example, which encodes a 3-phosphatase acting on PtdIns(3,4,5)P3, is a tumour suppressor gene that is often defective or missing in many human cancers such as gliomas (Taylor & Dixon, 2003). Myotubularins are 3-phosphatases that act on PtdIns3P, and are impaired in some forms of myotubular dystrophies (Taylor & Dixon, 2003). Clearly, the diversity and importance of the inositide phosphatases are further testaments to the fact that phosphoinositides are tightly controlled regulatory molecules with roles far beyond what was initially recognized as related to receptor-mediated calcium signalling.
Detection of compartmentalized phosphoinositide dynamics
The realization of the localized roles of phosphoinositides in distinct membrane compartments has created a need for methods by which these localized changes can be detected and followed. Antibodies against phosphoinositides have been introduced for some time (Miyazawa et al. 1988) but they have not been widely used because of technical difficulties during cell fixation and detection that caused inconsistent results between various laboratories. Interestingly, the alternative methods discussed below have raised the antibody work also to a different standard, and recently more successful efforts have been reported with their use (e.g. Hammond et al. 2006).
It was the significant developments in the identification and molecular and structural characterization of protein modules that recognize phosphoinositides that gave rise to the knowledge and the idea that these protein modules might be used to detect phosphoinositides in living cells (Hurley & Meyer, 2001). In the first reports the pleckstrin homology (PH) domain of selected proteins known to recognize PtdIns(4,5)P2 and PtdIns(3,4,5)P3 were used as GFP fusion proteins to detect these lipids in intact cells (Kontos et al. 1998; Stauffer et al. 1998; Várnai & Balla, 1998; Venkateswarlu et al. 1998; Varnai et al. 1999). Simultaneously, the FYVE domains (named after the first proteins in which this domain was identified: Fab1p, YOTB, Vac1p and EEA1) of the yeast Fab1p and mammalian EEA1 and Hrs proteins were used to show that these domains detect PtdIns3P (Burd & Emr, 1998; Gillooly et al. 2000). The first small-scale screen of mammalian PH domains (Dowler et al. 2000) followed by a thorough analysis of the whole identified yeast PH domain set (Yu et al. 2004) have identified several PH domains with unique lipid recognition specificities. However, the yeast study also showed that even with limited inositide binding specificity, a PH domain is able to display unique cellular localization patterns consistent with detection of a rather specific inositide pool. Over the last eight years the PH domain and FYVE domain based lipid probes have been widely and increasingly used with great success. These studies have concluded that PtdIns(4,5)P2 is mostly detected in the PM while PtdIns4P is mostly found in the Golgi, although PtdIns4P is also detected in the PM. PtdIns(3,4,5)P3 shows very low levels in quiescent cells and responds with a dramatic rise in the PM after stimulation of receptor tyrosine kinase receptors. PtdIns3P, on the other hand, is constitutively produced on early endosomes as detected by these fluorescent probes (Fig. 3). These studies have been covered in detail in several recent publications (Downes et al. 2005; Halet, 2005; Varnai & Balla, 2006) and are not further discussed here. What will be pointed out instead are the controversial points related to these data and their interpretations.
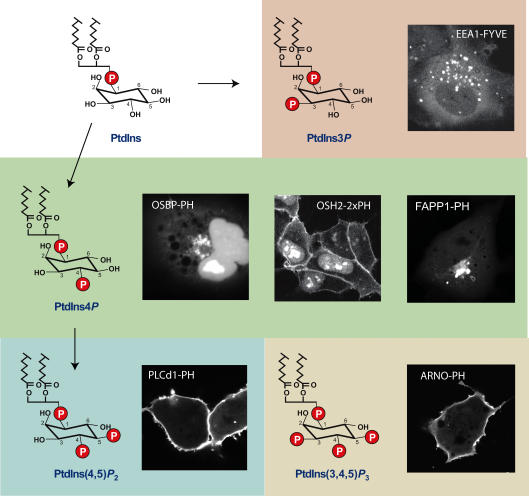
Figure 3. Steady-state distribution of inositide detecting probes visualized in live cells.
The structure of the inositide headgroup is shown along with the cellular distribution of the particular protein module recognizing the lipid and fused to the enhanced green fluorescent protein (GFP). Note that PtdIns4P can be detected either in the PM or in the Golgi, depending on the probe used. No such discrepancy has been found so far with probes detecting PtdIns(4,5)P2, PtdIns3P and PtdIns(3,4,5)P2 but it is still not certain that this means that these lipids are only found in those membranes. More probes are needed to investigate this question.
