Abstract
The various cardiac regions have specific action potential properties appropriate to their electrical specialization, resulting from a specific pattern of ion-channel functional expression. The present study addressed regionally defined differential ion-channel expression in the non-diseased human heart with a genomic approach. High-throughput real-time RT-PCR was used to quantify the expression patterns of 79 ion-channel subunit transcripts and related genes in atria, ventricular epicardium and endocardium, and Purkinje fibres isolated from 15 non-diseased human donor hearts. Two-way non-directed hierarchical clustering separated atria, Purkinje fibre and ventricular compartments, but did not show specific patterns for epicardium versus endocardium, nor left- versus right-sided chambers. Genes that characterized the atria (versus ventricles) included Cx40, Kv1.5 and Kir3.1 as expected, but also Cav1.3, Cav3.1, Cavα2δ2, Navβ1, TWIK1, TASK1 and HCN4. Only Kir2.1, RyR2, phospholamban and Kv1.4 showed higher expression in the ventricles. The Purkinje fibre expression-portrait (versus ventricle) included stronger expression of Cx40, Kv4.3, Kir3.1, TWIK1, HCN4, ClC6 and CALM1, along with weaker expression of mRNA encoding Cx43, Kir2.1, KChIP2, the pumps/exchangers Na+,K+-ATPase, NCX1, SERCA2, and the Ca2+-handling proteins RYR2 and CASQ2. Transcripts that were more strongly expressed in epicardium (versus endocardium) included Cav1.2, KChIP2, SERCA2, CALM3 and calcineurin-α. Nav1.5 and Navβ1 were more strongly expressed in the endocardium. For selected genes, RT-PCR data were confirmed at the protein level. This is the first report of the global portrait of regional ion-channel subunit-gene expression in the non-diseased human heart. Our data point to significant regionally determined ion-channel expression differences, with potentially important implications for understanding regional electrophysiology, arrhythmia mechanisms, and responses to ion-channel blocking drugs. Concordance with previous functional studies suggests that regional regulation of cardiac ion-current expression may be primarily transcriptional.
Cardiac function depends on the appropriate timing of contraction in each region, as well as on an appropriate beating rate. The various cardiac regions are characterized by specific action potential (AP) shapes, resulting from regionally defined complements of ionic currents (Schram et al. 2002). For example, it is well known that the density of the inward-rectifier K+-current IK1 is larger in ventricles than atria, whereas the density of the acetylcholine-activated K+-current IKACh is larger in atria (Schram et al. 2002). Accordingly, transcripts for the inwardly rectifying K+-channel subunit Kir2.1 (mediating IK1) are more strongly expressed in the ventricle (Dhamoon et al. 2004), whereas transcripts for Kir3.1 (encoding IKACh channels) are predominantly expressed in the atria (DePaoli et al. 1994). A higher density of the transient outward current, Ito, is characteristic of epicardium versus endocardium, in parallel with stronger expression of KChIP2 transcripts in the epicardium (Rosati et al. 2001). In addition to its role in normal cardiac electrical functioning, electrical heterogeneity also contributes to a variety of cardiac arrhythmia mechanisms (Schram et al. 2002; Antzelevitch, 2004).
To date, however, studies of ion-transporter subunit distribution within specific regions of the human heart have been limited to a small number of candidate subunits believed to be of importance. Additionally, limited access to healthy human cardiac tissue samples has forced most investigators to work with explanted cardiac-transplant recipient hearts, generally strongly affected by heart disease. Recent progress in genomics, particularly for mouse and man, has led to the identification of a detailed repertoire of ion-channel genes. In parallel, recently developed high-throughput quantitative molecular biology techniques permit simultaneous measurement of the expression of up to hundreds of genes, thereby permitting the creation of a global portrait of ion-channel expression (Marionneau et al. 2005). In the present study, we used a high-throughput quantitative approach to assess regionally defined expression of genes encoding ion-channel and transporter subunits in the non-diseased human heart.
Methods
Human tissue samples
Non-diseased human hearts that were technically unusable for transplantation (based on logistical, not patient-related, considerations) were obtained from organ donors. Before cardiac explantation, organ donor patients did not receive medication except dobutamine, furosemide and plasma expanders. The investigations conform to the principles outlined in the Declaration of Helsinki of the World Medical Association. All experimental protocols were approved by the Ethical Review Board of the Medical Center of the University of Szeged (No. 51-57/1997 OEJ). Human cardiac tissue was dissected from 15 donors (44 ± 12 years; mean ±s.d.; min 18; max 65; 7 women and 8 men) and stored in cardioplegic solution at 4°C for 4–8 h before being frozen in liquid nitrogen.
Left and right atrial (LA and RA) samples were dissected from the tip of the appendages. Transmural left and right ventricular (LV and RV) tissue samples were consistently obtained from the basal region of the heart. Selective left and right epicardial (Lepi and Repi) and endocardial (Lendo and Rendo) tissues were obtained by cutting 1 mm thick slices from the epicardial and endocardial surfaces of the base, respectively (Zicha et al. 2004). Purkinje fibre mRNA was extracted from false tendons dissected from the ventricles.
Comparisons of gene expression between cardiac compartments were conducted within sets of samples from individual patients (except for Purkinje fibres), thus controlling for interindividual variability. Seven matching samples were available for LA versus LV comparison, five for RA versus RV, seven for Lepi versus Lendo, eight for Repi versus Rendo, five for LA versus RA, five for LV versus RV, six for Lepi versus Repi and six for Lendo versus Rendo. Purkinje fibres were obtained from eight donors.
The sampled hearts were macroscopically and microscopically normal. Lack of hypertrophy was further confirmed by measuring the expression of atrial natriuretic factor (ANF) transcripts (data not shown).
RNA preparation
Total RNA from each cardiac sample was isolated and DNase-treated with the RNeasy Fibrous Tissue Mini Kit (Qiagen) following manufacturer's instructions. The quality of total RNA was assessed with polyacrylamide-gel microelectrophoresis (Agilent 2100 Bioanalyser) and by reverse transcriptase-polymerase chain reaction (RT-PCR). Lack of genomic DNA contamination was verified by PCR.
TaqMan real-time reverse transcriptase-polymerase chain reaction
The TaqMan Low-Density Array (TLDA, Applied Biosystems) technology was used in a two-step RT-PCR process, as previously reported (Marionneau et al. 2005). Briefly, first-strand cDNA was synthesized from 2 μg of total RNA using the High-Capacity cDNA Archive Kit (Applied Biosystems). PCR reactions were then performed on TLDA with the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). The 384 wells of each card were preloaded with 96 × 4 predesigned fluorogenic TaqMan probes and primers. Probes were labelled with the fluorescent reporter 6-carboxyfluorescein (FAM®, Applera Corp.) at the 5′-end and with non-fluorescent quencher on the 3′-end. The genes selected for their cardiac expression (listed in the online supplemental material) encode 72 ion channel α- and β-subunits, 17 Na+,K+-ATPase isoforms and proteins involved in calcium homeostasis, three hypertrophy markers (ANF, BNP, β-MHC) and four reference genes for normalization. Data were collected with instrument spectral compensation with Applied Biosystems SDS 2.1 software and analysed with the threshold cycle (Ct) relative-quantification method (Livak & Schmittgen, 2001). Ten genes (Cavγ4, Cavγ5, Cavγ6, CFTR, Kv1.1, Kv2.2, Kv3.1, Kv3.2, Kir1.1 and Nav1.4) were eliminated because their expression level frequently fell under the threshold for detection (> 50% undetermined data). Among the remaining genes, nine exhibited a maximum of 7% undetermined data and the remaining 70 exhibited no undetermined data point. We selected the hypoxanthine guanine phosphoribosyl transferase (HPRT) gene for data normalization, as the most uniformly distributed gene. The relative expression of each gene versus HPRT was calculated for each sample (ΔCt indicates normalized data).
Data analysis
Data were analysed in three independent ways. (i) Two-way hierarchical agglomerative clustering was applied to the gene-expression matrix consisting of grouped biopsies and the 79 genes with valid expression involved in electrical signalling. We applied average linkage clustering with Pearson correlation using the Cluster software (Eisen et al. 1998). Clusters were visualized using the Treeview software. (ii) For each compartment, the relative expression of each gene versus HPRT (2−ΔCt) was calculated and then averaged (data listed in the online supplemental material). (iii) For each gene and patient, the ratios between matching samples (atrium versus ventricle, epicardium versus endocardium, and left versus right compartments) were calculated and then averaged. For unmatched samples (Purkinje fibres versus RV) the ratio for each gene versus the mean RV was calculated and then averaged. Pair-wise comparisons between cardiac compartments were conducted on within-patient matched samples by Student's paired t test (P < 0.05 considered significant, n= 5–8). Unpaired t tests were used for unmatched samples.
Western blot
Proteins were extracted from four additional donor tissue samples, as previously described (Zicha et al. 2004). Proteins were fractionated on either 7.5% (Kv1.5, Kv4.3 and Cav1.3) or 10% (Cx40, KChIP2, and TWIK1) SDS-polyacrylamide gels and were transferred electrophoretically to Immobilon-P polyvinylidene fluoride membranes (Millipore). Membranes were blocked and incubated with antibodies as previously described (Zicha et al. 2004). Kv1.5 polyclonal antibody (1: 500 dilution) was purchased from Upstate; Cx40 (1: 1000) and Kv4.3 (1: 500) polyclonal antibodies were purchased from Chemicon Laboratories; Cav1.3 polyclonal antibody (1: 2000) was a kind gift from Dr Hiroshi Hibino (Osaka, Japan); TWIK1 polyclonal antibody (1: 3000) was a kind gift from Dr Jacques Barhanin (Valbonne, France); and KChIP2 monoclonal antibody (1: 1000) was a kind gift from Dr James Trimmer (Davis, USA). Protein loading was controlled by probing with anti-GAPDH antibody (Research Diagnostics). After washing, membranes were incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG secondary antibody (1: 10000, Santa Cruz). Bound antibodies were detected with Western Lightening Chemiluminescence Reagent Plus (Perkin-Elmer Life Sciences). For each antibody, time exposure was similar for membranes containing atrial and ventricle samples and for membranes containing epicardial and endocardial samples. Band densities were measured using Scion Image (Scion Corp.).
