Abstract
We have investigated the mycorrhizal associations of two nonphotosynthetic orchids from distant tribes within the Orchidaceae. The two orchids were found to associate exclusively with two distinct clades of ectomycorrhizal basidiomycetous fungi over wide geographic ranges. Yet both orchids retained the internal mycorrhizal structure typical of photosynthetic orchids that do not associate with ectomycorrhizal fungi. Restriction fragment length polymorphism and sequence analysis of two ribosomal regions along with fungal isolation provided congruent, independent evidence for the identities of the fungal symbionts. All 14 fungal entities that were associated with the orchid Cephalanthera austinae belonged to a clade within the Thelephoraceae, and all 18 fungal entities that were associated with the orchid Corallorhiza maculata fell within the Russulaceae. Restriction fragment length polymorphism and single-strand conformational polymorphism analysis of ectomycorrhizal tree roots collected adjacent to Cephalanthera showed that (i) the fungi associated internally with Cephalanthera also form typical external ectomycorrhizae and that (ii) ectomycorrhizae formed by other Basidiomycetes were abundant where the orchid grows but these fungi did not associate with the orchid. This is the first proof of ectomycorrhizal epiparasitism in nature by an orchid. We argue that these orchids are cheaters because they do not provide fixed carbon to associated fungi. This view suggests that mycorrhizae, like other ancient mutualisms, are susceptible to cheating. The extreme specificity in these orchids relative to other ectomycorrhizal plants agrees with trends seen in more conventional parasites.
Keywords: mycorrhiza, ecology, symbiosis, specificity, ribosomal DNA sequences
Ancient, widespread mutualisms are expected to attract cheaters, i.e., parasites of the mutualism that interact by mimicking a mutualist but do not provide the usual benefits to the other interactant (1, 2). Although such cheating may be a common occurrence, there are only a few well documented examples, such as the fig/fig wasp (3), yucca/yucca moth (4), and ant/plant (5, 6) systems.
Specificity is a critical parameter in all interactions, especially intimate, symbiotic ones. Parasites, under the narrow definition of Thompson (7), show a striking global pattern of extreme specialization (7, 8). Specificity varies widely in symbiotic mutualists (9), and the reasons for this variation are controversial (9, 10). Cheaters of nonsymbiotic (i.e., nonintimate) mutualisms, such as plants and pollinators, are predicted to become specialized in some circumstances and not in others (1). Specificity in cheaters of symbiotic mutualisms has been little studied.
Mycorrhizae are intimate symbioses between fungi and the underground organs of plants; the mutualism is based on the provisioning of minerals, and perhaps water, to the plant by the fungus in return for fixed carbon from the plant (11). Ectomycorrhizae (ECM) are the dominant mycorrhizal type formed by forest trees in temperate regions (12), and they are critical to nutrient cycling and to structuring of plant communities in these regions (13).
There are several indications that the ECM mutualism is not immune to cheating. For example, the fungus Entoloma saepiens forms an apparent ECM structure (mantle) on Rosa and Prunus roots but destroys the root epidermal cells (14). Similarly, Tuber melanosporum directly parasitizes the roots of some plants, causing brules, or patches of dead herbs and grasses, under its primary ECM host trees (15). Perhaps most startling are suggestions of cheating by mycorrhizal plants. The Monotropoideae is a subfamily consisting entirely of nonphotosynthetic, so-called myco-heterotrophic (16), plants that obtain all of their carbon through ECM associations (17). Although other benefits may accrue to mycorrhizal fungi, capture of fixed carbon from photosynthetic host plants is thought to be the most significant reward (11, 18). A recent analysis of the Monotropoideae (19) uncovered a high specificity of the plant toward the fungus in most species examined. Because of the high specificity, similar to that of many parasites, and the inverted direction of net carbon flow in the mycorrhizae of the Monotropoideae, it has been argued that these plants represent parasites of ECM mutualism (19). However, physiological studies of this interaction are limited to those of Bjorkman (17), and many believe that parasitism has not been proven in these plants.
Mycorrhizae are unique physiologically, anatomically, and in the identity of the fungal associates in the Orchidaceae. In addition, there are more myco-heterotrophic genera in the Orchidaceae than in any other family (20). Basidiomycetous fungi of the form-genus Rhizoctonia are the predominant mycorrhizal associates of the Orchidaceae (21), and most are soil saprophytes or pathogens (21), unlike the fungi involved in arbuscular mycorrhizal (AM) and ECM associations. Physiological studies have consistently failed to uncover the transport of organic carbon, in any form, from orchid to fungus, even in photosynthetic orchid species (21, 22). In contrast, movement of sugars from fungus to orchid has been documented repeatedly (23). For these reasons, Smith and Read concluded that “the symbiosis cannot be regarded as mutualistic” in orchid mycorrhizae (21). Thus, the fungal associations of orchids would appear to have little to do with mutualistic AM and ECM symbioses. However, associations between orchids and unidentified ECM fungi have been described (24–26). The associations of some orchids also appear to be highly specific in the wild (27, 28) although specificity in orchid mycorrhizae remains controversial (29, 30).