Binding of water-soluble versus membrane bound inositol phosphate headgroups
The specificity of inositide recognition lies in the stereospecific recognition of the inositol phosphate headgroup by the protein module, regardless of whether the latter is an antibody or a lipid-binding protein domain. This, by principle, assumes that a soluble inositol phosphate of the same phosphorylation status is also able to bind to the same binding site. However, if the localization of the protein domain to the membrane is also aided by additional interactions (electrostatic, hydrophobic or protein–protein) either with the diacylglycerol backbone or other membrane components, then the soluble inositol phosphate will be only partially active in displacing the lipid probe from the membrane inside the cells. In contrast, if the binding to the soluble inositol phosphate headgroup is tighter than that to the same inositide headgroup located in the membrane environment (e.g. because of spatial constraints, such as more difficult access to the phosphodiester 1-phosphate), then any increase in the level of the inositol phosphate will displace a disproportionally larger fraction of the probe from the membrane. This has been an important point often argued about in the case of the PLCδ1PH–GFP probe that binds Ins(1,4,5)P3 with higher affinity than PtdIns(4,5)P2 assessed in in vitro binding assays. Because of these apparent relative affinities, this probe is often considered a better sensor of InsP3 increase than of PtdIns(4,5)P2 decrease (Hirose et al. 1999). Several experimental and theoretical modelling approaches have addressed this question (reviewed recently in Varnai & Balla, 2006). The conclusion from these studies is that large InsP3 increases do contribute to the translocation of PLCδ1PH–GFP from the PM, but in most cells this probe follows PtdIns(4,5)P2 changes relatively faithfully and it is probably a greater mistake to consider the PLCδ1PH–GFP a pure InsP3 sensor than to consider it a PtdIns(4,5)P2 sensor. Given the fact that the contribution of the lipid decrease and the InsP3 increase to the redistribution of the PLCδ1PH–GFP can change from cell to cell and even from agonist to agonist in the same cell, it is best to treat these changes as signs of PLC activation until the contribution of the single components is tested in a controlled manner as elegantly done in Horowitz et al. (2005).
The lack of localization does not mean the lack of lipids
It is becoming more and more obvious that the cellular localization of a lipid probe depends on several factors in addition to its inositol lipid binding. While the reason for this is not clear for most lipid-binding domains, it most probably reflects additional interactions with other lipids or proteins in the membrane. This does not make these probes less useful as often the presence of the lipids is essential for probe localization and lipid changes can be followed as changes in localization. However, this also means that the lipid changes are monitored only in the context of a molecular complex in a specific membrane compartment as opposed to an overall change in every membrane where the lipid is found. The best examples to highlight this difference are shown by PH domains that recognize PtdIns4P. The PH domains of the oxysterol binding protein (OSBP) and FAPP1 (Four phosphate adaptor protein) are found in the Golgi/Tgn membranes and their localization appears to depend on both PtdIns4P and Arf1–GTP (Godi et al. 2004; Balla et al. 2005; Weixel et al. 2005). These probes can detect PtdIns4P in the PM but only under special conditions such as a recovery from a massive PLC activation (Balla et al. 2005). In contrast, the OSH2 (yeast oxysterol binding protein homologue 2) PH domain proves to be a better probe for PtdIns4P in the PM with poor detection of the lipid in the Golgi (Roy & Levine, 2004; Yu et al. 2004) (Fig. 3). It is not clear what other interactions make the latter probe prefer the PM, but this example clearly illustrates the notion that lack of localization of a probe cannot be taken as an argument for the lack of the lipid at a particular membrane. This question often comes up in the context of whether PtdIns(4,5)P2 is present in the Golgi, as none of the PH domains that recognize this lipid decorate the Golgi, yet several lines of evidence suggest that PtdIns(4,5)P2 has functional roles in this compartment (De Matteis et al. 2005). More studies will be needed to address these questions with alternative methods such as EM analysis of PH domain binding to fixed tissues or cells that should be less dependent on protein–protein interactions of the probes (Watt et al. 2002).
