Abstract
ATP-sensitive K+ channels (KATP channels) are metabolic sensors formed by association of a K+ channel, Kir6, and an ATP-binding cassette (ABC) protein, SUR, which allosterically regulates channel gating in response to nucleotides and pharmaceutical openers and blockers. How nucleotide binding to SUR translates into modulation of Kir6 gating remains largely unknown. To address this issue, we have used a novel conformational KATP channel inhibitor, rhodamine 123 (Rho123) which targets the Kir6 subunit in a SUR-dependent manner. Rho123 blocked SUR-less Kir6.2 channels with an affinity of ∼1 μm, regardless of the presence of nucleotides, but it had no effect on channels formed by the association of Kir6.2 and the N-terminal transmembrane domain TMD0 of SUR. Rho123 blocked SUR + Kir6.2 channels with the same affinity as Kir6.2 but this effect was antagonized by ATP. Protection from Rho123 block by ATP was due to direct binding of ATP to SUR and did not entail hydrolysis because it was not mimicked by AMP, did not require Mg2+ and was reduced by mutations in the nucleotide-binding domains of SUR. These results suggest that Rho123 binds at the TMD0–Kir6.2 interface and that binding of ATP to SUR triggers a change in the structure of the contact zone between Kir6.2 and domain TMD0 of SUR that causes masking of the Rho123 site on Kir6.2.
ATP-sensitive potassium (KATP) channels are potassium-selective channels that are present in many types of excitable cells where they serve to modulate resting membrane potential, and therefore excitability, as a function of changes in intracellular metabolic potential (Seino & Miki, 2003; Nichols, 2006). This role stems from their capacity to sense the balance between the cytoplasmic nucleotides ATP, which inhibits them, and ADP, which activates them (Tarasov et al. 2004). To achieve this task, KATP channels possess two subunits: the inward rectifier K+ channel Kir6.2 (or Kir6.1) and the sulphonylurea receptor (SUR) of the ATP-binding cassette (ABC) protein superfamily (Moreau et al. 2005b). In the channel complex, four Kir6.2 molecules assemble to form a K+ pore inhibited by internal ATP, and four SUR molecules surround this pore (Mikhailov et al. 2005) and regulate its gating allosterically, activating it in response to ADP (Nichols et al. 1996) and KATP channel openers such as cromakalim, pinacidil and diazoxide (Moreau et al. 2000), and inhibiting it in response to drugs such as the anti-diabetic sulphonylureas (Moreau et al. 2005b). The presence of endoplasmic reticulum (ER) retention signals in both Kir6.2 and SUR that are only masked in the octameric complex ensures that only fully assembled channels reach the plasma membrane (Zerangue et al. 1999).
SUR is unique among ABC proteins because its only known function is to regulate an ion channel and its only position is next to Kir6.2. Nonetheless, its sequence predicts a similar structure as that of most other members of the ABCC subfamily (Moreau et al. 2005b). SUR possess the core ABC protein domains, two transmembrane domains TMD1 and TMD2 and two cytosolic nucleotide-binding domains NBD1 and NBD2. In addition, like multidrug resistance protein 1 (MRP1) and a few other ABC proteins, it has a supplementary N-terminal transmembrane domain TMD0, predicted to be comprised of five transmembrane helices, attached by a cytoplasmic loop L0 to the rest of the protein. If the contribution of TMD0 to the targeting and function of MRP1 is not essential (Bakos et al. 1998), it appears to be fundamental for SUR and its interaction with Kir6.2. Experimental evidence (Chan et al. 2003; Babenko & Bryan, 2003) shows that TMD0 solidly attaches to Kir6.2 and could modulate its gating in conjunction with loop L0. These conclusions rely mainly on coexpression of various SUR fragments with Kir6.2 and observation of the activity of the resulting channels. Such experiments suggest that different fragments of SUR adopt different conformations with Kir6.2 but do not determine whether and when full-length wild-type channels adopt some of these conformations and dynamically switch among them in responses to various stimuli.
Here, we introduce a new tool able to probe the conformation of the SUR–Kir6.2 interface. This tool is the fluorescent dye rhodamine 123 (Rho123) which, like other rhodamine derivatives, is commonly used to locate mitochondria and monitor mitochondrial transmembrane potential, because it is a membrane-permeant fluorescent dye that is positively charged and therefore accumulates in the electronegative mitochondrial matrix (Johnson et al. 1981). Rho123 is known to inhibit ATP synthase by binding to an unknown site on the F1-ATPase (Modica-Napolitano & Aprille, 1987). It also binds to and is a substrate of the ABC transporters MRP1 (Daoud et al. 2000) and P-glycoprotein (Loetchutinat et al. 2003). Here we demonstrate that Rho123 blocks the KATP channel by interacting not with SUR but with Kir6.2. As this effect depends on the conformation of SUR, and more precisely on that of the TMD0–Kir6.2 interface, we show that Rho123 is a valuable probe of the different configurations of this interface induced by physiological and pharmacological ligands of SUR.
Methods
Experimental conditions were essentially as previously described (Moreau et al. 2005a). All constructs were derived from mouse Kir6.2 (GenBank accession no. D50581), hamster SUR1 (GenBank accession no. L40623) and rat SUR2A (GenBank accession no. D83598), and subcloned in Xenopus oocyte expression vectors derived from pGEMHE (Liman et al. 1992). The construct TMD0 consisted of residues 2–195 of SUR1 with a FLAG epitope (sequence DYKDDDDK) added at the N-terminal (identical to construct F195 of Chan et al. (2003)). Mutations were introduced by PCR (QuickChange Site-Directed Mutagenesis Kit, Stratagene). For the Kir6.2ΔC36 construct (i.e. Kir6.2 with the last C-terminal 36 amino acids deleted), a premature stop codon was introduced at the correct position. The entire coding sequence of each clone was verified by sequencing. After amplification and linearization, plasmid DNAs were transcribed in vitro by using the T7 mMessage mMachine Kit (Ambion) to produce cRNA for subsequent oocyte microinjection.
Female Xenopus laevis were anaesthetized with 3-aminobenzoic acid ethyl ester (1 g l(water)−1). Part of one ovary was removed with a minilaparotomy, the incision was sutured, and the animal was allowed to recover. Animal handling and experiments fully conformed with French regulations and were approved by local governmental veterinary services (authorization no. 28-03-15 from the Ministère de l'Agriculture, Direction des Services Vétérinaires to M.V.). Stage V or VI oocytes were defolliculated by a 60 min incubation at 19°C with 2 mg ml−1 type A collagenase (Sigma-Aldrich). Selected oocytes were injected the next day with cRNAs encoding wild-type or truncated Kir6.2 (0.2 or 2 ng) and, where applicable, TMD0 (2 ng) or wild-type or modified SURs (6 ng). Injected oocytes were stored at 19°C in Barth's solution containing (mm): KCl 1, MgSO4 0.82, NaCl 88, NaHCO3 2.4, CaCl2 0.41, Ca(NO3)2 0.3 and Hepes 16 (pH 7.4) supplemented with 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 100 μg ml−1 gentamycin. Three to five days after injection, oocytes were devitellinized and recombinant KATP channels were characterized by the patch-clamp technique in the excised inside-out configuration at room temperature (∼22°C). Patch pipettes contained (mm): K+ 154, Cl− 146, Mg2+ 5 and Pipes-KOH 10 (pH 7.1). The cytoplasmic face of the patch was bathed in solutions which, unless otherwise noted, contained (mm): K+ 174, Cl− 40, EGTA 1, Mg2+ 1, Pipes-KOH 10 (pH 7.1) and methanesulphonate− as the remaining anion. For experiments in 0 Mg2+, Mg2+ was omitted and EGTA was replaced by EDTA. ATP, AMP (potassium salt; Sigma-Aldrich), and rhodamine compounds (30 mm stock in ethanol; Sigma-Aldrich) were added as specified. Membrane potential was −50 mV except where noted. Applications of the various solutions to the patch were performed using RSC-100 or RSC-200 automated sewer pipes systems (Bio-Logic; Vivaudou & Forestier, 1995). Pipe switching time was set to 200 ms; however, the gravity-driven flow rate being slow, real solution switching at the membrane patch took longer and there were occasional artefacts due to mixing of the oocyte bathing solution containing 2 mm ATP.