In this report, we show that all of the fungi associated with the nonphotosynthetic orchids Cephalanthera austinae (A. Gray) Heller and Corallorhiza maculata Rafinesque belong to ectomycorrhizal clades and that symbionts of one these orchids form ECM with surrounding trees while simultaneously associating with the orchid in nature. Furthermore, specificity in both orchids is marked. These results demonstrate unexpected associations and mycorrhizal anatomies in two important clades of ECM fungi. They also provide support for the contention that the ECM symbiosis has been invaded by cheaters.
MATERIALS AND METHODS
Sample Collection.
Orchid samples were collected from May to September in 1992–1995 by excavating roots or rhizomes under the ephemeral flowering spikes at the locations shown in Table 1. A systematic search for tree roots surrounding C. austinae was made for 11 samples (Table 2) by removing 10-cm diameter soil cores of 20–40 cm depth that were centered on the inflorescences. The soil was then washed, and both orchid and tree roots were collected using sieves, with a final screen opening size of 0.5 mm. Orchid roots and rhizomes were washed and frozen, and ECM tree roots were recovered from sieves, sorted into mycorrhizal “morphotypes” under the dissecting microscope (33, 34), and then lyophilized.
Table 1.
Occurrence of all fungi associated with C. maculata and C. austinae
|
C. maculata
|
C. austinae
|
||
|---|---|---|---|
| Fungus | Counties/states* | Fungus | Counties/states* |
| Russula graveolens HDT54290 | El Dorado; Glenn; Marin; Mendocino; Plumas; Siskiyou; Tehama; Clark, WA; King, WA | ||
| Russula flaviceps LT51 | Contra Costa; San Mateo | ||
| Russula sp. DED5585 | Contra Costa; San Mateo | Thelephoraceae 1 | Humboldt |
| Russula murrillii HDT53368 | Mendocino; Lane, OR; Lewis, WA | Thelephoraceae 2 | Glenn |
| Russula amoenolens SNF63 | Contra Costa | Thelephoraceae 3 | Glenn |
| Russula sp. SNF288 | Glenn; Lane, OR; Pierce, WA | Thelephoraceae 4 | Tehama |
| Russula sp. LT37 | Glenn; Tehama | Thelephoraceae 5 | El Dorado |
| Russula integra HDT54375 | Mendocino | Thelephoraceae 6 | Lane, OR |
| Russula sp. LT40 | Tuolumne | Thelephoraceae 7-I | El Dorado; Trinity; Tuolumne; King, WA |
| Russula californiensis HDT54442 | Contra Costa | Thelephoraceae 8 | El Dorado, Tehama; Trinity; Tuolumne |
| Gymnomyces abietis SNF74 | Alpine; Mono; Sierra | Thelephoraceae 9 | El Dorado |
| Lactarius sp. LT80 | Contra Costa | Thelephoraceae 10 | Glenn; Tehama |
| Russulaceae 1 | El Dorado; Plumas | Thelephoraceae 11 | Humboldt |
| Russulaceae 2 | Glenn; Mendocino | Thelephoraceae 12 | Monterey |
| Russulaceae 3 | Tehama | Thelephoraceae 13-I | Monterey |
| Russulaceae 4 | Humboldt | Thelephoraceae 14 | Humboldt |
| Russulaceae 5 | Summit, OH | ||
| Russulaceae 6 | Sauk, WI | ||
C. maculata associates exclusively with fungi of the Russulaceae, and C. austinae associates exclusively with fungi of the Thelephoraceae over the entire geographic range sampled. In total, 26 individuals of C. austinae were sampled, covering essentially the entire range of this orchid (31), and 68 individuals of C. maculata were sampled over a wider area but covering less of the range of this orchid. Species of fungal associates are given where fungal ITS RFLP patterns obtained directly from orchid tissue, using three restriction enzymes, were exactly matched to patterns from fungal fruit bodies. The genus Lactarius is closely related to the genus Russula, and Gymnomyces is a hypogeous genus thought to be derived from Russula (32). Collection numbers following fruit bodies refer to the following herbaria: LT, SNF, private herbarium of Thomas D. Bruns, University of California, Berkeley; DED, HDT, the Harry D. Thiers Herbarium of the San Francisco State University. Family-level designations from unmatched fungal ITS–RFLPs are based on the sequence analyses presented in Fig. 1. The two Thelephoroid fungi that were isolated from orchid tissue are designated by “I.”
Counties are in California except as noted.
Table 2.