Results
The transcriptional expression levels of 72 α- and β-ion channel subunits and 17 Na+,K+-ATPase isoforms and proteins involved in calcium homeostasis were investigated by high-throughput real-time RT-PCR. For 10 genes, expression was undetectable after 40 cycles of PCR amplification in any cardiac compartment. These were excluded from further analysis. All fluorescent probes had 100% PCR efficiency (Applied Biosystems application note). This standardized method permits quantitative comparison of expression levels among cardiac compartments and among genes.
Hierarchical clustering of Purkinje fibre, atrial and ventricular tissues
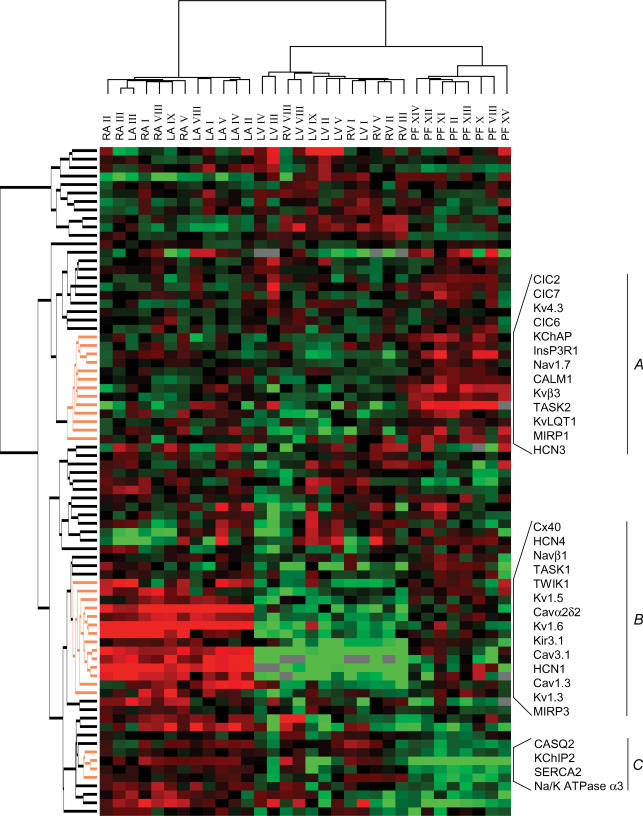
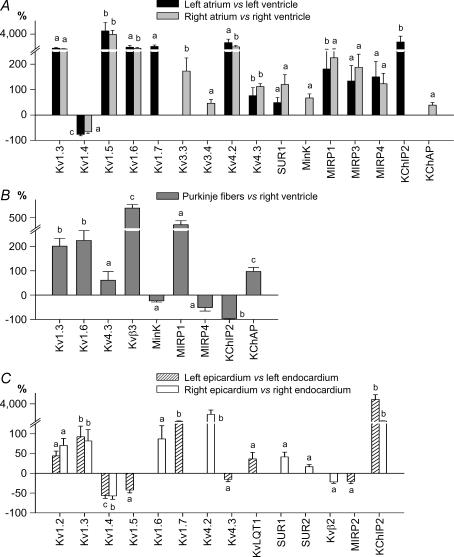
Two-way hierarchical clustering analysis groups samples according to gene-expression differences, with the most similar expression patterns located closest to each other and the most different patterns furthest apart. For visual appreciation, the samples are linked to a tree. The size of tree branches indicates the distance between clusters: larger branch sizes indicate greater separation among clusters. Hierarchical clustering analysis was applied to 32 atrial, ventricular and Purkinje fibre samples (Fig. 1). There was a first-order branch separation between atrial samples on one branch and ventricular and Purkinje fibre samples on the other, demonstrating full discrimination of atrial from ventricular compartments. In a second-order branch, Purkinje fibres clustered separately from ventricular myocardium. There was no overlap between primary compartments. That is, atrial, ventricular and Purkinje fibre samples clustered separately, indicating clearly different gene expression. The genes that were most relevant for clustering are indicated in Fig. 1. Group A contains genes that are more strongly expressed in Purkinje fibres versus working atrial or ventricular myocardium. These include Kv4.3, Kvβ3, MiRP1, KChAP, TASK2, ClC6, CALM1 and InsP3R1. Group B contains genes that are more abundant in atria and Purkinje fibres than in working ventricles, including, as expected for atria, Cx40, Kv1.5 and Kir3.1 (Schram et al. 2002), but also Cav1.3, Cav3.1, Cavα2δ2, Navβ1, MIRP3, TWIK1 and TASK1. No gene clusters with stronger ventricular-myocardial expression versus atrium were observed: i.e. atrial samples were distinct through over- rather than under-expression of ion-channel subunit genes. Group C contains genes with relatively weaker expression in Purkinje fibres compared to atria or working ventricular tissues, including KChIP2, SERCA2 and CASQ2. Within atrial and ventricular clusters, samples from the left- versus right-sided cavities did not separate.
Figure 1. Two way hierarchical agglomerative clustering applied to 79 genes (vertically) and to 5 left and right atria (LA and RA), 7 left and right ventricles (LV and RV) and 8 Purkinje fibre samples (horizontally).
The input consisted of the ratio for each patient and gene versus reference gene. Each gene is represented by a single row of coloured boxes and each patient by a single column. The entire gene clustering is shown on the left. Three selected gene clusters are shown on the right (A–C) containing relevant genes for Purkinje fibres and working myocardium discrimination (A, C), and for atrial–ventricular discrimination (B). Each colour patch in the map represents the gene expression level in one sample from one patient, with expression levels ranging from bright green (lowest) to bright red (highest). Missing values are coded as silver.
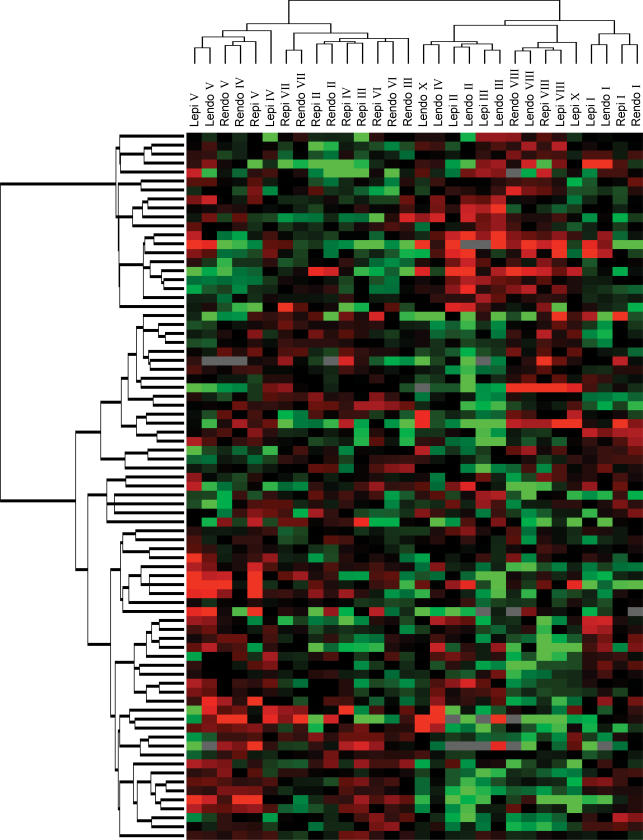
The same approach was applied to basal epicardial and endocardial samples. As shown in Fig. 2, epicardial and endocardial samples intermingled, with no distinct separation. Differences in ion-channel gene expression were thus not strong enough to discriminate epi- from the endocardial expression patterns. Since age and sex could affect cardiac electrophysiology by altering ion-channel gene expression, we performed hierarchical analysis with age and sex as covariates and saw no significant effects (data not shown). As for the tissue analysis shown in Fig. 1, there was no significant clustering of left versus right-sided transmural layer samples. The lack of left-right clustering in both Figs 1 and 2 indicates a lack of specific ion-channel gene signature for left- versus right-sided tissues.
Figure 2. Two-way hierarchical agglomerative clustering applied to 79 genes (vertically) and to 7 left epicardial (Lepi) and left endocardial (Lendo), and 8 right epicardial (Repi) and right endocardial (Rendo; horizontally) samples.
Same overall format as in Fig. 1.
Regional expression levels of ion-channel and transporter subunits
The method we used to quantify transcripts permits comparison of relative expression levels among genes (Tables 1–4). Among the Ca2+-channel genes, the L-type Ca2+-channel α-subunit Cav1.2 (α1c) was the most strongly expressed overall, whereas the other L-type α-subunit (Cav1.3) showed much lower expression (Table 1A). The T-type Ca2+-channel subunit Cav3.1 (α1G) was expressed predominantly in the atrium and Cav3.2 (α1H) showed lower-level expression. No significant expression of Cav3.3 (α1I) was detected. The expression of Ca2+-channel regulatory subunit transcripts (Cavα2δ1, Cavα2δ2, Cavβ2) was also strong.
Table 1.