Data acquisition and analysis were performed with in-house software. Slow fluctuations of the baseline were removed by interactive fitting with a spline curve and subtraction of this fit from the signal. Non-linear curve fitting was performed with Origin software (OriginLab).
The tracings shown in the illustrations originate from different patches, except where noted, and represent continuous records. To quantify the effects of rhodamine in 0 ATP, where currents are more prone to rundown, whenever possible current decline before application was extrapolated manually and this extrapolated value served as the control current. However, no obvious differences were noticed between overall results and observations limited to patches displaying little or no rundown. Results are displayed as means ±s.e.m.
Results
Rhod123, an unusual inhibitor of KATP channels
Xenopus oocytes injected either with cRNA of Kir6.2ΔC36 (Tucker et al. 1997), or with a mixture of cRNAs coding for SUR2A and Kir6.2 expressed exogenous K+ conductance inhibited by ATP. The effects of Rho123 were tested on these two KATP channel types. In certain conditions described below, Rho123 was found to cause channel inhibition. Block was gradually achieved within ∼30 s with an average half-time of 9.1 ± 1 s (n= 44) in all configurations and it reversed slowly within several minutes (see below). Thus, inhibition was quantified by comparing values of currents before and long enough (∼30 s) after application of Rho123 to approach steady-state.
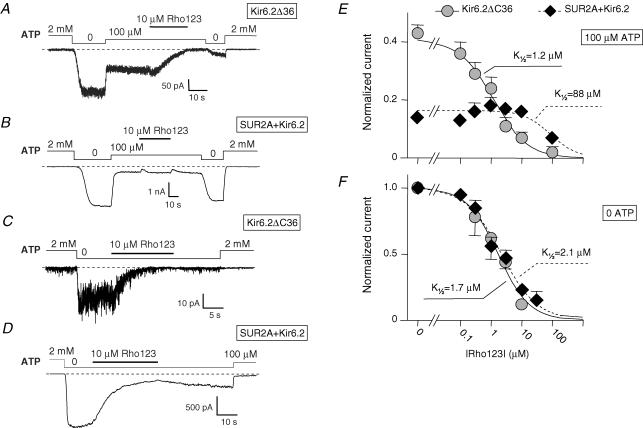
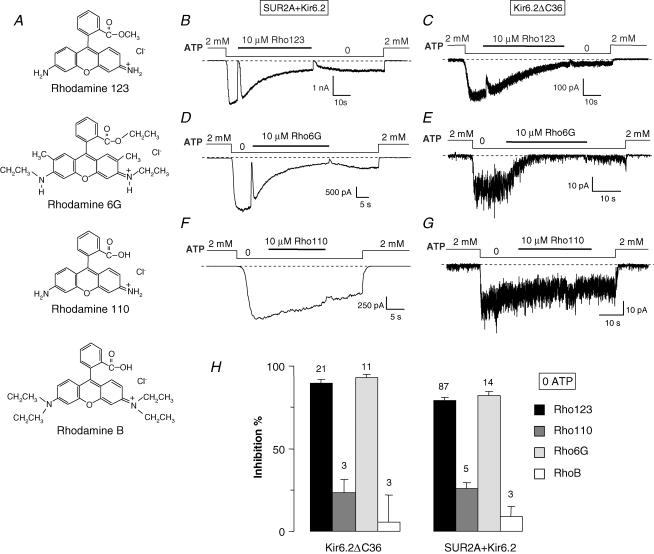
In the presence of 100 μm ATP (in the presence of 1 mm total Mg2+), 10 μm Rho123 blocked Kir6.2ΔC36 channels (Fig. 1A; inhibition of 88 ± 2%, n= 22) but had no significant effects on SUR2A + Kir6.2 channels (Fig. 1B; inhibition of 2 ± 5%, n= 57). At higher concentrations, Rho123 did induce a significant block of the latter channels. The concentration dependency of block in the presence of 100 μm ATP, illustrated in Fig. 1E, shows K½ values of 1.2 μm and 88 μm for Kir6.2ΔC36 and SUR2A + Kir6.2, respectively. The Hill coefficient for Kir6.2ΔC36 was close to unity, suggesting a bimolecular reaction. For the SUR2A + Kir6.2 data, the Hill coefficient could not be determined and was set to 1 because only one Rho123 concentration (100 μm) induced significant inhibition. As this concentration blocked 51% of the current in 12 patches, the K½ value could nonetheless be accurately estimated.
Figure 1. ATP-dependent inhibition by Rho123 of Kir6.2ΔC36 and SUR2A + Kir6.2 channels.
A–D, currents were recorded in inside-out patches from Xenopus oocytes expressing either Kir6.2ΔC36 (A and C) or SUR2A + Kir6.2 (B and D). Rho123 (10 μm) was applied in the presence of 100 μm ATP (A and B) or in the absence of nucleotides (C and D). E, average currents for Kir6.2ΔC36 (grey symbols) and SUR2A + Kir6.2 (⋄) measured in the presence of 100 μm ATP plotted against the concentration of Rho123. Currents were normalized to the current measured in 0 ATP before application of blocker. Each point represents data from n= 9–60 patches for SUR2A + Kir6.2 and n= 4–16 patches for Kir6.2ΔC36. Error bars represent s.e.m. and are not drawn when smaller than symbols. Hill equation fitting yielded ½= 1.2 μm and h= 0.8 for Kir6.2ΔC36 (continuous line) and ½= 88 μm and h= 1 for SUR2A + Kir6.2 (dashed line). F, equivalent data obtained in 0 ATP. Each point is an average from n= 5–88 patches. Curve fitting yielded ½= 1.7 μm and h= 0.85 for Kir6.2ΔC36 (continuous line) and ½= 2.1 μm and h= 0.75 for SUR2A + Kir6.2 (dashed line).
In the absence of nucleotides (Fig. 1C and D), Kir6.2ΔC36 and SUR2A + Kir6.2 channels were highly sensitive to block by 10 μm Rho123, with inhibition of 89 ± 2% (n= 21) and 79 ± 2% (n= 87), respectively. This similarity is seen in the concentration–inhibition data obtained in the absence of nucleotides (Fig. 1F) which yielded K½ values of 1.7 and 2.1 μm for Kir6.2ΔC36 and SUR2A + Kir6.2, respectively, with Hill coefficients near unity in both cases.