SSCP analysis of fungi associated with C. austinae and surrounding tree ECM
| California county/sample | Fungi in orchid | Zygosity | Thelephoroid ECM fungi on adjacent tree roots | Zygosity | Other ECM fungi on adjacent tree roots |
|---|---|---|---|---|---|
| Tehama 1 | Thelephoraceae 2 | hm | Thelephoraceae 2 (1) | hm | Russula + 2 |
| Tuolumne 1 | Thelephoraceae 8 | hm | Thelephoraceae 8 | het | Lactarius, Russula + 3 |
| 2 | Thelephoraceae 7 | het | Thelephoraceae 7 | het | 1 |
| Humboldt 1 | Thelephoraceae 1 | het | Thelephoraceae 1 | het | 0 |
| 2 | Thelephoraceae 11 | het | Thelephoraceae 11 | het | 3 |
| Monterey 1 | Thelephoraceae 12; Thelephoraceae 13 | hm | Thelephoraceae 13 (2) | hm | 4 |
| Trinity 1 | Thelephoraceae 7 | het | Thelephoraceae 7 (1) | het | 3 |
| 2 | Thelephoraceae 8 | hm | Thelephoraceae 8 | hm | 3 |
| El Dorado 1 | Thelephoraceae 7 | het | Thelephoraceae 7 | het | 3 |
| 2 | Thelephoraceae 7 | het | Thelephoraceae 7 | het | 5 |
| 3 | Thelephoraceae 7 | hm | Thelephoraceae 7 (1) | hm | 2 |
The Thelephoroid fungi that associate internally with C. austinae simultaneously form ECM on photosynthetic hosts in nature. Each row shows the fungal ITS–RFLP type of the fungi found in orchid roots and the type found in Thelephoroid fungi forming ECM on photosynthetic tree roots from a single 10-cm diameter soil core. Other Thelephoroid species, in addition to those associated with the orchid, often formed ECM, which were recovered from the same soil cores; numbers of these are given in parentheses. RFLPs also were generated from ECM that did not have Thelephoroid morphologies to estimate the diversity of ECM-forming Basidiomycetes present at each site. However, no attempt was made to identify these non-Thelephoroid fungi except in the case of several types known to belong to the Russulaceae. In addition to RFLP analysis, the ITS region amplified from Thelephoroid fungi on orchid and tree roots was compared by single-strand conformational polymorphism analysis. The migration patterns were different for each distinct RFLP type but were identical in orchid/tree pairs from each soil core except in one case, in which two bands migrated identically, but the ectomycorrhizal sample had two additional bands. However, ECM from other cores had the same two band patterns as the orchid in this core. Therefore, all of the orchid fungi were shown to form ECM as well. In general, if either the ITS 1 or ITS 2 region displayed three or four bands, the ITS was interpreted as heterozygous (44). het, heterozygous; hm, homozygous.
Fungal Isolation.
Fungal isolation was attempted from two samples of C. austinae and from numerous samples of C. maculata. After sterilizing the roots by immersion in 20% bleach for 10 minutes, intracellular coils of intact hyphae (pelotons) were plated onto modified Melin–Norkrans medium (35) with 50 μg/ml each streptomycin and tetracycline by serial transfer to sterile water using a pipetman under a laminar flow hood. Legitimate cultures from C. austinae had clamp connections and dark hyphae similar to those seen in the pelotons; these cultures grew extremely slowly (detectable within 2 weeks of plating) directly from pelotons. Isolates from single pelotons were subcultured and subject to DNA fingerprinting, as described below.
Internal Transcribed Spacer (ITS) Restriction Fragment Length Polymorphism (RFLP) Analysis.
Total DNA was extracted from orchids as follows. Root cross sections were freeze-thawed three times and then ground in SDS/EDTA buffer (36) and extracted with chloroform. DNAs were isolated and purified from this extract using Gene-Clean (Bio 101, Vista, CA) according to the manufacturer’s instructions. Total DNA was extracted from lyophilized ECM root tips and fungal fruit bodies following the protocol of Gardes and Bruns (33). PCR amplification of the ITS region of the nuclear ribosomal repeat with the Basidiomycete-specific primers ITS 1F together with either ITS 4 or ITS 4B followed by digestion and RFLP analysis was performed (33). ITS fragments that were weakly amplified from several orchid samples were gel purified and reamplified using the same primers and conditions. The ITS region was digested separately with the restriction enzymes AluI, HinfI, and MboI. Two independent ITS analyses, starting with DNA extraction, were performed for every sample of C. austinae and for the majority of C. maculata samples (exceptions were due to lack of material). Only samples (whether from orchids, fungal isolates, tree ECM, or fruit bodies) that displayed identical RFLP patterns on the same gel for all three enzymes, including any submolar bands, were considered to belong to the same ITS RFLP type.
Single-Strand Conformational Polymorphism Analysis.
PCR products (0.5 μl) amplified using ITS1F and ITS4B were used as templates for labeled PCRs containing 0.8 μl of [35S]dATPαS (Amersham; specific activity > 1000 Ci/mmol, 10 mCI/ml) and final concentrations of 0.67 μM for each dNTP/50 mM KCl/10 mM Tris⋅HCl, pH 8.3/2.5 mM MgCl/0.1 g/liter gelatin/0.25 units of Taq DNA polymerase/0.48 μM each of the primers ITS1F and ITS2 (ITS 1 region) or ITS 3 and ITS 4 (ITS 2 region)/9.1 μl H2O. Labeled reactions were carried out for 10 cycles using 95°C/55°C/72°C and 1 min for each step, followed by a final 7-min extension at 72°C. Samples (3 μl) generated from orchid roots and Thelephoroid ECM from the same soil core were combined with 3 μl of formamide stop dye (United States Biochemical), denatured at 95°C for 5 min, iced, and then run in adjacent lanes on a nondenaturing mutation detection enhancement gel (FMC) at 2–4 volts overnight at room temperature. The gel was then dried and exposed to Kodak X-Omat AR film for 3–5 days.