Tissue-related expression of Ca2+ channels, Na+ channels and connexins
| Subunit | RA | RV | PF |
|---|---|---|---|
| A. Calcium channels | |||
| Cav1.2 | 647 ± 78 | 749 ± 116 | 455 ± 60 |
| Cav1.3 | 7.7 ± 1.8 | 0.3 ± 0.1 | 1.2 ± 0.2 |
| Cav3.1 | 20.6 ± 3.1 | 1.1 ± 0.3 | 4.3 ± 1.1 |
| Cav3.2 | 7.4 ± 0.5 | 11.1 ± 1.6 | 8.9 ± 1.5 |
| Cavα2δ1 | 234 ± 21 | 291 ± 24 | 283 ± 26 |
| Cavα2δ2 | 188 ± 40 | 13.6 ± 2.2 | 30.7 ± 4.2 |
| Cavβ2 | 88.3 ± 18.0 | 89.4 ± 13.4 | 99.3 ± 12.5 |
| B. Connexins | |||
| Cx40 | 608 ± 186 | 177 ± 38 | 389 ± 71 |
| Cx43 | 438 ± 60 | 898 ± 125 | 430 ± 53 |
| Cx45 | 240 ± 22 | 226 ± 26 | 125 ± 9 |
| C. Sodium channels | |||
| Nav1.1 | 1.5 ± 0.5 | 2.7 ± 0.7 | 1.5 ± 0.5 |
| Nav1.3 | 3.0 ± 0.7 | 7.1 ± 1.8 | 6.7 ± 1.2 |
| Nav1.5 | 588 ± 53 | 731 ± 99 | 501 ± 51 |
| Nav1.7 | 0.4 ± 0.1 | 1.4 ± 0.1 | 6.9 ± 0.7 |
| Nav2.1 | 201 ± 32 | 286 ± 38 | 237 ± 46 |
| Navβ1 | 622 ± 108 | 316 ± 63 | 397 ± 56 |
| Navβ2 | 54.2 ± 6.9 | 91.1 ± 20.0 | 70.0 ± 18.6 |
| Navβ3 | 6.2 ± 0.9 | 13.3 ± 1.3 | 12.3 ± 2.2 |
Abbreviations: RA, right atrium; RV, right ventricle; PF, Purkinje fibre. Units for all expression values are 2−ΔCtversus reference gene (×100) expressed as means ±s.e.m.
Table 4.
Tissue-related expression of pumps, exchangers and calcium handling subunits
| Subunit | RA | RV | PF |
|---|---|---|---|
| A. Pumps and exchangers | |||
| Na/K ATPase α1 | 1010 ± 232 | 1272 ± 201 | 1502 ± 265 |
| Na/K ATPase α3 | 4100 ± 167 | 5446 ± 526 | 1345 ± 195 |
| Na/K ATPase β1 | 1519 ± 97 | 2219 ± 188 | 1631 ± 138 |
| NCX1 | 1401 ± 55 | 2208 ± 278 | 1189 ± 139 |
| PMCA1 | 48.7 ± 6.2 | 54.7 ± 4.3 | 55.8 ± 5.7 |
| PMCA4 | 720 ± 76 | 874 ± 128 | 934 ± 146 |
| SERCA2 | 7801 ± 746 | 8696 ± 840 | 2687 ± 194 |
| SERCA3 | 79.2 ± 6.5 | 61.4 ± 4.4 | 81.4 ± 13.7 |
| B. Calcium handling and signalling | |||
| CALM1 | 123 ± 11 | 182 ± 8 | 478 ± 38 |
| CALM3 | 1017 ± 56 | 1349 ± 119 | 1299 ± 79 |
| CAM-PRP | 63.1 ± 6.3 | 86.9 ± 9.7 | 71.9 ± 9.2 |
| CASQ1 | 96.8 ± 26.3 | 130 ± 29 | 157 ± 35 |
| CASQ2 | 3524 ± 286 | 4147 ± 416 | 1916 ± 236 |
| InsP3R1 | 117 ± 12 | 153 ± 16 | 246 ± 26 |
| InsP3R3 | 69.5 ± 7.9 | 106 ± 19 | 131 ± 16 |
| PLB | 4703 ± 871 | 9519 ± 1008 | 7622 ± 1343 |
| RYR2 | 12940 ± 1352 | 25054 ± 2571 | 8612 ± 397 |
Abbreviations: RA, right atrium; RV, right ventricle; PF, Purkinje fibre. Units for all expression values are 2−Δ Ctversus reference gene (×100) expressed as means ±s.e.m.
As expected, Cx43 was the predominant connexin subunit in the ventricle (Table 1B). Cx40 predominated in the atria, with significant expression in Purkinje fibres that was about equal that of Cx43. Cx45 transcripts were detectable in all tissues. Among the voltage-gated sodium channel α-subunits, Nav1.5 (SCN5A) exhibited the highest expression (Table 1C). Nav2.1 (SCN6A–SCN7A) transcripts were also abundant. Navβ1 was by far the most abundant β-subunit.
Within the Kv-channel α-subunit family, Kv1.5, Herg, Kv4.3 and KvLQT1 transcripts predominated (Table 2A). Kv1.5 transcripts were ∼45-fold more strongly expressed in atrial versus ventricular tissues. Among K+-channel β-subunits, KChIP2, MIRP3, SUR2, KChAP, MinK and MIRP2 were the most abundant mRNA species (Table 2B). KChIP2 exhibited the strongest intertissue variations, with very low-level expression in Purkinje fibres. SUR2 transcripts were much more abundant than SUR1.
Table 2.
Tissue-related expression of Kv channel α-subunits and K+ channel β-subunits
| Subunit | RA | RV | PF |
|---|---|---|---|
| A. Kv channel α-subunits | |||
| Herg | 379 ± 40 | 400 ± 36 | 403 ± 56 |
| Kv1.2 | 2.7 ± 0.4 | 4.1 ± 1.3 | 4.1 ± 1 |
| Kv1.3 | 4.1 ± 1.0 | 1.6 ± 0.3 | 3.6 ± 0.4 |
| Kv1.4 | 7.0 ± 1.8 | 34.0 ± 6.5 | 35.0 ± 3.8 |
| Kv1.5 | 652 ± 112 | 26.1 ± 5.3 | 37.2 ± 10.4 |
| Kv1.6 | 9.4 ± 0.7 | 3.5 ± 0.8 | 8.1 ± 1.0 |
| Kv1.7 | 19.3 ± 4.9 | 13.9 ± 5.7 | 15.0 ± 1.1 |
| Kv2.1 | 5.8 ± 3.1 | 8.9 ± 8.3 | 12.8 ± 1.4 |
| Kv3.3 | 2.9 ± 0.4 | 1.7 ± 0.5 | 0.8 ± 0.2 |
| Kv3.4 | 42.7 ± 2.3 | 30.0 ± 4.8 | 26.5 ± 2.4 |
| Kv4.1 | 7.0 ± 0.9 | 4.0 ± 1.1 | 9.5 ± 1.7 |
| Kv4.2 | 2.0 ± 0.7 | 1.5 ± 0.7 | 3.7 ± 1.0 |
| Kv4.3 | 148 ± 20 | 91.0 ± 14.8 | 181 ± 20 |
| KvLQT1 | 80.0 ± 9.8 | 116 ± 9 | 102 ± 5 |
| B. K+ channel β-subunits | |||
| KChAP | 44.3 ± 3.7 | 40.1 ± 1.4 | 66.8 ± 6.5 |
| KChIP2 | 611 ± 83 | 704 ± 195 | 13.5 ± 4.0 |
| Kvβ1 | 14.9 ± 3.3 | 25.3 ± 3.3 | 12.7 ± 1.8 |
| Kvβ2 | 22.8 ± 3.6 | 46.7 ± 4.6 | 82.9 ± 10.0 |
| Kvβ3 | 0.2 ± 0.1 | 0.2 ± 0.1 | 2.4 ± 0.3 |
| MinK | 28.2 ± 4.1 | 21.5 ± 1.7 | 12.0 ± 0.4 |
| MIRP1 | 2.3 ± 0.5 | 1.4 ± 0.4 | 3.0 ± 0.6 |
| MIRP2 | 23.8 ± 2.1 | 23.9 ± 1.7 | 35.0 ± 7.1 |
| MIRP3 | 57.5 ± 12.6 | 25.6 ± 4.5 | 34.5 ± 4.6 |
| MIRP4 | 3.3 ± 0.5 | 2.4 ± 0.4 | 0.8 ± 0.2 |
| SUR1 | 2.1 ± 0.3 | 1.7 ± 0.4 | 1.4 ± 0.3 |
| SUR2 | 55.3 ± 5.9 | 99.6 ± 20.2 | 134 ± 11 |
Abbreviations: RA, right atrium; RV, right ventricle; PF, Purkinje fibre. Units for all expression values are 2−Δ Ctversus reference gene (×100) expressed as means ±s.e.m.
Transcripts underlying the inwardly rectifying potassium current IK1 (Kir2.1, Kir2.2 and Kir2.3) were expressed in every cardiac region, whereas Kir3.1 (responsible for the muscarinic-gated K+-channel IKACh) predominated in the atria (Table 3A). Purkinje fibre expression of Kir3.1 was substantially greater than in ventricular muscle, consistent with the differential effects of muscarinic stimulation on Purkinje versus ventricular repolarization (Malfatto et al. 1996). Both Kir6.2 and Kir6.1 subunits were detectable and most strongly expressed in ventricles. Among the tandem two-pore domain K+-channels, TWIK1 was strongly expressed, particularly in atria, and TASK1 also showed atrial-selective expression.
Table 3.
Tissue-related expression of Kir channels, HCN channels and Cl− channels
| Subunit | RA | RV | PF |
|---|---|---|---|
| A. Kir channels | |||
| Kir2.1 | 49.8 ± 7.3 | 201 ± 26 | 72.6 ± 10.2 |
| Kir2.2 | 117 ± 25 | 125 ± 22 | 46.8 ± 6.6 |
| Kir2.3 | 213 ± 41 | 106 ± 26 | 61.6 ± 10.8 |
| Kir3.1 | 158 ± 24 | 3.9 ± 1.3 | 48.4 ± 9.2 |
| Kir3.4 | 39.2 ± 6.8 | 20.4 ± 33.4 | 24.6 ± 5.9 |
| Kir6.1 | 49.7 ± 7.6 | 158 ± 16 | 63.6 ± 9.8 |
| Kir6.2 | 125 ± 19 | 172 ± 40 | 55.9 ± 4.5 |
| TASK1 | 130 ± 34 | 8.9 ± 2.3 | 14.1 ± 2.1 |
| TASK2 | 7.0 ± 1.6 | 5.3 ± 0.6 | 9.8 ± 1.8 |
| TWIK1 | 745 ± 115 | 216 ± 31 | 350 ± 43 |
| B. HCN channels | |||
| HCN1 | 70.3 ± 9.6 | 0.4 ± 0.1 | 8.2 ± 3.1 |
| HCN2 | 159 ± 41 | 24.7 ± 6.0 | 11.4 ± 2.3 |
| HCN3 | 4.2 ± 0.7 | 5.7 ± 1.6 | 8.9 ± 1.2 |
| HCN4 | 214 ± 32 | 75.9 ± 17.9 | 98.2 ± 11.1 |
| C. Chloride channels | |||
| CIC2 | 10.7 ± 1.2 | 13.2 ± 1.1 | 18.6 ± 2.8 |
| CIC3 | 130 ± 11 | 153 ± 21 | 95.3 ± 4.0 |
| CIC6 | 132 ± 11 | 146 ± 12 | 175 ± 18 |
| CIC7 | 160 ± 19 | 183 ± 34 | 164 ± 20 |
Abbreviations: RA, right atrium; RV, right ventricle; PF, Purkinje fibre. Units for all expression values are 2−Δ Ctversus reference gene (×100) expressed as means ±s.e.m.
