Abstract
It is often taken for granted that the human species is divided in rather homogeneous groups or races, among which biological differences are large. Studies of allele frequencies do not support this view, but they have not been sufficient to rule it out either. We analyzed human molecular diversity at 109 DNA markers, namely 30 microsatellite loci and 79 polymorphic restriction sites (restriction fragment length polymorphism loci) in 16 populations of the world. By partitioning genetic variances at three hierarchical levels of population subdivision, we found that differences between members of the same population account for 84.4% of the total, which is in excellent agreement with estimates based on allele frequencies of classic, protein polymorphisms. Genetic variation remains high even within small population groups. On the average, microsatellite and restriction fragment length polymorphism loci yield identical estimates. Differences among continents represent roughly 1/10 of human molecular diversity, which does not suggest that the racial subdivision of our species reflects any major discontinuity in our genome.
Keywords: genetic variation, microsatellite loci, restriction polymorphisms, racial classification
In 1972, Richard Lewontin analyzed allele frequencies at 15 protein loci and concluded that 85% of the overall human genetic diversity is represented by individual diversity within populations (1). Differences among seven racial groups accounted for less than 7% of the total. Nei and Roychoudhury reached a similar apportionment of genetic diversity among populations from three continents (2). Although these results were repeatedly confirmed by studies of protein markers (3–5), the idea that the human species is deeply subdivided into races has not disappeared (6, 7). Reasons for this include some perceived discontinuity among populations, usually reported for quantitative traits (6), and the possibility that protein markers, including blood groups, may not exhaustively describe genetic variation, leaving open the possibility that the undetected variation might show a different pattern.
In this study, we analyzed how DNA variation is distributed at 109 loci (Fig. 1). Based on the lengths and frequencies of the microsatellite alleles and on the frequencies of allelic variants at restriction fragment length polymorphism (RFLP) loci, we quantified the differences among members of the same population, among populations of the same continent, and among four or five geographical groups.
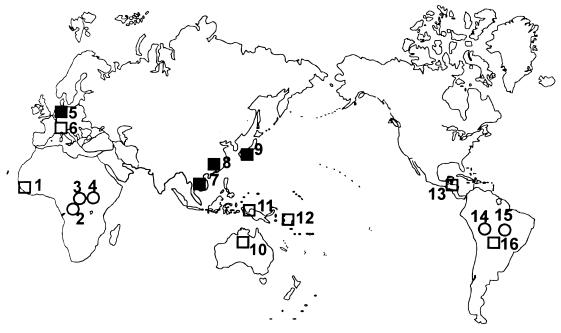
Figure 1.
Distribution of the samples considered. Samples and sample sizes for the three types of markers (microsatellites, RFLPs, RFLP–individual genotypes) are as follows: 1, Senegalese (0, 0, 28); 2, Mbuti pygmies from Zaire (10, 81, 28); 3, Lisongo from Central African Republic (9, 0, 0); 4, Biaka pygmies from Central African Republic (10, 67, 27); 5, North Europeans (15, 348, 28); 6, Northern Italians (14, 110, 0); 7, Cambodians born in Cambodia sampled in the San Francisco area (10, 124, 27); 8, Chinese born in China sampled in the San Francisco area (10, 110, 28); 9, Japanese born in Japan sampled in the San Francisco area (10, 74, 28); 10, Australians from the Northern Territory (10, 35, 15); 11, New Guineans (10, 127, 27); 12, Nasioi melanesians from Bougainville, Solomon Islands (10, 33, 21); 13, Maya from Yucatan peninsula, Mexico (10, 0, 14); 14, Karitiana from Rondonia, Brazil (10, 0, 0); 15, Suruì from Rondonia, Brazil (10, 0, 0); and 16, mixed Suruì and Karitiana (0, 50, 0). Different symbols refer to the expected levels of population complexity. Open circles, single villages or camps; open squares, groups of villages or localities; and solid squares, entire countries or subcontinental regions.
Materials and Methods
Three largely independent sets of genetic data were used in this study. The microsatellite database comprises individual allele lengths for 29 repeats and one tetranucleotide repeat of chromosomes 13 and 15. This is the set of data used by Bowcock et al. (8) from which we excluded nonhuman primates. The map distances between adjacent loci, except eight of them, are such that linkage disequilibrium can hardly be considered a major disturbing factor. Fourteen populations are included, for an overall sample size of 148. However, for no marker was a complete set of 296 chromosomes available. The missing values never exceed 10% at any given locus, and in no case were they replaced by interpolated data.