Binding and sequestering inositides distorts biology
Overexpression of protein domains that bind inositides by definition is expected to alter the availability of these lipids for enzymes and effectors. The only question is the extent to which these aberrations take place and limit our conclusions based on the use of these probes. FRAP (fluorescence recovery after photobleaching) experiments have shown that the steady-state membrane localization of the PLCδ1PH–GFP is the result of its very rapid cycling between a membrane (PtdIns(4,5)P2)-bound and cytosolic states (van Der Wal et al. 2001). PLCδ1PH–GFP overexpression is expected to increase the amount of PtdIns(4,5)P2 that is bound to the PH domain at any moment of time but this pool is not likely to be all taken from the total PtdIns(4,5)P2 that was present before expressing the construct, but partially represents ‘extra’ PtdIns(4,5)P2 that is in dynamic equilibrium with the rest of this lipid. Because of its rapid on–off rates, this extra pool is available for PLC-mediated hydrolysis as well as to 5-phosphatases as demonstrated by numerous studies, and therefore sequestration of the PtdIns(4,5)P2 pool by the PH domain is functionally not so prominent. Nonetheless, overexpression of this probe is clearly toxic to the cells as evidenced by rounding and detachment and accumulation of large vesicles inside cells expressing high levels of the protein. This effect is partly due to the interruption of membrane and cytoskeletal contacts (Raucher et al. 2000). In a recent study we analysed the dominant negative effects of several PH domains with PtdIns(3,4,5)P3 recognition on cellular responses known to be regulated by the same lipid. We found an unexpected specificity in their inhibitory profiles in that some of the PH domains preferentially inhibited PtdIns(3,4,5)P3-dependent pathways that were related to the function of the proteins from which the PH domain was obtained. These studies led us to conclude that the dominant negative effects in many cases are probably related to sequestration of putative protein binding partners rather than the lipids themselves since the lipids can be easily produced in extra amounts (Varnai et al. 2005). In most recent studies the protein binding partner for the Grp1/ARNO PH domain has been identified as Arf6 (Cohen et al. 2007). It is important to emphasize that at moderate expression levels the presence of these probes is well compensated by the cells, as most convincingly shown by the successful creation of transgenic mice expressing PH-domain GFP reporters without any obvious functional defects (Nishio et al. 2007). However, determination of the protein binding partners for the individual inositide binding domains will largely help our understanding of their biology as well as the critical evaluation of the lipid data obtained with their use.
How to quantify changes in probe distribution
One of the important questions during analysis of phosphoinositide dynamics monitored by the PH domain GFP probes is how to assess changes in localization. The simplest method is to monitor the cytoplasmic versus membrane-bound fluorescence using quantitative imaging. However, this may be problematic when cells change shape or the fluorescent probe distribution changes during the measurements unrelated to lipid changes (such as slow diffusion in or out of the nucleus). Moreover, intensity increases in the membrane are often caused by intense membrane ruffling when new membranes are added to the PM and these intensity increases – which mostly require PtdIns(3,4,5)P3– do not necessarily mean real membrane recruitment of the fluorescent probes. To overcome this problem it is advised that membrane markers are used as reference signals when assessing recruitment during intense membrane activity. FRET (fluorescence resonance energy transfer) measurements between two fluorophore pairs (usually CFP and YFP or YFP and mRFP) have been successfully used to monitor the membrane localization of the probes (van Der Wal et al. 2001; Balla et al. 2005). When expressed together, CFP- and YFP-fused PH domains, are within FRET distance producing high FRET signal when they are bound to the membrane. This signal drops as soon as the probes dissociate from the membrane and diffuse to the cytososol. Several controls are needed to ensure that the changes are indeed due to the translocation of the probes, but this method has been able to detect small changes that would otherwise escape detection (Varnai et al. 2006). An alternative method is the use of TIRF (total internal reflection fluorescence) microscopy to detect fluorescence only in the membrane region of the cell attached to the coverslip (Haugh et al. 2000; Schneider & Haugh, 2004) (see an example in Fig. 4). This method is fairly simple and straightforward (although it needs a special instrument) but it is also sensitive to artifacts originating from changes in cell shape and footprint or membrane additions during the experiment. The use of a fluorescent membrane marker can also help to filter out the changes unrelated to altered membrane localization of the lipid probe.
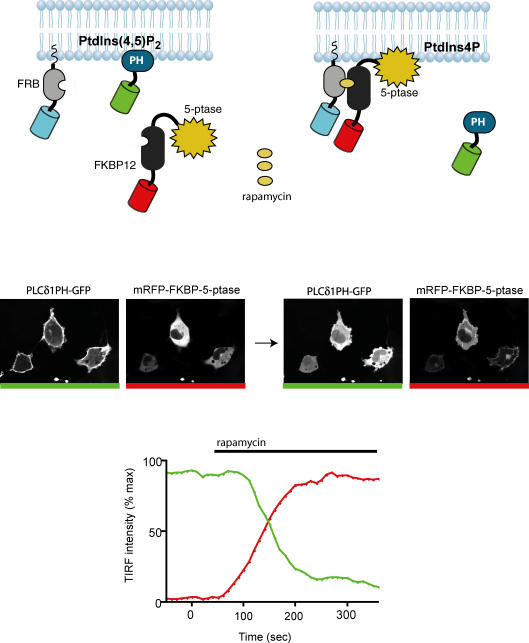
Figure 4. Chemically induced manipulation of PtdIns(4,5)P2 in the PM.