HCN4, HCN2 and HCN1 transcripts corresponding to the hyperpolarization-activated cyclic nucleotide-gated current (If), were detected in all tissues, with predominant atrial expression (Table 3B). Concerning Cl− channel transcripts (Table 3C), our data showed a rank order of expression ClC7 > ClC6 > ClC3 ≫ ClC2, without major tissue-based differences. As previously observed in the mouse (Marionneau et al. 2005), exchangers and intracellular Ca2+ handling proteins expressed at ∼10-times higher levels than ion-channel subunits (Table 4). The expression of SERCA2, RyR2 and phospholamban predominated over the other Ca2+-handling regulators.
Relative expression of atrium versus ventricle
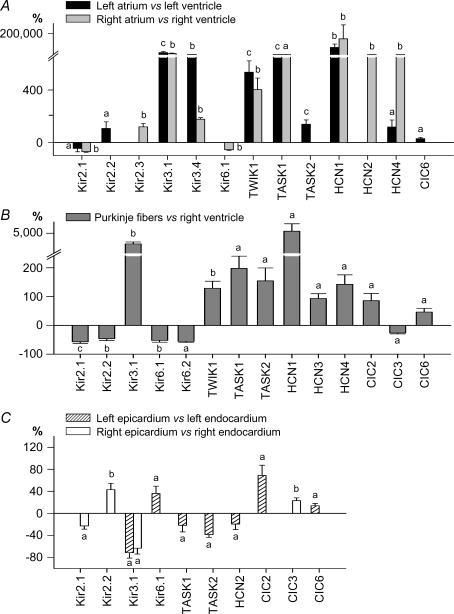
We next undertook in-depth comparisons of atrial versus ventricular expression levels. Because there is some consistency to overall function and conditions for right- versus left-sided chambers, we compared LA gene expression to that in LV, and RA expression to RV. Detailed comparisons between LA versus LV, as well as for RA versus RV, gene-expression are depicted in panels A of Figs 3–6. Only genes with significant differential expression between sets of matched atrial and ventricular samples from the same donors are shown.
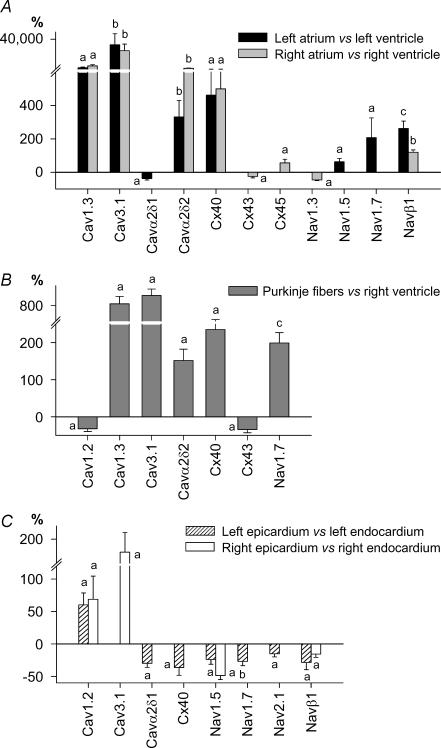
Figure 3. Expression profile of Ca2+ channels, connexins and Na+ channels in the human heart.
Differentially expressed genes in atria versus ventricles (A), in Purkinje fibres versus RV (B) and in epicardium versus endocardium (C). Data are expressed as percentage difference. a, P < 0.05; b, P < 0.01; and c, P < 0.001.
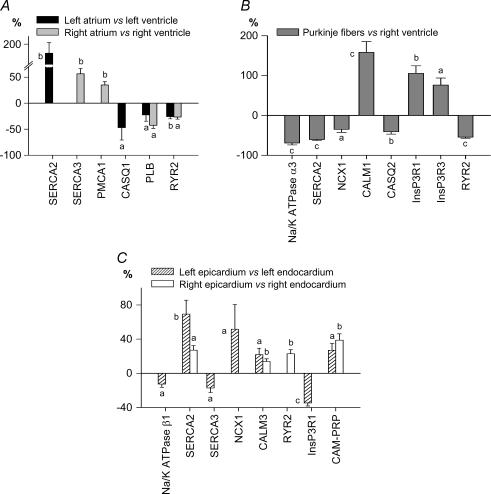
Figure 6. Expression profile of pumps, exchangers and Ca2+ handling proteins in the human heart.
Same presentation and symbols as in Fig. 3.
Among the cardiac intertissue comparisons, the atria exhibited the most specific signature, including not only the largest number of differential genes (37/79 versus ventricle), but also the largest differences (relative expression up to 200-fold versus ventricles) in accordance with the microarray analysis of Barth et al. (2005). All of the genes that were responsible for the atrial versus ventricular sample clustering on hierarchical analysis (Cav1.3, Cav3.1, Cavα2δ2, Cx40, Navβ1, Kv1.3, Kv1.5, Kv1.6, MIRP3, Kir3.1, TWIK1, TASK1, HCN1 and HCN4) were statistically significantly more strongly expressed at the individual-transcript level in both atria. Another subunit that was more strongly expressed in both atria than corresponding ventricles was Kv4.3. Kir3.4, Kv4.2, SUR1, MIRP1 and MIRP4 were also more strongly expressed in atria than ventricles, but their absolute expression level was very low in all regions. Only three genes (Kir2.1, phospholamban and RyR2) exhibiting relatively high overall expression were more weakly expressed in atria than ventricles, and only one with overall low-level expression (Kv1.4). Although in many cases atrial-ventricular differences were seen for both sides of the heart, for some genes, significant atrial-ventricular expression differences were seen only for one atrium.
The specific molecular signature of non-diseased human Purkinje fibres
There are no data available in the literature regarding ion-channel gene expression in human Purkinje fibres, and only one paper characterizing Purkinje fibre ion-channel subunit expression in an animal model (Han et al. 2002a). Panels B in Figs 3–6 shows the statistically significant differences that we observed between human Purkinje fibres and RV. Among the 38 differentially expressed genes, 23 (60%) were more strongly expressed in Purkinje fibres. Seventeen ion-channel genes differentially expressed in Purkinje fibres were also similarly differentially expressed in atrium versus ventricle, indicating similarities between Purkinje fibre and atrial portraits. These particularly include Cav1.3, Cav3.1, Cavα2δ2, Cx40, Kv4.3, Kv1.3, Kir3.1, MiRP1, TWIK1, TASK1, HCN1 and HCN4 (expression stronger than in ventricle) and Cx43, Kir2.1 and Kir6.1 (weaker than in ventricle). Differences specific to Purkinje fibres (i.e. not seen in atria) were much weaker expression of KChIP2 (−96%versus ventricle), weaker expression of Kir2.2 and Kir6.2 and stronger expression of Nav1.7, KChAP and HCN3. Differences in Ca2+-regulation genes also characterized Purkinje fibres versus ventricular myocardium, including stronger Purkinje fibre expression of CALM1, InsP3R1 and InsP3R3 and weaker expression of Na+,K+-ATPase α3, SERCA2, CASQ2, RyR2 and NCX1.
The specific molecular signature of non-diseased human epicardium versus endocardium
In a fashion similar to our analysis of atrial–ventricular differences, we analysed endocardium–epicardium differences in a chamber side-specific way, i.e. LV epicardium versus endocardium and RV epicardium versus endocardium, using matched samples from each heart. Only 10 genes were identified as statistically differing between epi- and endocardium for both right and left sides (panels C in Figs 3–6). Further, the relative differences between epicardium and endocardium were smaller compared to other interregional differences. As expected, KChIP2 exhibited the strongest transmural gradient, with ∼51-fold stronger expression in left epicardium versus left endocardium. For both Lepi and Repi, Cav1.2, SERCA2, CALM3 and calcineurin-α transcripts were concordantly more strongly expressed. Conversely, Nav1.5, Navβ1, Kv1.4 and Kir3.1 were more strongly expressed in the endocardium. Some genes showed differential epicardial-endocardial expression only on the right side (Cav3.1, Kir2.2, Kv4.2 and RYR2 were more strongly expressed in epicardium, Kvβ2 and Kir2.1 in endocardium). Similarly, KvLQT1, Kir6.1 and NCX1 were more strongly expressed in left epicardium versus endocardium, whereas Cavα2δ1, Cx40, Nav2.1, Kv1.5, TASK1, TASK2, HCN2, Na+,K+-ATPase α1 and InsP3R1 showed stronger expression in left endocardium.
Protein correlates
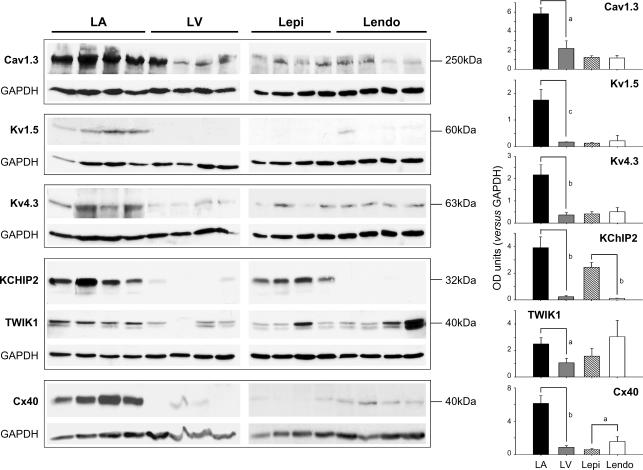
Western-blot experiments were conducted to relate protein expression to that of mRNA for selected ion-channel subunits. Figure 7 (left panels) shows original blots (4 samples for LA, LV, LV epicardium and LV endocardium all run on the same gel), as well as overall mean data (right panels). In agreement with transcriptional data, Cav1.3, Kv1.5, Kv4.3, KChIP2, TWIK1 and Cx40 were more strongly expressed in atria versus ventricular compartments. Cx40 and Kv1.5 expressed at very low levels in both ventricular endocardium and epicardium, whereas KChIP2 was more strongly expressed in epicardium. In general, protein expression was in good agreement with qRT-PCR results.