The RFLP database includes frequencies of the alternative alleles (presence or absence of cut) at 79 autosomal loci in 1109 individuals from 10 populations on 4 continents (extended data set). For 321 individuals from 12 populations on 5 continents, we also had full individual multilocus genotypes at 16 loci (reduced data set). Both data sets came from the analysis of cell lines that were used in a variety of other studies, in which sampling procedures are described in detail (9–12). As usual with molecular data, sample sizes were small. On the other hand, previous results show that the number of markers considered, rather than sample sizes, is crucial for population discrimination (13). Therefore, if some bias exists in the data of this study, the genetic differences between populations will tend to be overly emphasized.
A nonparametric method, analysis of molecular variance (1, 14), was used for hierarchically partitioning genetic diversity. At each locus, each individual allele was compared: (i) with the other alleles of the same sample; (ii) with the alleles of the other samples within the same continent; and (iii) with all of the alleles from other continents. This procedure was repeated independently for the multilocus genotypes of the reduced RFLP data set. The genetic variances thus computed reflected differences in allele frequencies and (for microsatellites) lengths. With a total sample size of n individuals from G continents and P populations, analysis of molecular variance generated a partition of the overall variance into G-1 df for the variance among continents, P-G for that among populations within continent, and N-P for that among individuals within a population. The significance of the estimated variances was tested by randomly assigning individuals to populations (according to two different randomization schemes) or populations to continents and repeating the randomizations 1000 times, each time recalculating the relevant variance. The observed variances were finally compared with the empirical expected distributions thus obtained.
Results and Discussion
Diversity among individuals of the same population (Table 1) was significant at 28 of 30 microsatellite loci and at all RFLP loci. It explained, on the average, 84.5% of the overall microsatellite variance and 83.6–84.5% of the overall RFLP variance (for the reduced and the extended data sets, respectively). Populations of the same continent tended to resemble each other; at the microsatellite level, their differences accounted only for 5% of diversity, reaching significance at nine loci; for RFLP loci, although significant in 54 cases, the comparisons between samples of the same continent accounted for less than 4% of the total molecular variance. Differences among continents accounted for the remaining fraction of variance, i.e., between 8 and 11.7%, and were significant at 12 microsatellite loci and 50 RFLP loci.
Table 1.
Analysis of molecular variance for 109 DNA loci
| Locus (probe name) | Variance components, %
|
||
|---|---|---|---|
| Within samples (P) | Among samples within continent (P) | Among continents (P) | |
| D13s270 (084xc5) | 92.8 (0.002) | 10.5 (0.018) | −3.3 (0.761) |
| D13s126 (1303) | 88.1 (<0.002) | 3.0 (0.152) | 8.9 (0.060) |
| D13s119 (1310) | 81.8 (<0.002) | 12.8 (0.012) | 5.5 (0.166) |
| D13s118 (1312) | 92.5 (0.002) | 6.9 (0.070) | 0.6 (0.359) |
| D13s125 (1320) | 80.6 (<0.002) | 11.4 (0.026) | 8.0 (0.080) |
| D13s144 (1348) | 94.7 (0.016) | 3.5 (0.134) | 1.8 (0.275) |
| Locus unknown (1523) | 77.5 (<0.002) | 8.6 (0.032) | 13.9 (0.060) |