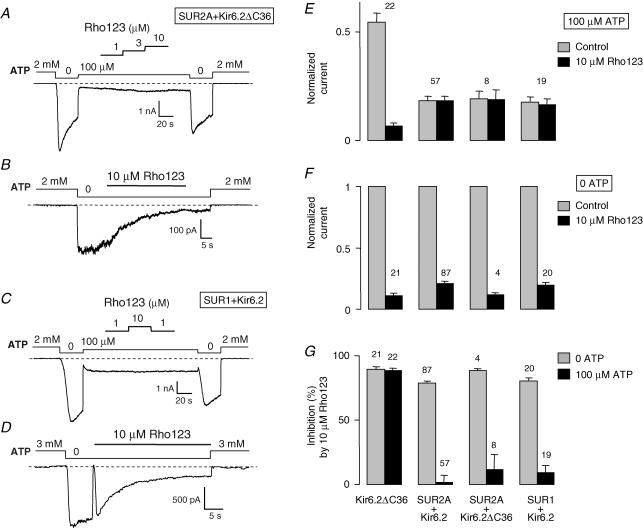
These results suggest that Rho123 acts on Kir6.2 and that this action is antagonized by SUR2A only when ATP is present. To rule out a role of the C-terminal deletion of Kir6.2 in these results, we coexpressed Kir6.2ΔC36 and SUR2A and observed that the response to Rho123 of the resulting SUR2A + Kir6.2ΔC36 channels was comparable to that of the SUR2A + Kir6.2 channels (Fig. 2). We also found that the pancreatic isoform SUR1 behaved like SUR2A as SUR1 + Kir6.2 channels were blocked by 10 μm Rho123 in absence of nucleotides (inhibition of 80 ± 2%, n= 20) but not in the presence of 100 μm ATP (inhibition of 9 ± 6%, n= 19).
Figure 2. ATP-dependent inhibition by Rho123 of SUR2A + Kir6.2ΔC36 and SUR1 + Kir6.2 channels.
A–D, currents were recorded in inside-out patches from Xenopus oocytes expressing either SUR2A + Kir6.2ΔC36 (A and B) or SUR1 + Kir6.2 (C and D). Rho123 (1–10 μm) was applied in the presence of 100 μm ATP (A and C) or in the absence of nucleotides (B and D). E, average currents for Kir6.2ΔC36, SUR2A + Kir6.2, SUR2A + Kir6.2ΔC36 and SUR1 + Kir6.2 measured before (grey bars) and after (black bars) application of 10 μm Rho123 in the presence of 100 μm ATP. Currents were normalized to the current measured in 0 ATP before application of blocker. F, equivalent data obtained in nucleotide-free solution. G, data from E and F expressed as percentage inhibition. Numbers above bars indicate the number of patches included in the average.
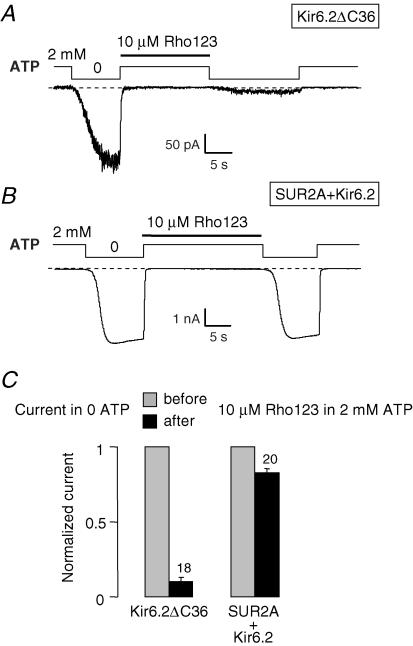
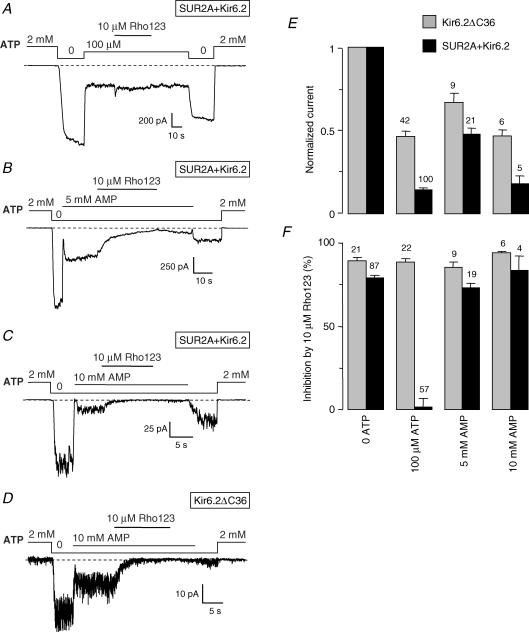
Inspection of records such as the one in Fig. 1B shows that ATP antagonized Rho123 block by preventing binding of Rho123 to SUR + Kir6.2 channels because, after washout of Rho123, rapid return to nucleotide-free, rhodamine-blocking conditions did not reveal any significant loss in the number of active channels indicative of the presence of channel-bound rhodamine. This is more clearly demonstrated in Fig. 3 where Rho123 was applied in the presence of 2 mm ATP. In these conditions, no block can be detected during Rho123 application because channels are already fully inhibited by ATP. However, it is seen in Fig. 3A that return to 0 ATP in the absence of Rho123 reveals a much decreased Kir6.2ΔC36 channel activity compared to the initial activity before Rho123 application. This reflects the persistent binding of Rho123 to the Kir6.2ΔC36 channels and demonstrates that Rho123 binds the closed channel in the same manner as the open channel (Fig. 1A and C). By contrast, the same experiment with SUR2A + Kir6.2 (Fig. 3B) shows no decrease in 0 ATP channel activity after application of Rho123 in 2 mm ATP. Because Rho123 dissociates slowly such a decrease would have been expected if Rho123 could bind to the channels in the presence of ATP.
Figure 3. ATP prevents binding of Rho123 to SUR2A + Kir6.2 channels.
A and B, inside-out patch clamp records from oocytes expressing Kir6.2ΔC36 (A) or SUR2A + Kir6.2 (B) illustrating the effect of application of 10 μm Rho123 in the presence of 2 mm ATP on channel activity in nucleotide-free solutions. C, average currents in nucleotide-free conditions for Kir6.2 and SUR2A + Kir6.2 channels measured before (grey bars) and after (black bars) application of 10 μm Rho123 in the presence of 2 mm ATP.
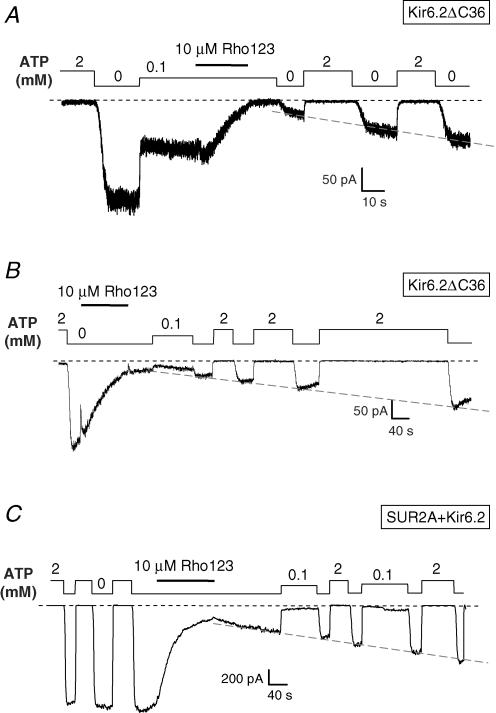
Recovery from Rho123 block
Block reversal was difficult to quantify because it happened on the same timescale as rundown, the rate of which was variable from patch to patch and over the recording period within the same patch. It was similarly difficult to assess whether nucleotides had a significant effect on block reversal because they also affect rundown. In spite of these limitations, we did not observe any systematic effect of channel type or nucleotide conditions on the block reversal rate. This is seen in Fig. 4A and C where reversal starts in 0 ATP and proceeds at the same rate independently of the absence of ATP or the presence of ATP at 100 μm or 2 mm. Dissociation is therefore clearly different from refreshment of rundown channels, which is strictly dependent on the presence of nucleotides (Furukawa et al. 1994). Furthermore, the rate of reversal was of the order of 10-fold less than the rate of block, as theoretically predicted for a bimolecular reaction with a concentration of Rho123 (10 μm) 10-fold higher than the dissociation constant (∼1 μm).