Figure 7. Western-blot analysis of key channel proteins.
Left, representative Western blots probed with anti-Cav1.3, anti-Kv1.5, anti-Kv4.3, anti-KChIP2, anti-TWIK1 and anti-Cx40 antibodies. GAPDH is shown as loading control. Expected molecular masses are indicated. Right, mean ±s.e.m. ion-channel subunit protein expression values normalized to that of GAPDH. a, P < 0.05; b, P < 0.01; and c, P < 0.001 for LA versus LV and for Lepi versus Lendo, n= 4 tissue samples per group.
Discussion
We have performed the first detailed analysis of regional ion-channel gene expression in the non-diseased human heart. Our study shows that human atria, ventricles and Purkinje fibres show distinct patterns of ion-channel and transporter expression. The intertissue differences are much greater than differences between left- and right-sided chambers and between endocardium and epicardium, which do not show distinct clustering on hierarchical analysis.
The challenge of ion-channel expression profiling and human cardiac spatial electrophysiological heterogeneity
The importance of regional heterogeneity in cardiac ion-channel function is well-recognized (Schram et al. 2002; Antzelevitch, 2004). Most previous studies have examined the molecular basis by quantifying the expression of limited numbers (generally 5 or fewer) of ion-channel genes and/or proteins in selected regions, e.g. right versus left atrium (Li et al. 2001), endocardium versus epicardium (Brahmajothi et al. 1999; Rosati et al. 2001; Zicha et al. 2004; Yamada et al. 2004) and atria versus ventricles (Melnyk et al. 2002). A problem with this approach is that only a limited number of subunits are assessed, usually including primarily subunits for which spatial heterogeneity is expected. Thus, the relative expression levels of these subunits compared to the overall spectrum of cardiac ion-channel subunits is unknown, subunits that may have important but unsuspected regional variability are not assessed, and a very limited range of ion channel function is examined. An emerging alternative approach is to establish a more global expression portrait with genomic methodology. Understanding ion-channel distribution in the human heart is clearly of great physiological and clinical importance, but presents additional difficulties in terms of the paucity of tissue sample availability, particularly for non-diseased human cardiac tissue. Genomic methodology has the additional advantage of obtaining a maximum amount of information from limited samples, like those available from non-diseased human hearts.
Figure 4. Expression profile of K+ channel α- and β-subunits in the human heart.
Same presentation and symbols as in Fig. 3.
Most previous large-scale studies of regional cardiac gene expression have used microarray-based profiling with pan-genomic arrays. Pan-genomic arrays are limited in their ability to detect differences in low-abundance gene products like those of most cardiac ion-channel subunits. In this study, we used high-throughput quantitative RT-PCR for a comprehensive panel of human cardiac ion channel and transporter subunit genes. This method has the advantages of being sensitive, specific and quantitative, and not requiring validation by other methods. Consequently, we were able to obtain a much more detailed regional ion-channel expression profile than previous studies. For example, Ellinghaus et al. (2005) described four human ion-channel genes with differential atrial–ventricular expression (Kv1.5, Kir2.1, TWIK1, TASK1) and Barth et al. (2005) described three such genes (Kir2.3, TWIK1, TWIK3) with the use of pan-genomic arrays, in contrast to the 24 we identified. Rosati et al. (2006) used pan-genomic arrays to study epicardial-endocardial gene-expression differences in rat hearts, identifying Nav1.5 and Kv4.2 as differentially expressed. We observed corresponding differences in Nav1.5 and KChIP2 (responsible for endocardial-epicardial Ito differences in mammalian hearts), but were also able to identify robust differential expression of nine other ion channel or transporter subunits.
Figure 5. Expression profile of Kir, HCN and Cl− channels in the human heart.
Same presentation and symbols as in Fig. 3.
Our approach differs from previous studies in that we analysed tissue compartments that have, to our knowledge, not previously been studied by wide-scale gene profiling (e.g. Purkinje fibres, right versus left sided atrium/ventricle and epicardium/endocardium). This is also to our knowledge the first study to apply gene-expression profiling simultaneously to such a large number of regional compartments, allowing us to compare the relative contributions of diverse regional factors within the same hearts. The use of apparently normal human hearts adds to the novelty and potential impact of our study.
Compatability with previously available information on regional ion-channel subunit expression variations and functional relevance
Among the differences that we observed, many are consistent with functionally relevant information in the previous literature and provide a useful confirmation of the validity of our approach. With respect to Ca2+-channel subunits, the predominant atrial expression of Cav1.3 has previously been reported in mouse, rabbit and human hearts (Mangoni et al. 2003; Marionneau et al. 2005; Qu et al. 2005). Among Na+-channel subunits, Nav1.5 and Navβ1 showed stronger endocardial versus epicardial expression, similar to a recent report in rat hearts (Rosati et al. 2006) and to observations that INa density is larger in rat and canine endocardium versus epicardium (Ashamalla et al. 2001; Ueda et al. 2004b).
With respect to K+-channel subunits, our findings are consistent with several known differences with great functional significance. Similar to Rosati et al. (2001, 2003), we found that KChIP2 is one of a small number of genes showing a strong epicardial–endocardial gradient, of great functional importance with respect to clinically relevant transmural repolarization gradients (Antzelevitch, 2004; Nabauer et al. 1996). The higher-level atrial versus ventricular expression levels of Kv1.5, TWIK1 and TASK1, and the lower atrial levels of Kir2.1, are consistent with previous observations of Ellinghaus et al. (2005). Kv1.5 encodes an important human atrial K+-channel that is an interesting potential target for new antiarrhythmic drug development (Nattel et al. 1999). TWIK1 and TASK1 encode two-pore background K+-channels with currently unclear but potentially substantial physiological importance (Duprat et al. 1997; Gaborit et al. 2005). Stronger ventricular Kir2.1 expression correlates with the more negative ventricular-myocyte resting potential (Schram et al. 2002) and the ventricular-selective arrhythmogenic consequences of Kir2.1 mutations in Andersen's syndrome (Plaster et al. 2001). The α-subunit Kv1.4 associated with the slow component, Itos, is more abundant in the endocardium, in agreement with studies showing larger Itos in endocardial cells (Kaab & Nabauer, 2001). Predominant atrial expression was previously found for HCN4 in humans and for HCN1 and HCN4 in the mouse (Ueda et al. 2004a; Marionneau et al. 2005), observations related to the pacemaking contribution of the If channels they encode (Thuringer et al. 1992; Michels et al. 2005).
The dominant epicardial expression of Cav1.2 is associated with preferential expression of RyR2, consistent with specific contractile properties of epicardium. Human epicardial cells display faster onset and peak of contraction than endocardial cells (Fulop et al. 2004). Epicardial cells also exhibit faster relaxation (Cordeiro et al. 2004). In agreement with these functional observations and previous findings (Prestle et al. 1999; Xiong et al. 2005), we observed that the sarcoplasmic-reticulum calcium-ATPase 2 (SERCA2) and Na+/Ca2+ exchanger responsible for cytoplasmic Ca2+ removal are predominantly expressed in the epicardium. Particularly strong inositol trisphosphate receptor (InsP3R) expression in Purkinje fibres is consistent with prior protein and in situ hybridization studies (Gorza et al. 1993), and may be important because of the emerging role of InsP3Rs in arrhythmias (Li et al. 2005).
It should be noted that not all of our findings were confirmatory. For example, the largest previous study of ion channel gene expression in cardiac Purkinje fibres noted lower-level expression of KvLQT1, HERG, KChIP2 and Cav1.2 mRNA, along with higher-level Kv3.4, Cav3.1, 3.2, 3.3 and HCN4, in canine Purkinje fibres compared to ventricular muscle (Han et al. 2002a). In addition, NCX1 protein was less strongly expressed in Purkinje fibres. In the present investigation, we observed lower-level transcript expression of KChIP2, Cav1.2 and NCX1, and higher-level Cav3.1 and HCN4, in human Purkinje fibres versus ventricular muscle. Our results confirm stronger HCN4 expression as a candidate mechanism for Purkinje fibre automaticity and weaker KChIP2 as an explanation for more slowly recovering Purkinje-fibre Ito. However, they leave open the mechanism for tetraethylammonium sensitivity of Purkinje Ito (previously suggested to be related to higher-level Kv3.4) and longer action potential duration (previously related to lower-level delayed-rectifier channel subunit expression). Rosati et al. (2007) recently reported several hundred-fold greater expression of Cav3.2 versus Cav3.1 in canine Purkinje fibres, whereas in the present study they were of the same order. The discrepancies may be due to species differences or to methodological issues. Because of the large number of observations in the present study, it would be impossible to catalogue all of the similarities and differences between the present findings and previous observations in the literature, but we have attempted to provide illustrative confirmatory observations of physiological significance as well as some potentially significant discrepancies.
Novel findings of potential functional significance
Because of the comprehensive and quantitative nature of our approach, a number of potentially significant and novel findings, many of them not predictable a priori, have emerged. They will be discussed in relation to specific tissue compartments and to overall regional ion-channel gene-expression profiles.
Atrium
Na+ current in human atrial cells has some specific inactivation properties not seen in ventricular tissue (Sakakibara et al. 1992). These may relate to the high-level expression and differential atrial distribution of the Navβ1 subunit (Makita et al. 1994) that we observed. We found high levels of Cavα2δ2 in the atria, which were ∼12- and 6-fold greater than in ventricle and Purkinje tissue, respectively. This subunit, which importantly modulates currents expressed by Cav1.2, 1.3 and 3.1 subunits (Gao et al. 2000), is therefore a likely contributor to atrial-specific properties of L- and T-type Ca2+-currents.