DNA Sequencing.
Direct fluorescent-labeled (DyeDeoxy system, Perkin–Elmer) sequencing of PCR products on an Applied Biosystems model 373 or 377 automated sequencer was performed. The primers ITS 1F (37), ITS 2, ITS 3, and ITS 4 (38) were used to produce bidirectional sequences for the entire ITS 1, 5.8S, and ITS 2 regions. The fungal-specific primers ML5 and ML6 were used similarly to amplify and sequence in two directions a ≈350-bp region of the mitochondrial large subunit ribosomal RNA gene (19, 34). Alignments were produced with Clustal V and corrected by hand using a color font. See Fig. 1 for phylogenetic methodology.
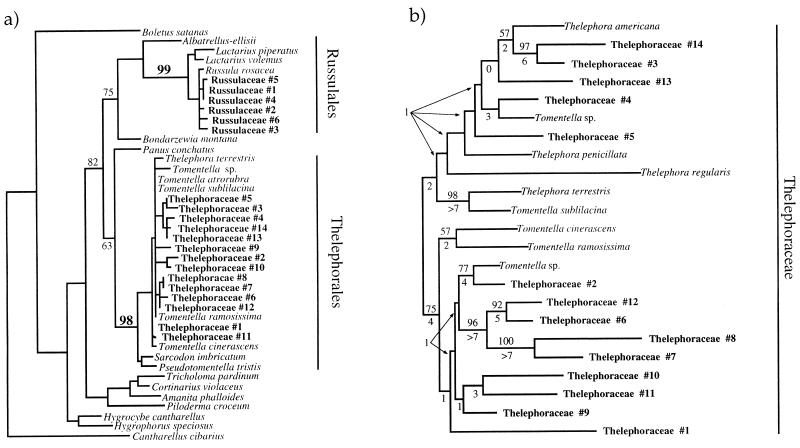
Figure 1.
Phylogenetic placement of orchid fungi. (a) Each orchid targets a distinct clade of ectomycorrhizal fungi. ML5–6 sequences from symbionts of both orchids were aligned with those from fruit bodies of 112 (mostly ECM) Basidiomycete species. This database has been used previously (19, 34); details will be presented elsewhere. Analysis by the distance method of neighbor-joining under a Jukes–Cantor one-parameter model using paup 4.0 (39) produced a tree in which all symbionts of C. austinae were grouped several nodes interior to the boundaries of the Thelephorales, and all unknown symbionts of C. maculata were similarly grouped well within the Russulales. These placements were strongly supported by a 1000 replicate neighbor-joining boot strap analysis (numbers near branches). Most taxa were then pruned from the tree, leaving a taxon from each major clade in the representation above. Cantharellus was used as the outgroup. (b) ITS sequences provide strong, independent evidence for the placement of the symbionts of C. austinae in a clade within the Thelephoraceae. ITS sequences were generated for each fungal ITS RFLP type associated with C. austinae and from fruit bodies of the Thelephoraceae. ITS sequences were obtained for Sarcodon imbricatum, Hydnum umbilicatum, Hydnellum peckii, and Pseudotomentella trisitis in addition to the taxa shown above. However, none of these could be aligned with the ITS sequences obtained from Cephalanthera associates or Thelephora and Tomentella fruit bodies. Parsimony analysis using 100 random addition replicates of the remaining taxa by paup produced the single tree, shown above. Midpoint rooting was used because of lack of an a priori choice for outgroup. The neighbor-joining tree also agreed with all well supported branches from the parsimony analysis. Parsimony boot strap values from 1000 replicates are shown above branches, and decay indices are shown below branches. Although taxa sampling was limited, this analysis provides evidence that all Cephalanthera associates are more closely related to Thelephora and Tomentella than to other sampled genera of the Thelephoraceae.
RESULTS
Diversity and Identity of Fungal Associates.
Microscopic inspection revealed that all C. austinae roots contained cortical cells with coils, or “pelotons,” of distinctively brown, thick-walled, clamped hyphae (Fig. 2d); the latter indicated that the fungi were Basidiomycetes. Three-enzyme ITS–RFLP analysis yielded 14 distinct combinations of RFLP patterns (Table 1). ITS RFLP patterns from the fungi isolated from C. austinae pelotons were identical to those produced directly from orchid tissue. ITS RFLP variants have been shown to be correlated with groups of closely related morpho-species in mushroom-forming Basidiomycetes (34, 40), implying that the number of ITS RFLP types may slightly underestimate the number of species. However, because our single-strand conformational polymorphism analysis (discussed below) frequently detected heterozygosity in the ITS and some corresponding homozygotes were distinguished as different ITS RFLP types, the number of ITS RFLP types also might overestimate the number of species involved. Therefore, we view the number of ITS RFLP patterns we have found in each orchid as the best current approximation of the number of fungal species.