The FRB fragment of the mTOR protein is targeted to the PM by an N-terminal palmitoylation sequence and is also tagged with CFP (this channel is not shown). The 5-phosphatase (5-ptase) domain of the type-IV phosphoinositide 5-phosphatase enzyme is fused to an mRFP–FKBP12 construct and this protein is cytosolic under basal conditions (see image on the left with red bar). Simultaneous expression of the PH domain of PLCδ1 fused to GFP shows PtdIns(4,5)P2 in the PM (image on the left with green bar). Addition of rapamycin heterodimerizes the FRB–FKBP12 pairs and thereby recruits the 5-phosphatase to the membrane (image on the right with red bar) which, in turn, rapidly eliminates PtdIns(4,5)P2 as indicated by the translocation of PLCδ1PH–GFP from the membrane to the cytosol (image on the right with green bar). These changes can be monitored by TIRF microscopy as indicated by the bottom graph. The fluorescence intensity of the PLCδ1PH–GFP in the membrane plane drops rapidly from its initial high value after rapamycin addition (green trace) as the 5-phosphatase is recruited to the PM (red trace).
How should inositides be manipulated in live cells?
Monitoring localized lipid changes is only one aspect of studying the spatially restricted signalling roles of phosphoinositides. It is also desirable that one should be able to induce changes in lipid levels within the cells. This has been achieved by adding exogenously the lipids to cells in such ways that the lipids enter the cells and reach internal membranes. Shuttling lipids into cells has been a popular way to test their effects (Ozaki et al. 2000). These techniques have the disadvantage of flooding the cell with a lipid and not being able to control where the lipid ends up and how it is degraded during its journey within the cell. This approach also takes away the spatially restricted aspect of the lipid changes and therefore may not be informative enough. An alternative and widely used approach has been to express the inositide kinases or phosphatases in the cell so that they will generate or eliminate the lipids in the locations where the enzymes are targeted by their natural mechanisms. The reverse of this is to use RNAi-mediated gene silencing to eliminate the kinase or phosphatase and study the consequences on the signalling process in question. While these techniques are still the most widely used and informative on the specific role of the enzymes in a particular cellular process, they have a big caveat, namely the time the cells are exposed to the lipid changes before they can be studied. Since phosphoinositides are key in the control of membrane flow and traffic between organelles, cells will undergo a lot of changes many of which are secondary to the altered inositides, making it difficult to determine the exact process they control.
The best solution would be to use specific inhibitors that selectively block the effects of the individual enzymes acting on phosphoinositides. This is clear when reviewing the literature on the successful use of PI 3-kinase inhibitors (wortmannin and LY 294002) or the inositol phosphatase inhibition by Li+ ions. Unfortunately, these inhibitors still inhibit too many inositide kinase enzymes and no specific inhibitor of the phosphatases has surfaced as yet. New efforts to find subtype-specific PI 3-kinase inhibitors are on their way (Knight et al. 2006) especially lately, as pharmaceutical companies have started to show an interest in this molecular target. Until we have a larger variety of inhibitors that are able to block selected pathways we needed to find alternative ways to manipulate inositide levels locally in an acutely regulated fashion.