We have obtained the first detailed comparison of putative Ito-forming subunits (Kv1.4, Kv4.3, Kv4.2 and KChIP2) in human atria versus ventricles. Kv4.3, the predominant Itoα-subunit, was expressed significantly more strongly in atrium than ventricle, potentially accounting (at least in part) for previously described functional and pharmacological differences between atrial and ventricular Ito (Nattel et al. 2000; Varro et al. 1993).
Purkinje fibres
The largest mass of new information emerging from the present study concerns cardiac Purkinje fibres. There are no previous data in the literature regarding ion-channel expression in human cardiac Purkinje fibres. Furthermore, the available information regarding Purkinje fibre ion-channel function and subunit expression is extremely limited, despite the substantial significance of Purkinje tissue in cardiac electrical function and arrhythmias (Schram et al. 2002).
Purkinje fibre INa displays tetrodotoxin (TTX) sensitivity, which is not shared with ventricular INa (Carmeliet, 1987) and is currently unexplained. Neuronal Nav1.7 subunits are TTX sensitive (Klugbauer et al. 1995) and were expressed ∼5- to 10-fold more strongly in human cardiac Purkinje fibres versus right atrium and ventricle, providing a potential explanation.
Transcripts encoding a variety of proteins involved in Ca2+ handling or coupled functions, including the principal Na+,K+-ATPase isoforms α3 and β1, NCX1, SERCA2, CASQ2 and RYR2, were expressed at substantially lower (27–75%) levels in Purkinje fibre versus ventricular muscle tissue, compatible with the limited contractile function of Purkinje fibres (Schram et al. 2002). Purkinje fibres classically display enhanced sensitivity to digitalis toxicity, despite reduced ouabain binding (Rhee, 1981). The low-level Purkinje-fibre Na+,K+-ATPase expression that we observed is likely to account for this previously puzzling finding, since Na+,K+-ATPase is both the functional target and receptor for digitalis.
We found Kv4.3-subunit expression to be abundant in human Purkinje fibres, whereas KChIP2 was sparse, consistent with the very slow recovery of human Purkinje fibre Ito from inactivation (Han et al. 2002b). Conversely, KChAP is more strongly expressed in Purkinje fibres than ventricular muscle and predominates over KChIP2. KChAP acts as a chaperone to enhance expression and current density of a subset of Kv channels, including Kv4.3 (Kuryshev et al. 2000). KChAP has not yet been shown to have a physiological role in ventricular tissue, but our results suggest the possibility that KChAP may participate in Purkinje fibre Ito. Consistent with previous findings in dogs (Pourrier et al. 2003), we found stronger expression of MiRP1 in Purkinje fibres than in ventricles, pointing to a possible role of Purkinje tissue in human arrhythmia syndromes associated with MiRP1 mutations (Abbott et al. 1999). Functional evidence suggests the absence of Kv1.5-based currents in human Purkinje fibres (Han et al. 2002b), paralleling previous observations for human ventricular muscle (Feng et al. 1997). Our observation of ∼20-fold lower expression levels of Kv1.5 in human Purkinje tissue versus atrium provides the first molecular data indicating atrial-selective expression of this subunit compared to Purkinje tissue. This finding is potentially important in view of the therapeutic interest of atrial-selective ion-channel blockade in atrial fibrillation (Goldstein & Stambler, 2005) and continuing controversy regarding the atrial-selective expression of Kv1.5-based currents (Sridhar et al. 2006). Lower Purkinje Kir2.x expression versus ventricular muscle is consistent with the smaller IK1 observed in Purkinje fibres (Verkerk et al. 1999). An acetylcholine-dependent K+-current (IKACh) has been described in rabbit Purkinje fibres (Carmeliet & Mubagwa, 1986) and stimulation of muscarinic receptors in canine Purkinje fibres reduces action potential duration (Malfatto et al. 1996), in contrast to the lack of IKACh in ventricular muscle and consistent with our observation of substantially greater Kir3.1 expression in Purkinje tissue.
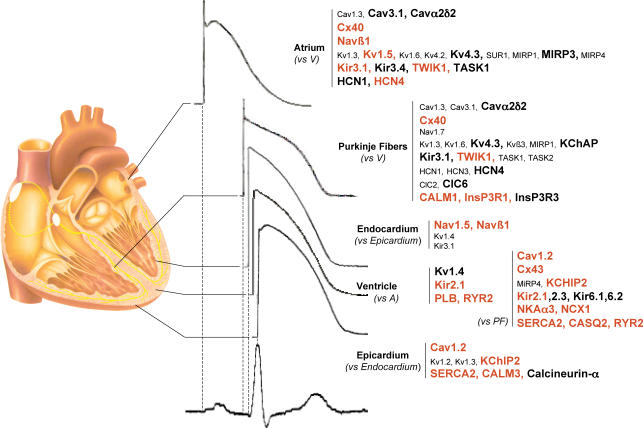
Overall interregional differences
One advantage of the type of broadly based expression analysis approach that we took is that it allows for the development of portraits of regionally specific gene expression. Figure 8 summarizes a number of regional expression portraits of interest. The absolute expression level of more strongly expressed genes in the region of interest is provided by colour coding: genes expressed > 20-fold the reference gene (HPRT) are indicated in bold and genes expressed at > 185 times the reference gene level are shown in red. Atrial-selective gene expression (versus ventricular) is shown for only the most robust differences, i.e. genes showing statistically significant differential expression in the same direction for both RA versus RV and LA versus LV. Of note, the atrial gene-expression signature shares many properties with that of Purkinje fibres. Of the 20 genes that are significantly different for atrium versus ventricle, 12 (60%) show the same directional statistically significant difference for Purkinje tissue versus ventricle. The basis for this similarity in ion-channel subunit expression between atrium and Purkinje fibres is unclear, but may relate to the strong contractile phenotype of working ventricular myocardium. One general difference between atrium and Purkinje tissue pertains to Ca2+-homeostasis proteins, which are much more strongly expressed in the atrium (as in the ventricles) compared to Purkinje tissue, possibly reflecting the primarily electrical (and non-contractile) specialization of the Purkinje system. The second difference lies in the weaker expression of Na+,K+-ATPase and stronger expression of inositol trisphosphate receptor genes in Purkinje fibres, which may be related to some of their specific arrhythmic properties.
Figure 8. Schematic diagram of the heart illustrating statistically significant gene-expression differences that create an ion-channel ‘signature’ for various cardiac regions.
The absolute expression levels of genes that are significantly more strongly expressed in a region of interest relative to the reference region indicated are provided by colour coding: genes expressed > 20-fold the reference gene (HPRT) in the tissue/region of interest are indicated in bold and genes expressed at > 185 times the reference gene level are shown in bold red. Values shown are differences that were statistically significant for both right and left-sided comparisons for atrium versus ventricle and epicardium versus endocardium.
We did not observe statistically significant overall gene expression clustering between right and left sided chambers or between ventricular epicardium and endocardium. This finding suggests that tissue differences (i.e. atrium versus ventricle versus Purkinje fibres) are much stronger determinants of ion-channel subunit expression pattern than regional differences. We were careful to analyse atrial–ventricular and epicardial–endocardial differences with matched samples for right versus left sided chambers. To our knowledge, this is the first time that an extensive analysis of this type has been performed. Despite the overall lack of distinct gene clustering for endocardium versus epicardium, there were specific differences (illustrated in Fig. 8) that appeared to be robust in that they were statistically different and in the same direction for both right epicardium versus endocardium and left epicardium versus endocardium. The endocardial gene-expression pattern is marked by stronger Na+-channel subunit expression, potentially reflecting a need for more rapid endocardial conduction. That this difference is not due to contamination by subendocardial Purkinje tissue is apparent by virtue of the lack of differential expression of the same subunits for Purkinje tissue versus ventricle. The epicardial expression pattern is marked by enhanced expression of genes related to Ca2+ handling (Cav1.2 and SERCA2), Ca2+ signalling (calmodulin and calcineurin) and Ito (KChIP2), which is believed to be an important regulator of Ca2+ entry, Ca2+ signalling and contractility (Sah et al. 2003; Lebeche et al. 2004).
Study limitations
Transcripts do not necessarily translate into functional proteins and although transcriptional regulation of ion-channel expression is a key regulatory mechanism (Rosati & McKinnon, 2004), translational and post-translational processes are also important. We confirmed corresponding changes for selected proteins of interest, but no technology is currently available to explore on a large scale the expression of a collection of membrane proteins such as those corresponding to the 79 genes we studied. For Western blot analyses, we selected proteins for study in order to contrast atrial with ventricular and epicardial with endocardial expression. All of the subunits selected had evidence for substantial atrial–ventricular mRNA differences, and two of them (KChIP2 and Cx40) significant endocardial–epicardial differences. This allowed us to test the specific hypothesis that A–V and Epi–Endo differences apparent with RT-PCR are also reflected in protein differences. In addition, Cav1.3, Kv1.5 and Cx40 are all important in the pathogenesis of atrial fibrillation (the most important clinical atrial electrical rhythm disturbance). TWIK1 was very strongly expressed as determined by RT-PCR, suggesting possible functional importance despite poorly understood physiology, so we felt that it would be worthwhile to determine whether protein differences parallel those in mRNA, supporting the potential relevance of the mRNA data for TWIK1.
Another limitation relates to the location of sampling. Heterogeneous expression exists within the different parts of the atrium and the base and apex of the ventricle. However, it is encouraging to note that atrial–ventricular–Purkinje fibre differences were much greater than those between right- and left-sided cavities or endocardium versus epicardium, suggesting that intertissue differences are likely to be robust across within-tissue regions. Finally, targeting of ion channel proteins to subcellular compartments is crucial for their function and cannot be approached by our global analysis. Despite these limitations, genomics represents a powerful means to improve our understanding of the complex mechanisms used by heart muscle to adapt to different functional requirements in diverse cardiac tissues and regions.