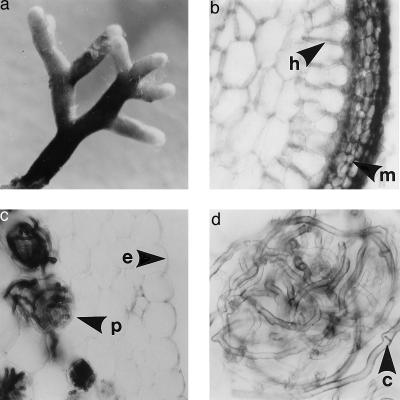
Figure 2.
The same fungal species forms contrasting mycorrhizal structures when associated with nonphotosynthetic orchid or photosynthetic tree host. All structures were formed simultaneously by the fungus Thelephoraceae 12 on tightly intermingled C. austinae and tree roots recovered from a single soil core. (a) Intact ECM tree root, ×25. (b) Cross section of an ECM tree root showing thick fungal mantle (arrow, m) and Hartig net (arrow, n) but no intracellular hyphae, ×400. (c) Cross section of a C. austinae root demonstrating the lack of a mantle surrounding the epidermis (arrow, e) or a Hartig net and the presence of intracellular coils or pelotons (arrow, p) typical of orchidaceous mycorrhizae, ×100. (d) Cephalanthera root cross section showing a single peloton and typical dark hyphae with clamp connections (arrow, c), ×400.
The peloton-forming hyphae seen in C. maculata, in contrast with Cephalanthera, were nearly hyaline and were never clamped. Eighteen ITS RFLP types were found across all collections of this orchid (Table 1). Hyphal growth from pelotons of this orchid was extremely limited, and the fungus could not be isolated in pure culture.
A combination of ITS RFLP matching between fungal patterns generated directly from orchid tissue and from fruit bodies (Table 1), ML5/6 sequence analysis (Fig. 1a), and ITS sequence analysis (Fig. 1b) shows that C. austinae associates exclusively with a clade of fungi within the Thelephoraceae and that C. maculata associates exclusively with fungi belonging to the Russulaceae.
Trophic Relations and Mycorrhizal Anatomy of the Fungal Symbionts.
Because some members of the Thelephoraceae only recently have been shown to be ECM (41, 42), we sought to determine whether the C. austinae Thelephoroid symbionts form ECM on photosynthetic hosts in nature. All 11 soil cores taken below C. austinae yielded tree ECM morphotypes with some of the following morphological features known to be typical of Thelephoroid ECM: cigar-brown color, clamped hyphae, cystidia, and a unique “polygon–synenchyma” mantle surface pattern (43). At least one of these Thelephoroid ECM from each soil core had an ITS–RFLP pattern matching that of the adjacent orchid roots from the same core (Table 2). We used single-strand conformational polymorphism analysis to confirm the match because this technique reveals the majority of sequence differences throughout a fragment whereas RFLP analysis surveys a more limited proportion of the bases. Both the ITS 1 and ITS 2 regions of the orchid fungi and Thelephoroid ECM from a single core had matching single-strand conformational polymorphism migration patterns in every case and differed in zygosity in only one instance (Table 2). ITS RFLP analysis of all other ECM from the soil cores, which were often in direct contact with orchid roots, showed that a wide variety of non-Thelephoroid, ECM-forming Basidiomycetes, including Russula species, were present (Table 2), yet C. austinae was never found to associate with a non-Thelephoroid fungus.
The Thelephoroid ECM formed on surrounding trees by Cephalanthera-associated fungi consistently possessed well developed fungal mantles at least three cells deep and intercellular Hartig nets penetrating one to three cell layers (Fig. 2b). Neither mantle nor Hartig net was ever observed in C. austinae roots. Rather, the dense fungal infection was almost entirely intracellular (Fig. 2, c and d); cortical cells from the third layer below the epidermis and extending to the vascular bundle were filled with loops of hyphae, or pelotons, characteristic of orchidaceous mycorrhizae (21).
The finding that fungi of the Thelephoraceae and Russulaceae associate intimately with orchids was unexpected because these fungi have never before been recorded as forming internal mycorrhizal structures and because a previous report indicated that Armillaria mellea was the symbiont of C. maculata (25). We are confident, however, that our identifications are correct for the following reasons: (i) independent fungal-specific amplification, RFLP comparison and sequencing, and phylogenetic analysis of a nuclear and a mitochondrial region each pinpointed the same fungal clades (Fig. 1, a and b; Table 1); (ii) few hyphae that might contaminate our amplifications were seen on the surface of orchid tissues whereas hyphae of the symbiotic fungi filled large volumes of the orchid cortex (Fig. 2c); (iii) the hyphal morphology seen in Cephalanthera cross sections and in fungal isolates was congruent, and the ITS RFLP patterns produced from the two sources were identical; and (iv) it is difficult to envision how either amplification of nonsymbiotic fungi from root or rhizome surfaces or widespread PCR contamination could produce the phylogenetically clustered, but nonhomogeneous, arrays of fungal entities found in each orchid.