Figure 4. Slow reversal of Rho123 block of Kir6.2ΔC36 and SUR2A + Kir6.2 channels.
Inside-out patch clamp records from oocytes expressing Kir6.2ΔC36 or SUR2A + Kir6.2 illustrating channel block by 10 μm Rho123 in the presence of 100 μm ATP for Kir6.2ΔC36 (A) and 0 ATP for Kir6.2ΔC36 (B) or SUR2A + Kir6.2 (C), and slow relief of block after washout of Rho123. The record of Fig. 1D is taken from the trace in C. The dashed line superimposed on the recovery phase of each trace was drawn manually to illustrate the progressive reversal of block.
Another implication of this observation of ATP-independent reversal concerns prevention by ATP of Rho123 block of SUR + Kir6.2 channels. If ATP does not increase the dissociation rate of the blocker, it must decrease its association rate. This conclusion is compatible with the hypothesis that ATP binding to SUR impairs blocker access to its binding site.
Comparison of the effects of Rho123 and related rhodamine compounds
Because rhodamine compounds are lipophilic, block by Rho123 could be caused by an indirect, non-specific perturbation of the membrane, possibly implicating phosphatidylinositol phosphates, which are strong modulators of Kir6.2 gating (Baukrowitz et al. 1998; Shyng & Nichols, 1998). To investigate the specificity of the effect of Rho123, we assayed three other rhodamine derivatives: rhodamine 6G (Rho6G), rhodamine 110 (Rho110) and rhodamine B (RhoB). Structurally, Rho110 is the closest analogue of Rho123, as it differs only by a single methyl group; Rho6G and RhoB are bulkier. Chemically, Rho6G is cationic like Rho123 whereas, at physiological pH, Rho110 and RhoB are zwitterionic. All three analogues display a higher membrane permeability than Rho123 (Loetchutinat et al. 2003).
Tests were performed in nucleotide-free conditions where Rho123 blocks both Kir6.2ΔC36 and SUR2A + Kir6.2 channels. At 10 μm, Rho110 and RhoB only weakly blocked Kir6.2ΔC36 (33 ± 8%, n= 3 and 6 ± 16%, n= 3, respectively) and SUR2A + Kir6.2 channels (26 ± 4%, n= 5 and 9 ± 6%, n= 3, respectively) (Fig. 5). These values are significantly less than those of ∼80% achieved by Rho123. By contrast, Rho6G was also a potent blocker (inhibition of 93 ± 2%, n= 11 and 82 ± 2%, n= 14 for Kir6.2ΔC36 and SUR2A + Kir6.2, respectively). Half-time of block by Rho6G was decreased (7.7 ± 2 s; n= 14) but statistically not different from the half-time of block (9.8 ± 1 s; n= 30) by Rho123. However, the net positive charge present on Rho123 and Rho6G, but not on Rho110 and RhoB, was not critical because block was little affected by membrane potential (data not shown), ruling out block by direct pore obstruction.
Figure 5. KATP channel inhibition by various rhodamine derivatives.
A, chemical structures of the tested rhodamine derivatives. B–G, currents were recorded in inside-out patches from Xenopus oocytes expressing either SUR2A + Kir6.2 (B, D and F) or Kir6.2ΔC36 (C, E and G). Indicated rhodamine compounds (10 μm) were applied in the absence of nucleotides. H, percentage inhibition by a 10 μm concentration of the indicated compounds of Kir6.2ΔC36 and SUR2A + Kir6.2 channels in the absence of nucleotides. Numbers above bars indicate the number of patches included in the average.
These results suggest that the presence of a neutral ester group in Rho123 and Rho6G, instead of a carboxyl group in Rho110 and RhoB, is crucial for block, and not the degree of hydrophobicity or the ionic charge.
KATP channel inhibition by rhodamine 123 is antagonized by nucleotide binding to the SUR subunit
The results so far suggest that Rho123 specifically interacts with Kir6.2 and that this interaction is antagonized by the SUR subunit, but only when ATP is present. As both subunits harbour nucleotide binding sites, the effects of ATP can be mediated by either one. As ATP has no effect on Rho123 block of SUR-less Kir6.2ΔC36 channels, the more straightforward interpretation of the data is that nucleotide binding to SUR determines the degree of block by Rho123. To further settle this point, we examined (1) whether AMP could replace ATP, (2) whether mutations of the nucleotide binding domains (NBDs) of SUR had any influence and (3) whether Mg2+ ions were important.
Although it has not been tested directly on SUR, AMP is unable to bind several related ABC proteins (Hou et al. 2000; Kern et al. 2000; Bouabe & Knittler, 2003; Wolters et al. 2005). As shown in Fig. 6, a high concentration of 10 mm AMP was necessary to cause an inhibition of Kir6.2 channels comparable to that achieved by 100 μm ATP. Inhibition by AMP, like ATP, was weaker for Kir6.2ΔC36 channels than for SUR2A + Kir6.2 channels. As summarized in Fig. 6F, unlike ATP, AMP did not protect the SUR2A + Kir6.2 channel against the inhibitory effect of Rho123. Indeed, 10 μm Rho123 in the presence of 10 mm AMP inhibited Kir6.2ΔC36 by 94 ± 1% (n= 6), which is almost the same value as in the presence of 100 μm ATP. By contrast, inhibition of SUR2A + Kir6.2 channels by 10 μm Rho123, which was insignificant in ATP, rose to 83 ± 9% (n= 4) in 10 mm AMP. These results were reproduced with 5 mm AMP. Thus, rhodamine block was equivalent in the presence of AMP and in nucleotide-free conditions.
Figure 6. AMP, unlike ATP, does not protect KATP channels from Rho123 block.
A–D, currents were recorded in inside-out patches from Xenopus oocytes expressing either SUR2A + Kir6.2 (A–C) or Kir6.2ΔC36 (D). Rho123 (10 μm) was applied in the presence of 100 μm ATP (A), 5 mm AMP (B) or 10 mm AMP (C and D). E, average currents for Kir6.2ΔC36 and SUR2A + Kir6.2 measured in the absence or the presence of ATP and AMP. Currents were normalized to the current measured in 0 ATP before application of blocker. F, percentage inhibition by 10 μm Rho123 of Kir6.2ΔC36 and SUR2A + Kir6.2 channels in the absence or the presence of ATP and AMP. Numbers above bars indicate the number of patches considered.