We cannot totally exclude the contamination of endocardial samples by subendocardial Purkinje fibre cells. However, the mass of the thin subendocardial Purkinje tissue layer is relatively small compared to that of underlying ventricular muscle. More importantly, important contamination seems unlikely based on the gene expression data themselves. For example, Kv1.4, Nav1.5 and Navβ1 are more strongly expressed in endocardium than epicardium, but these subunits were in fact not significantly more strongly expressed in Purkinje fibres than in ventricular muscle. If contamination of subendocardial muscle samples by Purkinje tissue were to account for the greater expression of these subunits in endocardium, they would have had to be expressed more strongly in pure Purkinje fibre tissue (like false tendons) than in ventricular muscle.
We examined a limited range of cardiac regions. There are many interesting additional regional differences in electrophysiological function that would be appropriate subjects for detailed expression-profiling analysis, e.g. apex versus base versus septum, various right atrial regions with distinct electrical function (pectinate muscles, AV ring, crista terminalis, appendage), various left atrial regions (pulmonary vein ostia, free wall, appendage), Bachmann's bundle, the nodes, etc. Practical considerations (time constraints, financial resources, tissue availability, etc.) precluded their inclusion in the present study but they are certainly an appropriate subject for future investigation.
Conclusions
This work has elucidated the regional expression of a large number of ion-channel subunit genes in the non-diseased human heart. We found unique ion channel and transporter subunit transcript expression signatures among atrial, working ventricle and Purkinje fibre compartments, in contrast to epicardial–endocardial and right–left chamber differences, which showed no distinct pattern on hierarchical clustering. This first detailed analysis that specifically and simultaneously characterizes the expression of a broad range of ion channel and transporter genes in multiple tissue compartments of the non-diseased human heart has significant potential implications for understanding regional electrophysiology, arrhythmia mechanisms, and responses to ion-channel blocking drugs. In addition, this study lays the groundwork for future investigations seeking to elucidate how ionic remodelling caused by heart disease alters the regional specificity of subunit expression and for studies of the functional importance of a number of currently poorly characterized subunits that show strong and/or highly differential regional expression.
Acknowledgments
This work was supported by grants from Ouest Genopole, Ministère Français de la Recherche (ACI), the Leducq Foundation, Canadian Institutes of Health Research, Bio-37 KPI and OTKA NI-61902. S.D. holds a tenured position from the French CNRS. The authors wish to thank Miklos Bitay, Jean-Marie Heslan and Marie-Jo Louerat for their excellent technical assistance, and France Thériault for expert secretarial assistance in manuscript preparation.
Supplemental material
Online supplemental material for this paper can be accessed at:
http://jp.physoc.org/cgi/content/full/jphysiol.2006.126714/DC1 and http://www.blackwell-synergy.com/doi/suppl/10.1113/jphysiol.2006.126714
References
- Abbott GW, Sesti F, Splawski I, Buck ME, Lehmann MH, Timothy KW, Keating MT, Goldstein SA. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97:175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- Antzelevitch C. Cellular basis and mechanism underlying normal and abnormal myocardial repolarization and arrhythmogenesis. Ann Med. 2004;36:5–14. doi: 10.1080/17431380410032553. [DOI] [PubMed] [Google Scholar]
- Ashamalla SM, Navarro D, Ward CA. Gradient of sodium current across the left ventricular wall of adult rat hearts. J Physiol. 2001;536:439–443. doi: 10.1111/j.1469-7793.2001.0439c.xd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kaab S, Pfeufer A, Uberfuhr P, Dugas M, Steinbeck G, Nabauer M. Functional profiling of human atrial and ventricular gene expression. Pflugers Arch. 2005;450:201–208. doi: 10.1007/s00424-005-1404-8. [DOI] [PubMed] [Google Scholar]
- Brahmajothi MV, Campbell DL, Rasmusson RL, Morales MJ, Trimmer JS, Nerbonne JM, Strauss HC. Distinct transient outward potassium current (Ito) phenotypes and distribution of fast-inactivating potassium channel α subunits in ferret left ventricular myocytes. J Gen Physiol. 1999;113:581–600. doi: 10.1085/jgp.113.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet E. Voltage-dependent block by tetrodotoxin of the sodium channel in rabbit cardiac Purkinje fibers. Biophys J. 1987;51:109–114. doi: 10.1016/S0006-3495(87)83315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet E, Mubagwa K. Desensitization of the acetylcholine-induced increase of potassium conductance in rabbit cardiac Purkinje fibres. J Physiol. 1986;371:239–255. doi: 10.1113/jphysiol.1986.sp015971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro JM, Greene L, Heilmann C, Antzelevitch D, Antzelevitch C. Transmural heterogeneity of calcium activity and mechanical function in the canine left ventricle. Am J Physiol Heart Circ Physiol. 2004;286:H1471–H1479. doi: 10.1152/ajpheart.00748.2003. [DOI] [PubMed] [Google Scholar]
- DePaoli AM, Bell GI, Stoffel M. G protein-activated inwardly rectifying potassium channel (GIRK1/KGA) mRNA in adult rat heart and brain by in situ hybridization histochemistry. Mol Cell Neurosci. 1994;5:515–522. doi: 10.1006/mcne.1994.1063. [DOI] [PubMed] [Google Scholar]
- Dhamoon AS, Pandit SV, Sarmast F, Parisian KR, Guha P, Li Y, Bagwe S, Taffet SM, Anumonwo JM. Unique Kir2.x properties determine regional and species differences in the cardiac inward rectifier K+ current. Circ Res. 2004;94:1332–1339. doi: 10.1161/01.RES.0000128408.66946.67. [DOI] [PubMed] [Google Scholar]
- Duprat F, Lesage F, Fink M, Reyes R, Heurteaux C, Lazdunski M. TASK, a human background K+ channel to sense external pH variations near physiological pH. EMBO J. 1997;16:5464–5471. doi: 10.1093/emboj/16.17.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus P, Scheubel RJ, Dobrev D, Ravens U, Holtz J, Huetter J, Nielsch U, Morawietz H. Comparing the global mRNA expression profile of human atrial and ventricular myocardium with high-density oligonucleotide arrays. J Thorac Cardiovasc Surg. 2005;129:1383–1390. doi: 10.1016/j.jtcvs.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Feng J, Wible B, Li GR, Wang Z, Nattel S. Antisense oligodeoxynucleotides directed against Kv1.5 mRNA specifically inhibit ultrarapid delayed rectifier K+ current in cultured adult human atrial myocytes. Circ Res. 1997;80:572–579. doi: 10.1161/01.res.80.4.572. [DOI] [PubMed] [Google Scholar]
- Fulop L, Banyasz T, Magyar J, Szentandrassy N, Varro A, Nanasi PP. Reopening of L-type calcium channels in human ventricular myocytes during applied epicardial action potentials. Acta Physiol Scand. 2004;180:39–47. doi: 10.1046/j.0001-6772.2003.01223.x. [DOI] [PubMed] [Google Scholar]
- Gaborit N, Steenman M, Lamirault G, Le Meur N, Le Bouter S, Lande G, Leger J, Charpentier F, Christ T, Dobrev D, Escande D, Nattel S, Demolombe S. Human atrial ion channel and transporter subunit gene-expression remodeling associated with valvular heart disease and atrial fibrillation. Circulation. 2005;112:471–481. doi: 10.1161/CIRCULATIONAHA.104.506857. [DOI] [PubMed] [Google Scholar]
- Gao B, Sekido Y, Maximov A, Saad M, Forgacs E, Latif F, Wei MH, Lerman M, Lee JH, Perez-Reyes E, Bezprozvanny I, Minna JD. Functional properties of a new voltage-dependent calcium channel α2δ auxiliary subunit gene (CACNA2D2) J Biol Chem. 2000;275:12237–12242. doi: 10.1074/jbc.275.16.12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein RN, Stambler BS. New antiarrhythmic drugs for prevention of atrial fibrillation. Prog Cardiovasc Dis. 2005;48:193–208. doi: 10.1016/j.pcad.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Gorza L, Schiaffino S, Volpe P. Inositol 1,4,5-trisphosphate receptor in heart: evidence for its concentration in Purkinje myocytes of the conduction system. J Cell Biol. 1993;121:345–353. doi: 10.1083/jcb.121.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Bao W, Wang Z, Nattel S. Comparison of ion-channel subunit expression in canine cardiac Purkinje fibers and ventricular muscle. Circ Res. 2002a;91:790–797. doi: 10.1161/01.res.0000039534.18114.d9. [DOI] [PubMed] [Google Scholar]
- Han W, Zhang L, Schram G, Nattel S. Properties of potassium currents in Purkinje cells of failing human hearts. Am J Physiol Heart Circ Physiol. 2002b;283:H2495–H2503. doi: 10.1152/ajpheart.00389.2002. [DOI] [PubMed] [Google Scholar]
- Kaab S, Nabauer M. Diversity of ion channel expression in health and disease. Eur Heart J. 2001;3(Suppl):K31–K40. [Google Scholar]
- Klugbauer N, Lacinova L, Flockerzi V, Hofmann F. Structure and functional expression of a new member of the tetrodotoxin-sensitive voltage-activated sodium channel family from human neuroendocrine cells. EMBO J. 1995;14:1084–1090. doi: 10.1002/j.1460-2075.1995.tb07091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryshev YA, Gudz TI, Brown AM, Wible BA. KChAP as a chaperone for specific K+ channels. Am J Physiol Cell Physiol. 2000;278:C931–C941. doi: 10.1152/ajpcell.2000.278.5.C931. [DOI] [PubMed] [Google Scholar]