DISCUSSION
Mycorrhizae are classified as mutualistic interactions, and little attention has been paid to possible nonmutualistic mycorrhizal associations (45). Our results clearly show that nonphotosynthetic orchids form associations with fungi that simultaneously form ECM with photosynthetic hosts in nature. All of the physiological evidence from orchids says that they are not mutualistic with their fungal associates (21). As we discuss further below, the associations of Cephalanthera and Corallorhiza are highly specialized relative to photosynthetic hosts of ECM fungi (46). Taken together, these data provide strong circumstantial evidence that the ECM symbiosis has, indeed, been invaded by cheating plants. The final test of this contention will be to measure fitness consequences of association with one of these myco-heterotrophic orchids to ECM fungi and their photosynthetic hosts. Because of problems in working with long lived plants and obligately symbiotic fungi, fitness measures are lacking for all ECM and for most putatively mutualistic symbioses (2).
If we accept that the ECM mutualism has been invaded by cheaters, several evolutionary hypotheses are then relevant to mycorrhizae. Reciprocal altruism models predict that mutualisms are evolutionarily unstable once they have been invaded by cheaters, unless checks and balances evolve to control them (47). Evidence for such controls is scarce but recently has been documented in the yucca/yucca moth system (48). The fact that some mycorrhizal plants reject the symbiosis when they are well fertilized (49) suggests that these plants are capable of exerting significant control. However, control mechanisms are only useful if cheaters can be distinguished from good mutualists (47). This would appear to be a ripe area for investigation in mycorrhizae.
Comparison of the Monotropoideae to the Orchidaceae shows that the ECM mutualism has been invaded by myco-heterotrophic plants through quite dissimilar internal and external routes. In the dicot Monotropoideae, the ancestors as well as the entire extant subfamily are ECM (19). Thus myco-heterotrophy arose from within the ECM symbiosis in this case. In contrast, monocot families closely allied to the Orchidaceae form internal, endomycorrhizal associations with Glomalean (AM) fungi (50). Therefore, the progenitors of orchids were presumably AM. Then, early in their history, orchids switched from AM to non-ECM Basidiomycete fungi but retained an AM-like internal mycorrhizal structure. Furthermore, Cephalanthera and Corallorhiza belong to quite divergent tribes within the Orchidaceae (51), each of which is dominated by photosynthetic species, many of which retain the primitive non-ECM association of most orchids (52). This implies that the combined phenomena of a jump to association with ECM fungi and a loss of photosynthesis occurred independently in these two orchids. Thus, the ECM fungus-associated orchids described here have invaded the mutualism through an intriguing set of jumps from outside of the ECM symbiosis. Further comparative study may reveal whether the jump to ECM fungi preceded or followed the loss of photosynthesis in these orchids.
The evolutionary history of the Orchidaceae may help to explain the endogenous mycorrhizal structures formed even with ECM fungi. However, the flexibility in mycorrhizal anatomy seen in members of the Russulaceae and Thelephoraceae is noteworthy. These fungal clades have broad host ranges and appear to be widespread and important components of temperate ECM communities (34, 53). The unexpected interaction of these fungi with orchids underlines our ignorance of the activities and functioning of important, widespread soil microbes.
Parasites are often more specialized than mutualist counterparts (8, 9), and our extensive sampling of C. austinae and C. maculata combined with phylogenetic analysis of the fungal associates provides strong evidence of specificity in these orchids. Associations in these orchids were restricted to two or three genera, each within a single Basidiomycete family (Fig. 1; Table 1). Furthermore, the 14 Thelephoroid fungi associated with Cephalanthera and the 18 Russuloid fungi associated with Corallorhiza are small fractions of the hundreds of North American species in each of these families. The associations of the orchids may then be restricted even within these fungal families. Further phylogenetic and geographic/population analysis is needed to address this. A similar level of specificity appears to hold for Corallorhiza trifida (26) although the fungi remain unidentified. The fact that numerous non-Thelephoroid ECM Basidiomycetes, including Russula species that we know associate with C. maculata, were found alongside C. austinae roots but were not associated with the orchid, suggests active targeting of specific fungi by the orchid rather than an alternative mechanism of avoidance of orchids by most ECM fungi.
A similar level of specificity is displayed in the myco-heterotrophic Monotropoideae, the most specific of which, Pterospora andromeda, was restricted to associations in the Rhizopogon subcaerulescens species group, and the least specific, Sarcodes sanguinea, was associated with species of at least three Basidiomycete families (19). In contrast, the highest specificity documented in a photosynthetic ECM host occurs in Alnus rubrus, in which 11 ECM morphotypes belonging to seven genera in six Basidiomycete families were seen over a wide geographic range (54). Most ECM plants associate much more broadly. For example, Pseudotsuga menziesii is estimated to associate with 2000 of the approximately 6000 worldwide species of ECM fungi (46). The extreme specificity in the Orchidaceae and the Monotropoideae relative to photosynthetic plants has arisen via independent evolutionary paths and is thus a striking example of convergence.