Chemically induced recruitment of enzymes to subcellular compartments
The rapamycin-induced heterodimerization of the FRB domain of mTOR and FKBP12 has been used to recruit proteins and induce molecular complex formation (Muthuswamy et al. 1999; Terrillon & Bouvier, 2004). Such heterodimerization system has been offered by Ariad (http://www.ariad.com). To eliminate the undesirable effects of rapamycin that inhibits the endogenous mTOR signalling pathway, Ariad has developed an analogue (AP21967) that does not act on endogenous mTOR but acts on a mutant form of the FRB that can be easily made in the FRB construct. This system has been applied to recruit small GTP binding proteins to the PM, but the AP21967 analogue was found to be too slow to mimic the physiological recruitment and activation process (Inoue et al. 2005). Therefore, another analogue (iRap) has been developed that induced much faster recruitment (Inoue et al. 2005). This system has recently been fashioned to alter phosphoinositide changes in the PM in a rapidly inducible manner (Heo et al. 2006; Suh et al. 2006; Varnai et al. 2006). Figure 4 shows the principle of this approach: the FRB fragment of mTOR – an ∼9 kDa size module – is targeted to the PM by an N-terminal palmitoylation/myristoylation sequence originated from Lyn or by the palmitoylation sequence of GAP43. This module can be also fused with a fluorescent protein to show its expression and localization or can be used untagged. Simultaneous expression of an inositol lipid 5-phosphatase (the yeast Inp54p in Suh et al. 2006; or a type-IV mammalian 5-phosphatase in Varnai et al. 2006), truncated to eliminate its own localization signals and fused to FKBP-12 and a fluorescent protein, yields a protein that remains in the cytosol and therefore has only limited (although not negligible) impact on membrane phosphoinositides. However, addition of rapamycin or iRap rapidly recruits the phosphatase to the membrane and causes the dephosphorylation of PtdIns(4,5)P2 as shown by the rapid decrease in the PM localization of the PLCδ1PH–GFP reporter. This method allows very quick reduction of PtdIns(4,5)P2 (and presumably PtdIns(3,4,5)P3 as the latter is synthesized from the former) levels the speed and extent of which depends on the concentration of the heterodimerizer (Varnai et al. 2006).
This method has allowed for the first time the unequivocal demonstration in intact cells that PtdIns(4,5)P2 regulates the KCNQ potassium channels (Suh et al. 2006) and also the Trpm8 channels (Varnai et al. 2006), and that elimination of PM PtdIns(4,5)P2 abolishes endocytosis of transferrin receptors (Varnai et al. 2006) and leads to the elimination of endocytic clathrin-coated pits (Zoncu et al. 2007). A similar approach was used to target the phosphoinositide 3-phosphatase, myotubularin, to the surface of Rab5 positive early endosomes to demonstrate the role of PtdIns3P in the morphogenesis of endosomes (Fili et al. 2006). This technique can be extended to targeting of other enzymes to other compartments and will allow analysis of the PtdIns(4,5)P2 regulation of several other processes in intact cells. This novel approach will also help us evaluate the accuracy and specificity of the lipid-imaging tools.
Concluding remarks and future perspectives
Analysis of cellular lipids has been lagging behind compared to the impressive progress made in genomics and proteomics, in spite of the well-recognized importance of various lipid classes in membrane biology and signalling. This is caused by the great difficulties in lipid analysis, especially at the single cell level. At the same time, it is increasingly obvious that lipid transport between membranes and local changes in lipid composition serves the basis of every process in the cell that is related to biological membranes. Phosphoinositides are regulatory lipids and should not be viewed through the same glass as the structural lipids that have major impact on the physico-chemical properties of membranes. However, because of their compact high charge density, these molecules have a significant local impact on membranes and are ideally suited to dock proteins to the membranes. In this regard, they resemble tyrosine phosphorylated motifs on the cytoplasmic aspect of membrane proteins. The universal role of these lipids is signified by the number of enzymes a cell commits to control their formation and elimination, all in a highly regulated manner. It appears that while several of these enzymes catalyse the same biochemical reaction, they clearly differ in their localization and regulatory partners thereby assigning their activity to control a biological process linked to a distinct cellular location. These signalling blocks are most likely to contain both inositide kinase and phosphatase enzymes as well as effector molecules that bind and respond to lipid changes. Combination of these factors provides the signalling specificity tailored to the needs of the cell.
If we want to better understand this organization we will have to continue to improve the methods that enable us to follow the lipid changes within the cell. We need to test additional protein domains that recognize the same lipid (for example PtdIns(4,5)P2) but perhaps are less sensitive to InsP3 changes and/or capable of detecting the lipid in other membranes. Development of intramolecular FRET sensors that change their FRET efficiency upon PtdIns(4,5)P2 (or other lipid) binding and probing lipid production by targeting the probe to different membranes would be a significant step forward. To limit distortion of lipid changes and distribution caused by the domains, we would need fluorophores with higher quantum efficiency and perhaps of smaller size to minimize interference from the fluorescent protein tag. Application of quantum dots conjugated to protein domains might be a promising approach, although it would be desirable to conjugate to the expressed proteins containing a small reactive tag within the cells to avoid the need for microinjection and production of recombinant proteins. We have to understand more about the interactions that contribute to the localization of the inositol lipid probes in order to improve their specificity and decrease their dominant negative effects. Manipulation of the lipids is clearly a complementary approach as it helps to uncover the limitations of the probes and also aids our understanding of the localized roles of inositides. While targeting of recruiting domains to additional compartments and generating recruitable active enzymes is an attractive possibility, greater emphasis should be placed in finding inhibitors that can specifically block the key enzymes of phosphoinositide metabolism. This would not only help to identify the processes for which these enzymes are important but also would aid discoveries of new potential pharmaceutical targets. Lipid research has now infiltrated cell biology as well as cellular signalling and it will surely contribute to a better understanding of the organization of the cell and of the conditions that lead to its pathological aberrations.