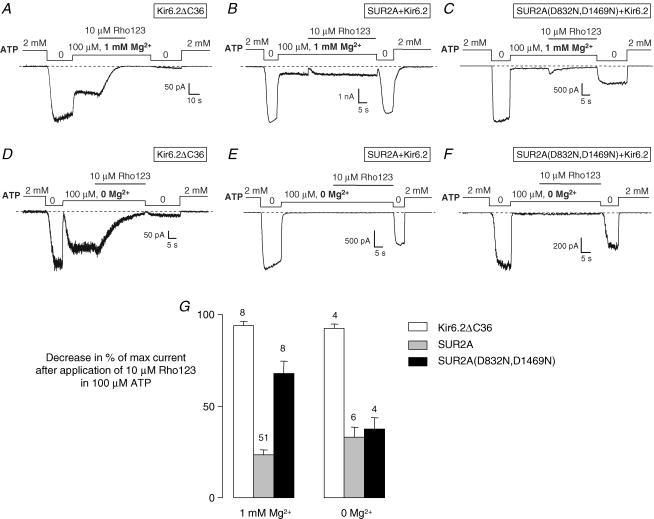
For more direct evidence of the involvement of SUR in mediating the effect of ATP, we have mutated the key Walker B aspartate residue (Walker et al. 1982) of both NBDs of SUR2A to glutamine (D832N and D1469N). Equivalent mutations have been reported to reduce azido-ATP photolabelling of SUR1 (Ueda et al. 1997) and MDR1 (Hrycyna et al. 1999). In order to compare wild-type and mutated channels, experiments were conducted as above with 10 μm Rho123 in the presence of 100 μm ATP. In these conditions the mutated channel is barely open as it displays a greater sensitivity to ATP inhibition than the wild-type channel as already reported by Gribble et al. (1998). To quantify inhibition by Rho123, we therefore resorted to comparing the maximal current in the absence of nucleotides before and after application of Rho123 in the presence of ATP as illustrated in Fig. 7. This is a valid approach as long as the rate of rundown is assumed to be comparable in different patches. Except in rare patches that were not processed, current decline was not significant in the absence of rhodamine over the short durations (∼2 min) of our tests. The decrease in maximal current (Fig. 7G) serves as an indicator of the Rho123-bound fraction of channels. After Rho123 application in 100 μm ATP and 1 mm Mg2+, this value was close to 100% (94 ± 3%, n= 8) for Kir6.2ΔC36 and much lower (23 ± 3%, n= 51) for SUR2A + Kir6.2. It was significantly (Student's t test, P < 0.0001) increased by the D832N/D1469N SUR2A mutations (68 ± 7%, n= 8). This result demonstrates that functional NBDs are required for ATP to protect Kir6.2 from Rho123 block.
Figure 7. Rho123 block protection by ATP does not require Mg2+ and is reduced by nucleotide-binding domain (NBD) mutations.
A–F, currents were recorded in inside-out patches from Xenopus oocytes expressing either Kir6.2ΔC36 (A and D), SUR2A + Kir6.2 (B and E) or SUR2A(D832N,D1469N) + Kir6.2 (C and F). Rho123 (10 μm) was applied in the presence of 100 μm ATP with 1 mm added Mg2+ (A–C) or no added Mg2+ and 1 mm EDTA to chelate residual divalent cations (D–F). G, percentage decrease in channel activity after application of Rho123 in the presence of 100 μm ATP with and without Mg2+. Rho123 inhibition in 100 μm ATP, which cannot be directly computed in some cases because of nearly complete channel block by ATP (as shown in C, E and F), was evaluated by comparing channel activity in absence of nucleotides before and after application of 10 μm Rho123 with and without Mg2+ for Kir6.2ΔC36, SUR2A + Kir6.2 and SUR2A(D832N,D1469N) + Kir6.2. Numbers above bars indicate the number of experiments considered.
Is ATP hydrolysis required for Rho123 block protection? Mutational analysis cannot provide an answer because NBD mutations can impair both nucleotide binding and hydrolysis. Further experiments were therefore performed in the absence of Mg2+ and other divalent cations that are necessary cofactors of the ATP hydrolytic reaction without being required for ATP binding (Matsuo et al. 2000). In these conditions (Fig. 7E and G), ATP was still able to confer significant protection from Rho123 block to the wild-type SUR2A + Kir6.2 channels: Rho123 inhibition was 33 ± 6% (n= 6) in 0 Mg2+, which is slightly greater than in the presence of Mg2+, although the difference was not statistically significant (Student's t test). Therefore, the effect of ATP on Rho123 block does not appear to require hydrolysis of the nucleotide. It is interesting that removal of Mg2+ enabled ATP to protect the SUR2A(D832N,D1469N) mutant as well as the wild-type (Fig. 7G; no statistically significant difference). This finding reinforces the notion that ATP binding is sufficient for protection because the role of the Walker B Asp residues in coordinating Mg2+ (Walker et al. 1982) suggests that the mutant would bind ATP better in the absence of divalent cations, which has been experimentally confirmed with MDR1 (Lerner-Marmarosh et al. 1999; Booth et al. 2000).
TMD0 domain of SUR decreases the rhodamine sensitivity to Kir6.2
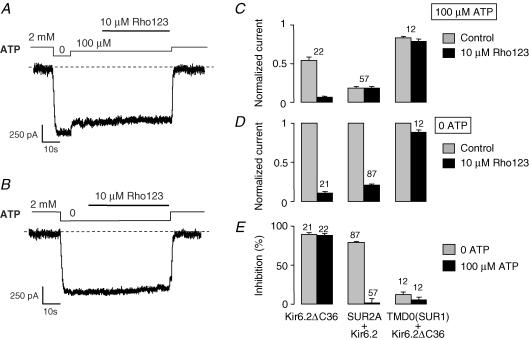
The domain TMD0 has been identified as a region of contact between SUR and Kir6.2 (Chan et al. 2003). TMD0 alone and Kir6.2ΔC36, when coexpressed, associate to form channels that have a reduced affinity for inhibitory nucleotides (Chan et al. 2003). In our hands, at 100 μm, ATP closed only 17 ± 2% (n= 12) of the TMD0 + Kir6.2ΔC36 channels compared to 54 ± 3% (n= 42) and 85 ± 2% (n= 80) observed with Kir6.2ΔC36 and SUR2A + Kir6.2, respectively.
As shown in Fig. 8, coexpression of TMD0 of SUR1 and Kir6.2ΔC36 produced channels that were almost insensitive to Rho123: in the presence and in the absence of ATP, inhibition by Rho123 was 6 ± 3% (n= 12) and 12 ± 3% (n= 12), respectively. These values were not statistically different (Student's t test).
Figure 8. Channels formed by the assembly of N-terminal domain TMD0 of SUR1 and Kir6.2ÄC36 are largely insensitive to block by Rho123.
A and B, inside-out patch clamp records from Xenopus oocytes coexpressing TMD0(SUR1) and Kir6.2ΔC36 illustrating the response to 10 μm Rho123 in the presence of 100 μm ATP (A) and in nucleotide-free (B) solutions. C, average currents for Kir6.2, SUR2A + Kir6.2, and TMD0(SUR1) + Kir6.2ΔC36 channels measured before (grey bars) and after (black bars) application of 10 μm Rho123 in the presence of 100 μm ATP. Currents were normalized to the current measured in 0 ATP before application of blocker. D, equivalent data but in nucleotide-free conditions E, data from C and D expressed as percentage inhibition. Numbers above each bar indicate the number of patches included in the average.
TMD0 + Kir6.2ΔC36 channels present therefore an unconditional resistance to block by Rho123 which distinguishes them from both SUR2A + Kir6.2 channels (resistant only in the presence of ATP) and Kir6.2ΔC36 channels (sensitive in all conditions).