This convergent specificity among myco-heterotrophic plants agrees with trends seen in parasites, particularly intimate ones that remain associated with a single host for prolonged periods (7). However, these plants do not fit the conventional picture in which the parasite lives on or in its host. Instead, the fungal host invades and proliferates within the parasitic plant. Furthermore, these highly specialized Monotrope and orchid species are quite rare relative to their fungal “prey,” suggesting that these plants exert little selective pressure on the fungi and that specific fungal defenses are unlikely to have evolved.
In conclusion, we have shown that two nonphotosynthetic orchids have independently evolved specialized associations with particular ECM fungi. Furthermore, these orchids somehow exclude Basidiomycetes outside their target clades. The evolution of specificity in Monotropes and orchids differs in that (i) it has evolved through narrowing from inside vs. jumps from outside the symbiosis, and (ii) it has led to the targeting of narrow vs. broad host range fungi (19). Monotropes and orchids also differ in their external vs. internal mycorrhizal structures. The internal mycorrhizae formed by ECM fungi associated with orchids illustrate fundamental plant control over the anatomy of the symbiotic organ. The reversal of carbon flow in these plants suggests that the ECM symbiosis has been invaded by cheaters, as have other ancient symbioses. The specialization in these plants agrees with that found in intimate parasites, despite the contrasts between these plants and other parasites. Several other distantly related nonphotosynthetic orchids associate with pathogenic Basidiomycetes (20), which also have a direct connection to the large carbon pool of a living host, unlike most saprophytic Rhizoctonia spp. We propose that the apparent predisposition of the Orchidaceae to loss of photosynthesis is a consequence of the unique capacity of these plants to become effective carbon robbers through specialization on various Basidiomycetes that are linked to large carbon resources.
Acknowledgments
We appreciate orchid samples or locations provided by Ronald Coleman, Holley Duffer, Jeffrey Hapeman, David Isle, Warren Stoutamire, Ian Sussex, Jon Titus, and Jape Taylor. We thank Michael Larsen, Harry Thiers, and James Trappe for fruit body identifications, Ignacio Chapela, Tim Szaro, John Thompson, and Detlev Vogler for comments on the manuscript, and members of the Bruns lab, especially Pierluigi Bonello and Tim Szaro, for technical cooperation. Funding was provided by the American Orchid Society, the Hardman Foundation, and National Science Foundation Grants DEB 9307150 and DEB 9628852.
ABBREVIATIONS
- ECM
ectomycorrhizae
- ITS
internal transcribed spacer region
- RFLP
restriction fragment length polymorphism
- AM
arbuscular mycorrhizae
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. U83466–U83487U83466U83467U83468U83469U83470U83471U83472U83473U83474U83475U83476U83477U83478U83479U83480U83481U83482U83483U83484U83485U83486U83487, U86838–U86864U86838U86839U86840U86841U86842U86843U86844U86845U86846U86847U86848U86849U86850U86851U86852U86853U86854U86855U86856U86857U86858U86859U86860U86861U86862U86863U86864, and U92537U92537).
References
- 1.Mainero J S, Martinez del Rio C. In: The Biology of Mutualism: Ecology and Evolution. Boucher D H, editor. New York: Oxford Univ. Press; 1985. pp. 192–216. [Google Scholar]
- 2.Boucher D H, James S, Keeler K H. Annu Rev Ecol Syst. 1982;13:315–347. [Google Scholar]
- 3.Bronstein J L. Oikos. 1991;61:175–186. [Google Scholar]
- 4.Pellmyr O, Leebens-Mack J, Huth C J. Nature (London) 1996;380:155–156. doi: 10.1038/380155a0. [DOI] [PubMed] [Google Scholar]
- 5.Janzen D H. Science. 1975;188:936–937. doi: 10.1126/science.188.4191.936. [DOI] [PubMed] [Google Scholar]
- 6.Letourneau D K. Science. 1990;248:215–217. doi: 10.1126/science.248.4952.215. [DOI] [PubMed] [Google Scholar]
- 7.Thompson J N. The Coevolutionary Process. Chicago: Univ. of Chicago Press; 1994. [Google Scholar]
- 8.Price P W. Evolutionary Biology of Parasites. Princeton: Princeton Univ. Press; 1980. [Google Scholar]
- 9.Law R, Koptur S. Biol J Linn Soc. 1986;27:251–267. [Google Scholar]
- 10.Borowicz V A, Juliano S A. Evol Ecol. 1991;5:385–392. [Google Scholar]