Acknowledgments
The author's research was supported by the Intramural Research Program of the National Institute of Child Health and Human Development of the National Institutes of Health. The author wishes to thank all current and previous members of the Section on Molecular Signal Transduction for their tremendous contribution to this line of research.
References
- Astle MV, Seaton G, Davies EM, Fedele CG, Rahman P, Arsala L, Mitchell CA. Regulation of phosphoinositide signaling by the inositol polyphosphate 5-phosphatases. IUBMB Life. 2006;58:451–456. doi: 10.1080/15216540600871159. [DOI] [PubMed] [Google Scholar]
- Attree O, Olivos IM, Okabe I, Bauiley LC, Nelson DL, Lewis RA, Mcinnes RR, Nussbaum RL. The Lowe's oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 1992;358:239–242. doi: 10.1038/358239a0. [DOI] [PubMed] [Google Scholar]
- Balla A, Balla T. Phosphatidylinositol 4-kinases; old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III α: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou CE, Lee YC. The structure of a myoinositol mannoside from Myobacterium tuberculosis glycolipid. Biochemistry. 1964;3:682–685. doi: 10.1021/bi00893a014. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Irvine RF. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 1984;312:315–321. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- Boronenkov IV, Anderson RA. The sequence of phosphatidylinositol-4-phosphate 5-kinase defines a novel family of lipid kinases. J Biol Chem. 1995;270:2881–2884. doi: 10.1074/jbc.270.7.2881. [DOI] [PubMed] [Google Scholar]
- Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnai P, Brown FB, Balla T, Donaldson JG. Active Arf6 recruits ARNO/Cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell. 2007 doi: 10.1091/mbc.E06-11-0998. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis MA, Di Campli A, Godi A. The role of the phosphoinositides at the Golgi complex. Biochim Biophys Acta. 2005;1744:396–405. doi: 10.1016/j.bbamcr.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Divecha N, Truong O, Hsuan JJ, Hinchliffe KA, Irvine RF. The cloning and sequence of the C isoform of PtdIns4P 5-kinase. Biochem J. 1995;309:715–719. doi: 10.1042/bj3090715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doughman RL, Firestone AJ, Anderson RA. Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J Membr Biol. 2003;194:77–89. doi: 10.1007/s00232-003-2027-7. [DOI] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes CP, Gray A, Lucocq JM. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 2005;15:259–268. doi: 10.1016/j.tcb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Eberhard DA, Cooper CL, Low MG, Holz RW. Evidence that the inositol phospholipids are necessary for exocytosis. Biochem J. 1990;268:15–25. doi: 10.1042/bj2680015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fili N, Calleja V, Woscholski R, Parker PJ, Larijani B. Compartmental signal modulation: Endosomal phosphatidylinositol 3-phosphate controls endosome morphology and selective cargo sorting. Proc Natl Acad Sci U S A. 2006;103:15473–15478. doi: 10.1073/pnas.0607040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier LM, Parton GP, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudici ML, Hinchliffe KA, Irvine RF. Phosphatidylinositol phosphate kinases. J Endocrinol Invest. 2004;27:137–142. [PubMed] [Google Scholar]
- Godi A, Di Campi A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Halet G. Imaging phosphoinositide dynamics using GFP-tagged protein domains. Biol Cell. 2005;97:501–518. doi: 10.1042/BC20040080. [DOI] [PubMed] [Google Scholar]
- Hammond GR, Dove SK, Nicol A, Pinxteren JA, Zicha D, Schiavo G. Elimination of plasma membrane phosphatidylinositol (4,5)-bisphosphate is required for exocytosis from mast cells. J Cell Sci. 2006;119:2084–2094. doi: 10.1242/jcs.02912. [DOI] [PubMed] [Google Scholar]
- Haugh JM, Codazzi F, Teruel M, Meyer T. Spatial sensing in fibroblasts mediated by 3′ phosphoinositides. J Cell Biol. 2000;151:1269–1280. doi: 10.1083/jcb.151.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K, Kadowaki S, Tanabe M, Takeshima H, Iino M. Spatiotemporal dynamics of inositol 1,4,5-trisphosphate that underlies complex Ca2+ mobilization patterns. Science. 1999;284:1527–1530. doi: 10.1126/science.284.5419.1527. [DOI] [PubMed] [Google Scholar]