| ACTC–chr. 15 | 98.3 (0.220) | 1.3 (0.274) | −0.4 (0.408) |
| D15s171 (B11MW) | 87.9 (<0.002) | −0.4 (0.418) | 12.4 (0.002) |
| D15s169 (B22MW) | 91.7 (0.002) | 2.1 (0.180) | 6.2 (0.042) |
| D13s133 (CA006) | 81.5 (<0.002) | 6.9 (0.054) | 11.5 (0.062) |
| D13s137 (CA010) | 90.5 (<0.002) | 4.3 (0.148) | 5.2 (0.126) |
| D13s227 (CA20) | 84.7 (<0.002) | −2.3 (0.744) | 17.6 (<0.002) |
| FES–chr. 15 | 97.2 (0.128) | −0.3 (0.390) | 3.1 (0.090) |
| GABRB3–chr. 15 | 91.5 (0.004) | 7.6 (0.040) | 0.9 (0.388) |
| D13s192 (HKCA3) | 87.7 (<0.002) | 3.3 (0.180) | 9.0 (0.002) |
| D13s193 (HKCA5) | 79.2 (<0.002) | −1.2 (0.564) | 22.0 (<0.002) |
| LIPC (HLIP) | 94.5 (0.030) | −3.5 (0.922) | 9.0 (<0.002) |
| D15s98 (MS112) | 93.4 (0.006) | 7.8 (0.064) | −1.1 (0.596) |
| D15s97 (MS14) | 89.6 (<0.002) | 3.3 (0.146) | 7.1 (0.070) |
| D15s100 (MS164) | 58.3 (<0.002) | 6.2 (0.050) | 35.5 (0.004) |
| D15s101 (MS178) | 54.4 (<0.002) | 2.9 (0.102) | 42.7 (0.002) |
| D13s115 (MS34) | 83.6 (<0.002) | 6.8 (0.058) | 9.7 (0.068) |
| D15s108 (Mfd102) | 73.5 (<0.002) | 2.7 (0.200) | 23.9 (0.012) |
| D13s71 (Mfd44) | 94.6 (0.002) | 5.2 (0.100) | 0.2 (0.466) |
| D15s95 (MX8) | 79.1 (<0.002) | 17.0 (0.006) | 3.9 (0.285) |
| D15s102 (N130) | 70.3 (<0.002) | 12.5 (0.008) | 17.2 (0.036) |
| D15s117 (PCR21) | 83.0 (<0.002) | 16.4 (0.002) | 0.6 (0.444) |
| D15s148 (PCR22) | 77.4 (<0.002) | 4.3 (0.098) | 18.3 (0.010) |
| D15s11 (P4-3R) | 81.6 (<0.002) | 8.4 (0.028) | 10.0 (0.042) |
| Average 30 microsatellite loci | 84.5 | 5.5 | 10.0 |
| 16 RFLP loci* | 83.6 (<0.001) | 8.4 (<0.001) | 8.0 (<0.050) |
| Average 79 RFLP loci† | 84.5 | 3.9 | 11.7 |
P is the fraction of randomization tests that gave variances equal to or higher than the observed one; significant values are in bold type. chr., chromosome.
Reduced database: 321 individuals, 5 continents.
Extended database: 1109 individuals, 4 continents.
The results of this study of DNA and the results of comparable analyses of protein polymorphism are remarkably similar (Table 2). The similarity of within-population diversity estimates in our three DNA data sets may have been increased by the fact that many individuals were typed for both microsatellites and RFLPs; therefore, the results obtained for the two kinds of markers may not be completely independent. Also, this study was based on a limited number of small samples; wider assemblages of data and a more regular distribution in space may somewhat modify the picture. However, if the geographic dispersion of samples distorted our estimates, it was by enhancing diversity among populations and continents and not within populations. Thus, our results suggest that at least four-fifths of human genetic variation reflects individual differences, no matter whether the variation is inferred from allele frequencies for moderately polymorphic protein markers or from allele lengths and frequencies at highly polymorphic DNA loci.
Table 2.
A summary of the results of this study and of previous comparable studies
| Polymorphism | Loci, n | Groups, n | Average variance components, %
|
||
|---|---|---|---|---|---|
| Within samples | Among samples within groups | Among groups | |||
| Protein (Lewontin 1972; ref. 1) | 17 | 7 | 85.4 | 8.3 | 6.3 |
| Protein (Latter 1980; ref. 3)* | 18 | 6 | 83.8–87.0 | 5.5–6.6 | 7.5–10.4 |
| Protein (Ryman et al. 1983; ref. 5)† | 25 | 3 | 86.0 | 2.8 | 11.2 |
| DNA (this study)‡ | 109 | 4–5 | 84.4 | 4.7 | 10.8 |
Latter used three different statistical techniques.
The study by Ryman et al. incorporates the data of ref. 4.
Average of three data sets weighted for the number of loci.