Discussion
Rhodamine 123, a novel KATP channel blocker targeting the TMD0–Kir6.2 interface
To the list of KATP channel blockers, we add yet another entry, Rho123, which distinguishes itself by its unique mode of action. Rho123 interacts with Kir6.2 as demonstrated by its effect on SUR-less Kir6.2ΔC36 channels. Experiments with other rhodamine compounds indicate that this interaction is structure-specific and does not arise simply from the amphiphile nature of rhodamines and a possible membrane perturbation. The zwitterionic Rho110, closest in structure to Rho123, and RhoB produced little channel block. The cationic Rho6G was slightly more potent than Rho123 (which is cationic as well), and its blocking rate constant was equivalent in spite of a greater membrane permeability. Thus, block does not depend on lipophilicity but on chemical structure. Charge appears to be important: Rho123 and Rho6G have an overall positive charge because they lack the negative carboxyl group present in Rho110 and RhoB. However, block was little affected by membrane potential (data not shown), ruling out a direct pore obstruction and suggesting that, if a specific binding site for rhodamines exists on the Kir6.2 protein, it is located outside the ion conduction pore.
The blocking effect of Rho123 could be related to an indirect alteration of the regulation by phospholipids of the channel. Phospholipids activate the KATP channel and, consequently, their chelation or screening by polyvalent cations such as neomycin results in a decrease in channel activity (Fan & Makielski, 1997). Rho123 is also cationic but it is monovalent and is unlikely to screen phospholipids when applied at micromolar concentrations in saline solutions containing much higher concentrations of divalent and monovalent cations. Furthermore, phospholipids act similarly on both SUR + Kir6.2 and SUR-less Kir6.2 channels (Baukrowitz et al. 1998) but Rho123 does not. Therefore, our results are more consistent with the hypothesis of a binding site for Rho123 on the Kir6.2 protein. Access to this site was slow and not linked to blocker lipophilicity, therefore it would probably be buried on the cytoplasmic face of the protein and allosterically linked to the channel gate (Fig. 9A).
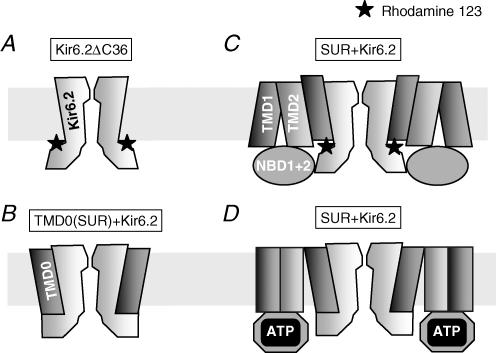
Figure 9. Structural rearrangements at the TMD0–Kir6.2 interface suggested by the observed changes in Rho123 sensitivity as a function of channel composition and modification.
A, block of Kir6.2 by Rho123 applied to its cytoplasmic face is observed in the presence and absence of nucleotides. As it is independent of blocker membrane permeability, voltage and channel open probability, the Rho123 binding site is likely to be outside the conduction pathway on the cytoplasmic side of the protein. B, the lack of effects of Rho123 on TMD0-Kir6.2 channel suggests that TMD0, expressed alone, masks the Rho123 site. C, in the absence of nucleotides or in the presence of AMP, the wild-type SUR + Kir6.2 channel is blocked by Rho123, suggesting that TMD0 adopts a conformation, different from the conformation when expressed alone, where the Rho123 site is accessible. D, wild-type channels become insensitive to Rho123 block upon binding of ATP. The phenotype induced by ATP (reduced sensitivity to block by Rho123 as well as by nucleotides) resembles that of TMD0-Kir6.2 channels: this suggests a common topology of the subunit interface between TMD0-Kir6.2 and ATP-bound SUR-Kir6.2.
The N-terminal domain TMD0 of SUR can associate with Kir6.2 (Chan et al. 2003). When we coexpressed TMD0 with Kir6.2ΔC36, Rho123 no longer inhibited the resulting channels. Thus, TMD0 protects Kir6.2 from inhibition by rhodamine. Although we may envision several mechanisms for this protection, we shall consider the simplest explanation, prevention of blocker binding by site masking, over more complex mechanisms such as allosteric modification of the blocker site or disruption of the linkage between blocker site and channel gate. This hypothetical scheme is illustrated in Fig. 9B. The exact mechanism might differ from the one depicted, but this will not affect the conclusion that rhodamine block is dependent on the interaction between TMD0 and Kir6.2.
ATP-bound SUR protects the KATP channel against block by rhodamine 123
When full KATP channels were reconstituted by expression of SUR1 or SUR2A with Kir6.2, inhibition by Rho123 became strongly dependent on cytosolic ATP levels. Without nucleotides, SUR + Kir6.2 responded to Rho123 like Kir6.2 alone with K½ of ∼2 μm in both cases. In the presence of ATP, SUR + Kir6.2 became much less sensitive to Rho123 inhibition (40-fold increase in K½ by 100 μm ATP) whereas Kir6.2ΔC36 remained as sensitive as in the absence of ATP. This difference was most striking with 10 μm Rho123 in the presence of 100 μm ATP (Fig. 1A and B) where Rho123had no effect on SUR + Kir6.2 but almost completely blocked Kir6.2ΔC36.
How does ATP prevent binding of Rho123? Most probably by binding to the NBDs of SUR and inducing a change in conformation affecting the SUR–Kir6.2 interface, because the combined presence of SUR and ATP is necessary to make the rhodamine site inaccessible. The alternative is that protection from Rho123 block occurs when ATP binds to the nucleotidic inhibitory site of Kir6.2. Nevertheless, in the presence of AMP, a nucleotide not known to interact with SUR but able to block Kir6.2 (Tucker et al. 1998; Ribalet et al. 2003; Matsuo et al. 2005), a high sensitivity to rhodamine was observed. Moreover, mutations of Walker B aspartate residues of SUR2A, which are expected to impair ATP binding and hydrolysis, significantly reduced the protective action of ATP. To what extent NBD mutations of ABC proteins affect ATP binding compared to ATP hydrolysis cannot be predicted with certainty given the divergent opinions found in the literature (e.g. Ueda et al. 1997; Urbatsch et al. 1998; Hrycyna et al. 1999; Lerner-Marmarosh et al. 1999). We therefore performed additional experiments in the absence of divalent cations which are necessary for hydrolysis. In these conditions, ATP still provided an effective protection of wild-type channels against block by Rho123. ATP also became effective on the double Walker B mutant, consistent with the role of the mutated aspartate in the coordination of Mg2+ (Smith et al. 2002) and the ability of this mutant to bind ATP better in the absence of Mg2+ (Weber et al. 1998; Lerner-Marmarosh et al. 1999; Booth et al. 2000).
These results suggest that channel resistance to Rho123 is acquired when ATP binds to SUR without requiring hydrolysis.
Mechanistic implications
Whereas Kir6.2ΔC36 is always sensitive to Rho123 and TMD0 + Kir6.2ΔC36 is always insensitive, the sensitivity of SUR + Kir6.2 is modulated by ATP binding to SUR. It appears that ATP binding, presumably to the cytosolic NBDs, induces a global change in the conformation of SUR and, in particular, of its TMD0 domain which drastically reduces access of Rho123 to its site. Therefore, with respect to Rho123, ATP-bound SUR + Kir6.2 resembles TMD0 + Kir6.2ΔC36, and ATP-free SUR + Kir6.2 resembles Kir6.2ΔC36. One possible interpretation of these observations is that ATP binding to SUR causes a functional uncoupling of TMD0 from the rest of SUR which renders ATP-bound SUR indistinguishable from TMD0 alone from the point of view of Kir6.2. A possible schematic representation of such a mechanism is shown in Fig. 9C and D.