- Hokin MR, Hokin LE. Enzyme secretion and incorporation of P32 into phospholipides of pancreas slices. J Biol Chem. 1953;203:967–977. [PubMed] [Google Scholar]
- Horowitz LF, Hirdes W, Suh BC, Hilgemann DW, Mackie K, Hille B. Phospholipase C in living cells: activation, inhibition, Ca2+ requirement, and regulation of M current. J Gen Physiol. 2005;126:243–262. doi: 10.1085/jgp.200509309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Meyer T. Subcellular targeting by membrane lipids. Curr Opin Cell Biol. 2001;13:146–152. doi: 10.1016/s0955-0674(00)00191-5. [DOI] [PubMed] [Google Scholar]
- Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, Balla T, Weiss WA, Williams RL, Shokat KM. A pharmacological map of the PI3-K family defines a role for p110α in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontos CD, Stauffer TP, Yang WP, York JD, Huang L, Blanar MA, Meyer T, Peters KG. Tyrosine 1101 of Tie2 is the major site of association of p85 and is required for activation of phosphatidylinositol 3-kinase and Akt. Mol Cell Biol. 1998;18:4131–4140. doi: 10.1128/mcb.18.7.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscovitch M, Chalifa V, Pertile P, Chen CS, Cantley LC. Novel function of phosphatidylinositol 4,5-biphosphate as a cofactor for brain membrane phospholipase D. J Biol Chem. 1994;269:21403–21406. [PubMed] [Google Scholar]
- Maeda N, Kawasaki T, Nakade S, Yokota N, Taguchi T, Kasai M, Mikoshiba K. Structural and functional characterization of inositol 1,4,5-trisphosphate receptor channel from mouse cerebellum. J Biol Chem. 1991;266:1109–1116. [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, Dewald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- Michell RH. Inositol phospholipids and cell surface receptor function. Biochim Biophys Acta. 1975;415:81–147. doi: 10.1016/0304-4157(75)90017-9. [DOI] [PubMed] [Google Scholar]
- Michell RH, Harwood JL, Coleman R, Hawthorne JN. Characteristics of rat liver phosphatidylinositol kinase and its presence in the plasma membrane. Biochim Biophys Acta. 1967;144:649–658. doi: 10.1016/0005-2760(67)90053-7. [DOI] [PubMed] [Google Scholar]
- Michell RH, Hawthorne JN. The site of diphosphoinositide synthesis in rat liver. Biochem Biophys Res Commun. 1965;21:333–338. doi: 10.1016/0006-291x(65)90198-1. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Umeda M, Horikoshi T, Yanagisawa K, Yoshioka T, Inoue K. Production and characterization of monoclonal antibodies that bind to phosphatidylinositol 4,5-bisphosphate. Mol Immunol. 1988;25:1025–1031. doi: 10.1016/0161-5890(88)90010-7. [DOI] [PubMed] [Google Scholar]
- Muthuswamy SK, Gilman M, Brugge JS. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol Cell Biol. 1999;19:6845–6857. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Catt KJ, Balla T. A wortmannin-sensitive phosphatidylinositol 4-kinase that regulates hormone-sensitive pools of inositolphospholipids. Proc Natl Acad Sci U S A. 1995;92:5317–5321. doi: 10.1073/pnas.92.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M, Watanabe KI, Sasaki J, Taya C, Takasuga S, Iizuka R, Balla T, Yamazaki M, Watanabe H, Itoh R, Kuroda S, Horie Y, Forster I, Mak TW, Yonekawa H, Penninger JM, Kanaho Y, Suzuki A, Sasaki T. Control of cell polarity and motility by the PtdIns(3,4,5)P3 phosphatase SHIP1. Nat Cell Biol. 2007;9:36–44. doi: 10.1038/ncb1515. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. The molecular heterogeneity of protein kinase C and its implications for cellular regulation. Nature. 1988;34:661–665. doi: 10.1038/334661a0. [DOI] [PubMed] [Google Scholar]
- Ozaki S, Dewald DB, Shope JC, Chen J, Prestwich GD. Intracellular delivery of phosphoinositides and inositol phosphates using polyamine carriers. Proc Natl Acad Sci U S A. 2000;97:11286–11291. doi: 10.1073/pnas.210197897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Kahn RA. GTP hydrolysis by ADP-ribosylation factor is dependent on both an ADP-ribosylation factor GTPase-activating protein and acidic phospholipids. J Biol Chem. 1994;269:10758–10763. [PubMed] [Google Scholar]