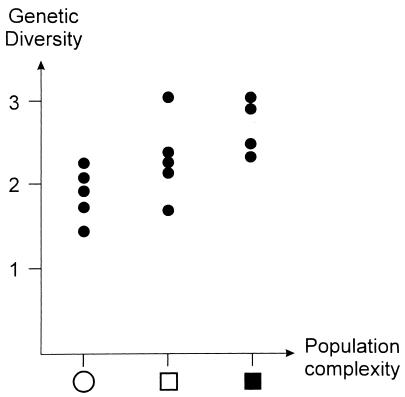
A different class of problems may have arisen from the fact that the populations examined in our study have different complexities, ranging from a camp of a few dozens of hunter–gatherers to a large country, such as China, or a wide region, such as Northern Europe. Might the latter samples, presumably heterogeneous, have inflated artificially our overall estimates of within-population variances? One way to answer is to see whether small isolated groups also show greater DNA homogeneity. For that purpose, we classified the populations studied into three groups, from small communities to very large regions (see legend to Fig. 1). We were aware that such a subdivision is somewhat arbitrary, but we were not using it for any exact calculation. At the microsatellite loci, genetic variation among individuals of the same population increases regularly with the population’s complexity, from 1.44 differences in repeat number for Suruì of Brazil to three or more for Cambodians, Italians, and Northern Europeans (averaged across the 30 microsatellite loci; Fig. 2). Although the latter samples came from large subcontinental regions, even small populations retained a substantial fraction of the global human variation. It seems reasonable to conclude that our large estimates of within-population diversity do not simply reflect the presence in this study of samples coming from large or ill-defined populations. In addition, the limited differences among spatially distant populations strongly suggest that groups occupying widely different geographical areas and ecological niches must have separated recently (15), as the often cited scenario of expansion of modern humans out of Africa suggests (16–19).
Figure 2.
Within-population diversity, i.e., average pairwise length differences at 30 microsatellite loci (y axis), as a function of population complexity (x axis). For symbols summarizing population complexity, see the legend to Fig. 1.
Implications for the Existence of Races in Humans
But what do these results imply for the race concept? Although no consensus has ever been reached on how many races exist in our species, with proposed figures ranging from 3 to 200 (20), in general a species is divided in races when it can be regarded as an essentially discontinuous set of individuals (21). Studies on a limited number of populations, like ours, cannot exclude that there are true discontinuities in the distribution of some genetic markers all over the world. However, only for one of the 109 loci studied was the within-population component of variance less than 50% of the total. If loci showing a discontinuous distribution across continents exist, they have not been observed in this study, and so the burden of the proof is now on the supporters of a biological basis for human racial classification.
Further support for the conclusions of this study comes from the observation that, almost without exception, gene frequencies form smooth clines over all continents (22). Zones of discontinuity in human gene frequency distributions are present, but the local gradients are so small that they can be identified only by simultaneously studying many loci using complex statistical techniques (23). In addition, such regions of relatively sharp genetic change do not surround large clusters of populations, on a continental or nearly continental scale. On the contrary, they occur irregularly, within continents and even within single countries (24, 25), often overlapping with geographic and linguistic barriers (26–29). Genetic enclaves seem to be mostly limited to islands. Probably any two populations compared at a sufficient number of loci may be shown to differ, as suggested by the fact that several variances among populations, although low in relative terms, are statistically significant in this study. However, this has little to do with the subdivision of the human population into a small number of clearly distinct, racial or continental, groups. The existence of such broad groups is not supported by the present analysis of DNA.
Even with the present, limited sample sizes, this study shows that previous findings of large individual diversity within populations were not due to the particular nature of the markers chosen, normally frequencies of protein variants at biallelic loci. Microsatellite loci are among the most polymorphic in the genome, yet they yield variance estimates in excellent agreement with the previous ones and with variances estimated from other DNA markers. The differences among human groups, even very distant ones and no matter whether the groups are defined on a racial or on a geographical basis, represent only a small fraction of the global genetic diversity of our species.
Acknowledgments
We thank Giorgio Bertorelle and Ayse Ergüven for their comments on an earlier version of this manuscript. This study was supported by Italian Consiglio Nazionale delle Ricerche Grant 95-0889 to G.B.