- 11.Allen M F. The Ecology of Mycorrhizae. Cambridge, U.K.: Cambridge Univ. Press; 1991. [Google Scholar]
- 12.Malloch D W, Pirozynski K A, Raven P H. Proc Natl Acad Sci USA. 1980;77:2113–2118. doi: 10.1073/pnas.77.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francis R, Read D J. Plant Soil. 1994;159:11–25. [Google Scholar]
- 14.Agerer R, Waller K. Mycorrhiza. 1993;3:145–154. [Google Scholar]
- 15.Plattner I, Hall I R. Mycol Res. 1995;99:1367–1370. [Google Scholar]
- 16.Leake J R. New Phytol. 1994;127:171–216. doi: 10.1111/j.1469-8137.1994.tb04272.x. [DOI] [PubMed] [Google Scholar]
- 17.Bjorkman E. Physiol Plant. 1960;13:308–327. [Google Scholar]
- 18.Lewis D H. In: Endomycorrhizas. Sanders F E, Mosse B, Tinker P B, editors. New York: Academic; 1975. pp. 119–148. [Google Scholar]
- 19.Cullings K W, Szaro T M, Bruns T D. Nature (London) 1996;379:63–66. [Google Scholar]
- 20.Furman T E, Trappe J M. Q Rev Biol. 1971;46:219–225. [Google Scholar]
- 21.Smith S E, Read D J. Mycorrhizal Symbiosis. San Diego: Academic; 1997. [Google Scholar]
- 22.Purves S, Hadley G. In: Endomycorrhizas. Sanders F E, Mosse B, Tinker P B, editors. New York: Academic; 1975. pp. 175–194. [Google Scholar]
- 23.Hadley G. New Phytol. 1984;96:263–273. [Google Scholar]
- 24.Warcup J H. New Phytol. 1985;99:273–280. [Google Scholar]
- 25.Campbell E O. Mich Bot. 1970;9:108–113. [Google Scholar]
- 26.Zelmer C D, Currah R S. Can J Bot. 1995;73:862–866. [Google Scholar]
- 27.Warcup J H. New Phytol. 1971;70:41–46. [Google Scholar]
- 28.Masuhara G, Katsuya K, Yamaguchi K. Mycol Res. 1993;97:746–752. [Google Scholar]
- 29.Hadley G. New Phytol. 1970;69:1015–1023. [Google Scholar]
- 30.Clements M A. Lindleyana. 1988;3:73–86. [Google Scholar]
- 31.Luer C A. The Native Orchids of the United States and Canada. Ipswitch, NY: The New York Botanical Garden; 1975. [Google Scholar]
- 32.Heim R. In: Evolution in the Higher Basidiomycetes. Petersen R H, editor. Knoxville, TN: Univ. of Tennessee Press; 1971. pp. 505–534. [Google Scholar]
- 33.Gardes M, Bruns T D. In: Methods in Molecular Biology. Species Diagnostics Protocols: PCR and Other Nucleic Acid Methods. Clapp J P, editor. Vol. 50. Clifton, NJ: Humana; 1996. pp. 177–186. [DOI] [PubMed] [Google Scholar]
- 34.Gardes M, Bruns T D. Can J Bot. 1996;74:1572–1583. [Google Scholar]
- 35.Marx D H. Phytopathology. 1969;59:153–163. [PubMed] [Google Scholar]
- 36.Lee S B, Taylor J W. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 282–287. [Google Scholar]
- 37.Gardes M, Bruns T D. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 38.White T J, Bruns T, Lee S, Taylor J. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 315–322. [Google Scholar]
- 39.Swofford, D. L. PAUP: Phylogenetic Analysis Using Parsimony (Illinois Natural History Survey, Champaign, IL), Ver. 4.0 beta.
- 40.Vilgalys R, Sun B L. Proc Natl Acad Sci USA. 1994;91:4599–4603. doi: 10.1073/pnas.91.10.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koljalg U. Mycol Res. 1992;96:215–220. [Google Scholar]
- 42.Agerer R. Z Mykol. 1994;60:143–158. [Google Scholar]
- 43.Haug I, Oberwinkler F. Trees. 1987;1:172–188. [Google Scholar]
- 44.Lessa E P, Applebaum G. Mol Ecol. 1993;2:119–129. doi: 10.1111/j.1365-294x.1993.tb00006.x. [DOI] [PubMed] [Google Scholar]
- 45.Francis R, Read D J. Can J Bot. 1995;73:S1301–S1309. [Google Scholar]
- 46.Molina R, Massicotte H, Trappe J M. In: Mycorrhizal Functioning: An Integrative Plant-Fungal Process. Allen M F, editor. New York: Chapman & Hall; 1992. pp. 357–423. [Google Scholar]
- 47.Axelrod R, Hamilton W D. Science. 1981;211:1390–1396. doi: 10.1126/science.7466396. [DOI] [PubMed] [Google Scholar]
- 48.Pellmyr O, Huth C J. Nature (London) 1994;372:257–260. [Google Scholar]
- 49.Marx D H, Hatch A B, Mendicino J F. Can J Bot. 1977;55:1569–1574. [Google Scholar]
- 50.Trappe J M. In: Ecophysiology of VA Mycorrhizal Plants. Safir G R, editor. Boca Raton, FL: CRC; 1987. pp. 5–25. [Google Scholar]
- 51.Dressler R L. Phylogeny and Classification of the Orchid Family. Portland, OR: Dioscorides; 1993. [Google Scholar]
- 52.Rasmussen H N. Terrestrial Orchids: From Seed to Mycotrophic Plant. New York: Cambridge Univ. Press; 1995. [Google Scholar]
- 53.Taylor A F S, Alexander I J. Mycol Res. 1991;95:381–384. [Google Scholar]
- 54.Miller S L, Koo C D, Molina R. Can J Bot. 1991;69:516–531. [Google Scholar]