- Raucher D, Stauffer T, Chen W, Shen K, Guo S, York JD, Sheetz MP, Meyer T. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell. 2000;100:221–228. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- Roy A, Levine TP. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J Biol Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- Sale GJ, Fujita-Yamaguchi Y, Kahn CR. Characterization of phosphatidylinositol kinase activity associated with the insulin receptor. Eur J Biochem. 1986;155:345–351. doi: 10.1111/j.1432-1033.1986.tb09497.x. [DOI] [PubMed] [Google Scholar]
- Schneider IC, Haugh JM. Spatial analysis of 3′ phosphoinositide signaling in living fibroblasts. II. Parameter estimates for individual cells from experiments. Biophys J. 2004;86:599–608. doi: 10.1016/S0006-3495(04)74138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- Smrcka AV, Sternweis PC. Regulation of purified subtypes of phosphatidylinositol- specific phospholipase C β by G protein α and βγ subunits. J Biol Chem. 1993;268:9667–9674. [PubMed] [Google Scholar]
- Stauffer TP, Ahn S, Meyer T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- Suh BC, Inoue T, Meyer T, Hille B. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh PG, Ryu SH, Moon KH, Suh HW, Rhee SG. Cloning and sequence of multiple forms of phospholipase-C. Cell. 1988;54:161–169. doi: 10.1016/0092-8674(88)90548-x. [DOI] [PubMed] [Google Scholar]
- Taylor GS, Dixon JE. PTEN and myotubularins: families of phosphoinositide phosphatases. Methods Enzymol. 2003;366:43–56. doi: 10.1016/s0076-6879(03)66004-0. [DOI] [PubMed] [Google Scholar]
- Terrillon S, Bouvier M. Receptor activity-independent recruitment of βarrestin2 reveals specific signalling modes. EMBO J. 2004;23:3950–3961. doi: 10.1038/sj.emboj.7600387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A, Cantley LC. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- van Der Wal J, Habets R, Varnai P, Balla T, Jalink K. Monitoring phospholipase C activation kinetics in live cells by FRET. J Biol Chem. 2001;276:15337–15344. doi: 10.1074/jbc.M007194200. [DOI] [PubMed] [Google Scholar]
- Várnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Balla T. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim Biophys Acta. 2006;1761:957–967. doi: 10.1016/j.bbalip.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Varnai P, Bondeva T, Tamas P, Toth B, Buday L, Hunyady L, Balla T. Selective cellular effects of overexpressed pleckstrin-homology domains that recognize PtdIns(3,4,5)P3 suggest their interaction with protein binding partners. J Cell Sci. 2005;118:4879–4888. doi: 10.1242/jcs.02606. [DOI] [PubMed] [Google Scholar]
- Varnai P, Rother KI, Balla T. Phosphatidylinositol 3-kinase–dependent membrane association of the Bruton's tyrosine kinase pleckstrin homology domain visualized in single living cells. J Biol Chem. 1999;274:10983–10989. doi: 10.1074/jbc.274.16.10983. [DOI] [PubMed] [Google Scholar]
- Varnai P, Thyagarajan B, Rohacs T, Balla T. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J Cell Biol. 2006;175:377–382. doi: 10.1083/jcb.200607116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswarlu K, Oatey PB, Tavare JM, Cullen PJ. Insulin-dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3-kinase. Curr Biol. 1998;8:463–466. doi: 10.1016/s0960-9822(98)70181-2. [DOI] [PubMed] [Google Scholar]
- Volinia S, Dhand R, Vanhaesebroeck B, Macdougall LK, Stein R, Zvelebil MJ, Domin J, Panaretou C, Waterfield MD. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14:3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt SA, Kular G, Fleming IN, Downes CP, Lucocq JM. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C δ1. Biochem J. 2002;363:657–666. doi: 10.1042/0264-6021:3630657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixel KM, Blumental-Perry A, Watkins SC, Aridor M, Weisz OA. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J Biol Chem. 2005;280:10501–10508. doi: 10.1074/jbc.M414304200. [DOI] [PubMed] [Google Scholar]
- Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Type-I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Perera RM, Sebastian R, Nakatsu F, Chen H, Balla T, Ayala G, Toomre D, De Camilli PV. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc Natl Acad Sci U S A. 2007;104:3793–3798. doi: 10.1073/pnas.0611733104. [DOI] [PMC free article] [PubMed] [Google Scholar]