Our results confirm that TMD0 of SUR is not just a passive anchor between SUR and Kir6.2 but constitutes a dynamic element that could serve as an allosteric linkage between the regulatory subunit SUR and the catalytic subunit Kir6.2. Based on the gating properties of deletion constructs, it has been proposed that TMD0 and the loop L0 connecting it to the rest of SUR modulates the gating of Kir6.2 (Babenko & Bryan, 2003; Fang et al. 2006). The present study does demonstrate that TMD0 motion and change in channel activity occur concomitantly; it does not establish a definite causal relationship. TMD0 motion could be a mere consequence of a rearrangement of the Kir6.2 gating elements mediated by other domains of SUR (Bryan et al. 2004). That TMD0 is not the only region of SUR interacting with Kir6.2 is not debatable given the compact geometry of the channel (Mikhailov et al. 2005). However, TMD0 by itself changes drastically the gating properties of Kir6.2 and coimmunoprecipitation experiments have shown that Kir6.2 associates more readily with TMD0 than with other domains (Chan et al. 2003; Babenko & Bryan, 2003). Therefore the evidence favours a scheme as outlined in Fig. 9 where modification of the core of SUR through binding of ATP is transduced into opening or closing of the Kir6.2 pore via TMD0. There is now evidence that the NBDs of ABC proteins can form dimers and that dimerization is triggered by ATP binding (Hopfner et al. 2000; Smith et al. 2002) and may cause large conformational changes (Chen et al. 2003). Such a process has been postulated to apply to the KATP channel (Yamada et al. 2004; Yamada & Kurachi, 2005). Our conclusions are consistent with a similar mechanism whereby ATP binding to the NBDs could drive, through NBD dimerization, a global structural change transmitted to Kir6.2 in part by TMD0. This change would precede hydrolysis which appears necessary to produce a significant change in the gating mechanism (Gribble et al. 1998; Zingman et al. 2001).
Perspectives
The rhodamine protection assay enables one to follow the binding to SUR of ATP, which switches the channel from a rhodamine-sensitive to a new rhodamine-resistant state. This permits to dissect the respective effects on SUR and Kir6.2 of a ligand such as ATP that is capable of interactions with both subunits. In this work, the concentration used – 100 μm ATP – appeared greater than its affinity for SUR because it was sufficient to switch a large fraction of the channels to a rhodamine-insensitive state. Preliminary experiments indicate that lowering the ATP concentration reduced the rhodamine-insensitive fraction in line with a concentration-dependent binding.
Finally, besides its use as a reporter of the conformation of the KATP channel, Rho123 could serve as a tool to identify the presence of SUR-less Kir6.2 channels in vitro or in vivo because of its ability to selectively block incomplete channels rather than well-assembled SUR + Kir6 channels. Such incomplete channels could find their way to the plasma membrane when channels are overexpressed in heterologous systems or possibly when introduced or natural mutations alter the assembly or trafficking of KATP channels (Chan et al. 2003).
Acknowledgments
We are grateful to Dr J. Bryan (Houston, TX, USA) for hamster SUR1, Dr S. Seino (Chiba, Japan) for mouse Kir6.2 and rat SUR2A, and Dr K. Chan (Cleveland, OH, USA) for the construct TMD0(SUR1). This work was made possible by grants to M.V. and postdoctoral fellowships to E.H. and R.D. from Commissariat à l'Energie Atomique; programme toxicologie nucléaire environnementale. Parts of this work have been published in abstract form (Hosy et al. 2006).
References
- Babenko AP, Bryan J. SUR domains that associate with and gate KATP pores define a novel gatekeeper. J Biol Chem. 2003;278:41577–41580. doi: 10.1074/jbc.C300363200. [DOI] [PubMed] [Google Scholar]
- Bakos E, Evers R, Szakacs G, Tusnady GE, Welker E, Szabo K, et al. Functional multidrug resistance protein (MRP1) lacking the N-terminal transmembrane domain. J Biol Chem. 1998;273:32167–32175. doi: 10.1074/jbc.273.48.32167. [DOI] [PubMed] [Google Scholar]
- Baukrowitz T, Schulte U, Oliver D, Herlitze S, Krauter T, Tucker SJ, et al. PIP2 and PIP as determinants for ATP inhibition of KATP channels. Science. 1998;282:1141–1144. doi: 10.1126/science.282.5391.1141. [DOI] [PubMed] [Google Scholar]
- Booth CL, Pulaski L, Gottesman MM, Pastan L. Analysis of the properties of the N-terminal nucleotide-binding domain of human P-glycoprotein. Biochemistry. 2000;39:5518–5526. doi: 10.1021/bi992931x. [DOI] [PubMed] [Google Scholar]
- Bouabe H, Knittler MR. The distinct nucleotide binding states of the transporter associated with antigen processing (TAP) are regulated by the nonhomologous C-terminal tails of TAP1 and TAP2. Eur J Biochem. 2003;270:4531–4546. doi: 10.1046/j.1432-1033.2003.03848.x. [DOI] [PubMed] [Google Scholar]
- Bryan J, Vila-Carriles WH, Zhao GL, Babenko AP, Aguilar-Bryan L. Toward linking structure with function in ATP-sensitive K+ channels. Diabetes. 2004;53:S104–S112. doi: 10.2337/diabetes.53.suppl_3.s104. [DOI] [PubMed] [Google Scholar]
- Chan KW, Zhang H, Logothetis DE. N-terminal transmembrane domain of the SUR controls trafficking and gating of Kir6 channel subunits. EMBO J. 2003;22:3833–3843. doi: 10.1093/emboj/cdg376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Daoud R, Kast C, Gros P, Georges E. Rhodamine 123 binds to multiple sites in the multidrug resistance protein (MRP1) Biochemistry. 2000;39:15344–15352. doi: 10.1021/bi0020574. [DOI] [PubMed] [Google Scholar]
- Fan Z, Makielski JC. Anionic phospholipids activate ATP-sensitive potassium channels. J Biol Chem. 1997;272:5388–5395. doi: 10.1074/jbc.272.9.5388. [DOI] [PubMed] [Google Scholar]
- Fang K, Csanady L, Chan KW. The N-terminal transmembrane domain (TMD0) and a cytosolic linker (L0) of sulphonylurea receptor define the unique intrinsic gating of KATP channels. J Physiol. 2006;576:379–389. doi: 10.1113/jphysiol.2006.112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Virag L, Furukawa N, Sawanobori T, Hiraoka M. Mechanism for reactivation of the ATP-sensitive K+ channel by MgATP complexes in guinea-pig ventricular myocytes. J Physiol. 1994;479:95–107. doi: 10.1113/jphysiol.1994.sp020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble FM, Tucker SJ, Haug T, Ashcroft FM. MgATP activates the β cell KATP channel by interaction with its SUR1 subunit. Proc Natl Acad Sci U S A. 1998;95:7185–7190. doi: 10.1073/pnas.95.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Hosy E, Derand R, Vivaudou M. A new molecular probe of the interactions between the two subunits of the KATP channel. Biophys J. 2006;90:1950. pos. [Google Scholar]
- Hou YX, Cui LY, Riordan JR, Chang XB. Allosteric interactions between the two non-equivalent nucleotide binding domains of multidrug resistance protein MRP1. J Biol Chem. 2000;275:20280–20287. doi: 10.1074/jbc.M001109200. [DOI] [PubMed] [Google Scholar]