- Lebeche D, Kaprielian R, del Monte F, Tomaselli G, Gwathmey JK, Schwartz A, Hajjar RJ. In vivo cardiac gene transfer of Kv4.3 abrogates the hypertrophic response in rats after aortic stenosis. Circulation. 2004;110:3435–3443. doi: 10.1161/01.CIR.0000148176.33730.3F. [DOI] [PubMed] [Google Scholar]
- Li D, Zhang L, Kneller J, Nattel S. Potential ionic mechanism for repolarization differences between canine right and left atrium. Circ Res. 2001;88:1168–1175. doi: 10.1161/hh1101.091266. [DOI] [PubMed] [Google Scholar]
- Li X, Zima AV, Sheikh F, Blatter LA, Chen J. Endothelin-1-induced arrhythmogenic Ca2+ signaling is abolished in atrial myocytes of inositol-1,4,5-trisphosphate (IP3)-receptor type 2-deficient mice. Circ Res. 2005;96:1274–1281. doi: 10.1161/01.RES.0000172556.05576.4c. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Makita N, Bennett PB, Jr, George AL., Jr Voltage-gated Na+ channel β1 subunit mRNA expressed in adult human skeletal muscle, heart, and brain is encoded by a single gene. J Biol Chem. 1994;269:7571–7578. [PubMed] [Google Scholar]
- Malfatto G, Zaza A, Vanoli E, Schwartz PJ. Muscarinic effects on action potential duration and its rate dependence in canine Purkinje fibers. Pacing Clin Electrophysiol. 1996;19:2023–2026. doi: 10.1111/j.1540-8159.1996.tb03274.x. [DOI] [PubMed] [Google Scholar]
- Mangoni ME, Couette B, Bourinet E, Platzer J, Reimer D, Striessnig J, Nargeot J. Functional role of L-type Cav1.3 Ca2+ channels in cardiac pacemaker activity. Proc Natl Acad Sci U S A. 2003;100:5543–5548. doi: 10.1073/pnas.0935295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marionneau C, Couette B, Liu J, Li H, Mangoni ME, Nargeot J, Lei M, Escande D, Demolombe S. Specific pattern of ionic channel gene expression associated with pacemaker activity in the mouse heart. J Physiol. 2005;562:223–234. doi: 10.1113/jphysiol.2004.074047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk P, Zhang L, Shrier A, Nattel S. Differential distribution of Kir2.1 and Kir2.3 subunits in canine atrium and ventricle. Am J Physiol Heart Circ Physiol. 2002;283:H1123–H1133. doi: 10.1152/ajpheart.00934.2001. [DOI] [PubMed] [Google Scholar]
- Michels G, Er F, Khan I, Sudkamp M, Herzig S, Hoppe UC. Single-channel properties support a potential contribution of hyperpolarization-activated cyclic nucleotide-gated channels and If to cardiac arrythmias. Circulation. 2005;111:399–404. doi: 10.1161/01.CIR.0000153799.65783.3A. [DOI] [PubMed] [Google Scholar]
- Nabauer M, Beuckelmann DJ, Uberfuhr P, Steinbeck G. Regional differences in current density and rate-dependent properties of the transient outward current in subepicardial and subendocardial myocytes of human left ventricle. Circulation. 1996;93:168–177. doi: 10.1161/01.cir.93.1.168. [DOI] [PubMed] [Google Scholar]
- Nattel S, Matthews C, De Blasio F, Han W, Li D, Yue L. Dose-dependence of 4-aminopyridine plasma concentrations and electrophysiological effects in dogs: potential relevance to ionic mechanisms in vivo. Circulation. 2000;101:1179–1184. doi: 10.1161/01.cir.101.10.1179. [DOI] [PubMed] [Google Scholar]
- Nattel S, Yue L, Wang Z. Cardiac ultrarapid delayed rectifiers: a novel potassium current family of functional similarity and molecular diversity. Cell Physiol Biochem. 1999;9:217–226. doi: 10.1159/000016318. [DOI] [PubMed] [Google Scholar]
- Plaster NM, Tawil R, Tristani-Firouzi M, Canun S, Bendahhou S, Tsunoda A, Donaldson MR, Iannaccone ST, Brunt E, Barohn R, Clark J, Deymeer F, George AL, Jr, Fish FA, Hahn A, Nitu A, Ozdemir C, Serdaroglu P, Subramony SH, Wolfe G, Fu YH, Ptacek LJ. Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen's syndrome. Cell. 2001;105:511–519. doi: 10.1016/s0092-8674(01)00342-7. [DOI] [PubMed] [Google Scholar]
- Pourrier M, Schram G, Nattel S. Properties, expression and potential roles of cardiac K+ channel accessory subunits: MinK, MiRPs, KChIP, and KChAP. J Membr Biol. 2003;194:141–152. doi: 10.1007/s00232-003-2034-8. [DOI] [PubMed] [Google Scholar]
- Prestle J, Dieterich S, Preuss M, Bieligk U, Hasenfuss G. Heterogeneous transmural gene expression of calcium-handling proteins and natriuretic peptides in the failing human heart. Cardiovasc Res. 1999;43:323–331. doi: 10.1016/s0008-6363(99)00119-4. [DOI] [PubMed] [Google Scholar]
- Qu Y, Baroudi G, Yue Y, El-Sherif N, Boutjdir M. Localization and modulation of α1D (Cav1.3), L-type Ca channel by protein kinase A. Am J Physiol Heart Circ Physiol. 2005;288:H2123–H2130. doi: 10.1152/ajpheart.01023.2004. [DOI] [PubMed] [Google Scholar]
- Rhee HM. Accumulation of [3H]-ouabain in functionally different canine cardiac tissues: differential Rb+ uptake. Br J Pharmacol. 1981;73:81–86. doi: 10.1111/j.1476-5381.1981.tb16774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati B, Dun W, Hirose M, Boyden PA, McKinnon D. Molecular basis of the T- and L-type Ca2+ currents in canine Purkinje fibres. J Physiol. 2007;579:465–471. doi: 10.1113/jphysiol.2006.127480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati B, Grau F, McKinnon D. Regional variation in mRNA transcript abundance within the ventricular wall. J Mol Cell Cardiol. 2006;40:295–302. doi: 10.1016/j.yjmcc.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Rosati B, Grau F, Rodriguez S, Li H, Nerbonne JM, McKinnon D. Concordant expression of KChIP2 mRNA, protein and transient outward current throughout the canine ventricle. J Physiol. 2003;548:815–822. doi: 10.1113/jphysiol.2002.033704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati B, McKinnon D. Regulation of ion channel expression. Circ Res. 2004;94:874–883. doi: 10.1161/01.RES.0000124921.81025.1F. [DOI] [PubMed] [Google Scholar]
- Rosati B, Pan Z, Lypen S, Wang HS, Cohen I, Dixon JE, McKinnon D. Regulation of KChIP2 potassium channel β subunit gene expression underlies the gradient of transient outward current in canine and human ventricle. J Physiol. 2001;533:119–125. doi: 10.1111/j.1469-7793.2001.0119b.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah R, Ramirez RJ, Oudit GY, Gidrewicz D, Trivieri MG, Zobel C, Backx PH. Regulation of cardiac excitation-contraction coupling by action potential repolarization: role of the transient outward potassium current (Ito. J Physiol. 2003;546:5–18. doi: 10.1113/jphysiol.2002.026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara Y, Wasserstrom JA, Furukawa T, Jia H, Arentzen CE, Hartz RS, Singer DH. Characterization of the sodium current in single human atrial myocytes. Circ Res. 1992;71:535–546. doi: 10.1161/01.res.71.3.535. [DOI] [PubMed] [Google Scholar]
- Schram G, Pourrier M, Melnyk P, Nattel S. Differential distribution of cardiac ion channel expression as a basis for regional specialization in electrical function. Circ Res. 2002;90:939–950. doi: 10.1161/01.res.0000018627.89528.6f. [DOI] [PubMed] [Google Scholar]
- Sridhar A, Lacombe VA, Hamlin RL, Carnes CA. Ultrarapid delayed rectifier current (IKur) in canine ventricle: Constitutive role in repolarization and reverse use dependence. Heart Rhythm. 2006;3:S219–S220. [Google Scholar]
- Thuringer D, Lauribe P, Escande D. A hyperpolarization-activated inward current in human myocardial cells. J Mol Cell Cardiol. 1992;24:451–455. doi: 10.1016/0022-2828(92)91833-q. [DOI] [PubMed] [Google Scholar]
- Ueda K, Nakamura K, Hayashi T, Inagaki N, Takahashi M, Arimura T, Morita H, Higashiuesato Y, Hirano Y, Yasunami M, Takishita S, Yamashina A, Ohe T, Sunamori M, Hiraoka M, Kimura A. Functional characterization of a trafficking-defective HCN4 mutation, D553N, associated with cardiac arrhythmia. J Biol Chem. 2004a;279:27194–27198. doi: 10.1074/jbc.M311953200. [DOI] [PubMed] [Google Scholar]
- Ueda N, Zipes DP, Wu J. Functional and transmural modulation of M cell behavior in canine ventricular wall. Am J Physiol Heart Circ Physiol. 2004b;287:H2569–H2575. doi: 10.1152/ajpheart.00526.2004. [DOI] [PubMed] [Google Scholar]
- Varro A, Nanasi PP, Lathrop DA. Potassium currents in isolated human atrial and ventricular cardiocytes. Acta Physiol Scand. 1993;149:133–142. doi: 10.1111/j.1748-1716.1993.tb09605.x. [DOI] [PubMed] [Google Scholar]
- Verkerk AO, Veldkamp MW, Abbate F, Antoons G, Bouman LN, Ravesloot JH, van Ginneken AC. Two types of action potential configuration in single cardiac Purkinje cells of sheep. Am J Physiol Heart Circ Physiol. 1999;277:H1299–H1310. doi: 10.1152/ajpheart.1999.277.4.H1299. [DOI] [PubMed] [Google Scholar]
- Xiong W, Tian Y, DiSilvestre D, Tomaselli GF. Transmural heterogeneity of Na+–Ca2+ exchange: evidence for differential expression in normal and failing hearts. Circ Res. 2005;97:207–209. doi: 10.1161/01.RES.0000175935.08283.27. [DOI] [PubMed] [Google Scholar]
- Yamada KA, Kanter EM, Green KG, Saffitz JE. Transmural distribution of connexins in rodent hearts. J Cardiovasc Electrophysiol. 2004;15:710–705. doi: 10.1046/j.1540-8167.2004.03514.x. [DOI] [PubMed] [Google Scholar]
- Zicha S, Xiao L, Stafford S, Cha TJ, Han W, Varro A, Nattel S. Transmural expression of transient outward potassium current subunits in normal and failing canine and human hearts. J Physiol. 2004;561:735–748. doi: 10.1113/jphysiol.2004.075861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.