- Hrycyna CA, Ramachandra M, Germann UA, Cheng PW, Pastan I, Gottesman MM. Both ATP sites of human P-glycoprotein are essential but not symmetric. Biochemistry. 1999;38:13887–13899. doi: 10.1021/bi991115m. [DOI] [PubMed] [Google Scholar]
- Johnson LV, Walsh ML, Bockus BJ, Chen LB. Monitoring of relative mitochondrial membrane potential in living cells by fluorescence microscopy. J Cell Biol. 1981;88:526–535. doi: 10.1083/jcb.88.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern A, Felfoldi F, Sarkadi B, Varadi A. Expression and characterization of the N- and C-terminal ATP-binding domains of MRP1. Biochem Biophys Res Commun. 2000;273:913–919. doi: 10.1006/bbrc.2000.3040. [DOI] [PubMed] [Google Scholar]
- Lerner-Marmarosh N, Gimi K, Urbatsch IL, Gros P, Senior AE. Large scale purification of detergent-soluble P-glycoprotein from Pichia pastoris cells and characterization of nucleotide binding properties of wild-type, Walker A, and Walker B mutant proteins. J Biol Chem. 1999;274:34711–34718. doi: 10.1074/jbc.274.49.34711. [DOI] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Loetchutinat C, Saengkhae C, Marbeuf-Gueye C, Garnier-Suillerot A. New insights into the P-glycoprotein-mediated effluxes of rhodamines. Eur J Biochem. 2003;270:476–485. doi: 10.1046/j.1432-1033.2003.03403.x. [DOI] [PubMed] [Google Scholar]
- Matsuo M, Kimura Y, Ueda K. KATP channel interaction with adenine nucleotides. J Mol Cell Cardiol. 2005;38:907–916. doi: 10.1016/j.yjmcc.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Matsuo M, Tanabe K, Kioka N, Amachi T, Ueda K. Different binding properties and affinities for ATP and ADP among sulfonylurea receptor subtypes, SUR1, SUR2A, and SUR2B. J Biol Chem. 2000;275:28757–28763. doi: 10.1074/jbc.M004818200. [DOI] [PubMed] [Google Scholar]
- Mikhailov MV, Campbell JD, de-Wet H, Shimomura K, Zadek B, Collins RF, et al. 3-D structural and functional characterization of the purified KATP channel complex Kir6.2-SUR1. EMBO J. 2005;24:4166–4175. doi: 10.1038/sj.emboj.7600877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modica-Napolitano JS, Aprille JR. Basis for the selective cytotoxicity of rhodamine 123. Cancer Res. 1987;47:4361–4365. [PubMed] [Google Scholar]
- Moreau C, Gally F, Jacquet-Bouix H, Vivaudou M. The size of a single residue of the sulfonylurea receptor dictates the effectiveness of KATP channel openers. Mol Pharmacol. 2005a;67:1026–1033. doi: 10.1124/mol.104.008698. [DOI] [PubMed] [Google Scholar]
- Moreau C, Jacquet H, Prost AL, D'Hahan N, Vivaudou M. The molecular basis of the specificity of action of KATP channel openers. EMBO J. 2000;19:6644–6651. doi: 10.1093/emboj/19.24.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau C, Prost AL, Derand R, Vivaudou M. SUR, ABC proteins targeted by KATP channel openers. J Mol Cell Cardiol. 2005b;38:951–963. doi: 10.1016/j.yjmcc.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Nichols CG. KATP channels as molecular sensors of cellular metabolism. Nature. 2006;440:470–476. doi: 10.1038/nature04711. [DOI] [PubMed] [Google Scholar]
- Nichols CG, Shyng SL, Nestorowicz A, Glaser B, Clement JP, Gonzalez G, et al. Adenosine diphosphate as an intracellular regulator of insulin secretion. Science. 1996;272:1785–1787. doi: 10.1126/science.272.5269.1785. [DOI] [PubMed] [Google Scholar]
- Ribalet B, John SA, Weiss JN. Molecular basis for Kir6.2 channel inhibition by adenine nucleotides. Biophys J. 2003;84:266–276. doi: 10.1016/S0006-3495(03)74847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino S, Miki T. Physiological and pathophysiological roles of ATP-sensitive K+ channels. Prog Biophys Mol Biol. 2003;81:133–176. doi: 10.1016/s0079-6107(02)00053-6. [DOI] [PubMed] [Google Scholar]
- Shyng SL, Nichols CG. Membrane phospholipid control of nucleotide sensitivity of KATP channels. Science. 1998;282:1138–1141. doi: 10.1126/science.282.5391.1138. [DOI] [PubMed] [Google Scholar]
- Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov A, Dusonchet J, Ashcroft F. Metabolic regulation of the pancreatic β-cell ATP-sensitive K+ channel – a pas de deux. Diabetes. 2004;53:S113–S122. doi: 10.2337/diabetes.53.suppl_3.s113. [DOI] [PubMed] [Google Scholar]
- Tucker SJ, Gribble FM, Proks P, Trapp S, Ryder TJ, Haug T, et al. Molecular determinants of KATP channel inhibition by ATP. EMBO J. 1998;17:3290–3296. doi: 10.1093/emboj/17.12.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker SJ, Gribble FM, Zhao C, Trapp S, Ashcroft FM. Truncation of Kir6.2 produces ATP-sensitive K+ channels in the absence of the sulphonylurea receptor. Nature. 1997;387:179–183. doi: 10.1038/387179a0. [DOI] [PubMed] [Google Scholar]
- Ueda K, Inagaki N, Seino S. MgADP antagonism to Mg2+-independent ATP binding of the sulfonylurea receptor SUR1. J Biol Chem. 1997;272:22983–22986. doi: 10.1074/jbc.272.37.22983. [DOI] [PubMed] [Google Scholar]
- Urbatsch IL, Beaudet L, Carrier I, Gros P. Mutations in either nucleotide-binding site of P-glycoprotein (Mdr3) prevent vanadate trapping of nucleotide at both sites. Biochemistry. 1998;37:4592–4602. doi: 10.1021/bi9728001. [DOI] [PubMed] [Google Scholar]
- Vivaudou M, Forestier C. Modification by protons of frog skeletal muscle KATP channels: effects on ion conduction and nucleotide inhibition. J Physiol. 1995;486:629–645. doi: 10.1113/jphysiol.1995.sp020840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J, Hammond ST, Wilke-Mounts S, Senior AE. Mg2+ coordination in catalytic sites of F1-ATPase. Biochemistry. 1998;37:608–614. doi: 10.1021/bi972370e. [DOI] [PubMed] [Google Scholar]
- Wolters JC, Abele R, Tampe R. Selective and ATP-dependent translocation of peptides by the homodimeric ATP binding cassette transporter TAP-like (ABCB9) J Biol Chem. 2005;280:23631–23636. doi: 10.1074/jbc.M503231200. [DOI] [PubMed] [Google Scholar]
- Yamada M, Ishii M, Hibino H, Kurachi Y. Mutation in nucleotide-binding domains of sulfonylurea receptor 2 evokes Na-ATP-dependent activation of ATP-sensitive K+ channels: implication for dimerization of nucleotide-binding domains to induce channel opening. Mol Pharmacol. 2004;66:807–816. doi: 10.1124/mol.104.002717. [DOI] [PubMed] [Google Scholar]
- Yamada M, Kurachi Y. A functional role of the C-terminal 42 amino acids of SUR2A and SUR2B in the physiology and pharmacology of cardiovascular ATP-sensitive K+ channels. J Mol Cell Cardiol. 2005;39:1–6. doi: 10.1016/j.yjmcc.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Zerangue N, Schwappach B, Jan YN, Jan LY. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane KATP channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- Zingman LV, Alekseev AE, Bienengraeber M, Hodgson D, Karger AB, Dzeja PP, et al. Signaling in channel/enzyme multimers: ATPase transitions in SUR module gate ATP-sensitive K+ conductance. Neuron. 2001;31:233–245. doi: 10.1016/s0896-6273(01)00356-7. [DOI] [PubMed] [Google Scholar]