ABBREVIATION
- RFLP
restriction fragment length polymorphism
References
- 1.Lewontin R C. Evol Biol. 1972;6:381–398. [Google Scholar]
- 2.Nei M, Roychoudhury A K. Am J Hum Genet. 1974;26:421–443. [PMC free article] [PubMed] [Google Scholar]
- 3.Latter B D H. Am Nat. 1980;116:220–237. [Google Scholar]
- 4.Nei M, Roychoudhury A K. Mol Biol Evol. 1993;10:927–943. doi: 10.1093/oxfordjournals.molbev.a040059. [DOI] [PubMed] [Google Scholar]
- 5.Ryman N, Chakraborty R, Nei M. Hum Hered. 1983;33:93–102. doi: 10.1159/000153357. [DOI] [PubMed] [Google Scholar]
- 6.Harrison G A, Tanner J M, Pilbeam D R, Baker P T. Human Biology. 4th Ed. Oxford: Oxford Univ. Press; 1989. [Google Scholar]
- 7.Stein P L, Rowe B M. Physical Anthropology. 4th Ed. New York: McGraw–Hill; 1989. [Google Scholar]
- 8.Bowcock A M, Ruiz-Linares A, Tomfohrde J, Minch E, Cavalli-Sforza L L. Nature (London) 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 9.Bowcock A M, Bucci C, Hebert J M, Kidd J R, Kidd K K, Friedlaender J S, Cavalli-Sforza L L. Gene Geogr. 1987;1:47–64. [PubMed] [Google Scholar]
- 10.Bowcock A M, Hebert J M, Mountain J L, Kidd J R, Rogers J, Kidd K K, Cavalli-Sforza L L. Gene Geogr. 1991;5:151–173. [PubMed] [Google Scholar]
- 11.Lin A A, Hebert J M, Mountain J L, Cavalli-Sforza L L. Gene Geogr. 1994;8:191–214. [PubMed] [Google Scholar]
- 12.Poloni E S, Excoffier L, Mountain J L, Langaney A, Cavalli-Sforza L L. Ann Hum Genet. 1995;59:43–61. doi: 10.1111/j.1469-1809.1995.tb01605.x. [DOI] [PubMed] [Google Scholar]
- 13.Pamilo P, Nei M. Mol Biol Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- 14.Excoffier L, Smouse P E, Quattro J M. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein D B, Ruiz-Linares A, Cavalli-Sforza L L, Feldman M. Proc Natl Acad Sci USA. 1995;92:6723–6727. doi: 10.1073/pnas.92.15.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stringer C B, Andrews P. Science. 1988;239:1263–1268. doi: 10.1126/science.3125610. [DOI] [PubMed] [Google Scholar]
- 17.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson A C. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 18.Takahata N. Mol Biol Evol. 1993;10:2–22. doi: 10.1093/oxfordjournals.molbev.a039995. [DOI] [PubMed] [Google Scholar]
- 19.Mirazon Lahr M, Foley R. Evol Anthropol. 1994;3:48–60. [Google Scholar]
- 20.Armelagos G J. Am J Phys Anthropol. 1994;93:381–383. doi: 10.1002/ajpa.1330930308. [DOI] [PubMed] [Google Scholar]
- 21.Mayr E. Animal Species and Evolution. Cambridge, MA: Belknap; 1963. [Google Scholar]
- 22.Cavalli-Sforza L L, Menozzi P, Piazza A. History and Geography of Human Genes. Princeton: Princeton Univ. Press; 1994. [Google Scholar]
- 23.Barbujani G, Sokal R R. Proc Natl Acad Sci USA. 1990;87:1816–1819. doi: 10.1073/pnas.87.5.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbujani G, Sokal R R. Am J Hum Genet. 1991;48:398–411. [PMC free article] [PubMed] [Google Scholar]
- 25.Calafell F, Bertranpetit J. Am J Phys Anthropol. 1994;93:201–215. doi: 10.1002/ajpa.1330930205. [DOI] [PubMed] [Google Scholar]
- 26.Barbujani G, Nasidze I S, Whitehead G N. Hum Biol. 1994;66:639–668. [PubMed] [Google Scholar]
- 27.Sajantila A, Lahermo P, Anttinen T, Lukka M, Sistonen P, Savontaus M, Aula P, Beckman L, Tranebjaerg L, Gedde-Dahl T, Issel-Tarver L, DiRienzo A, Pääbo S. Genome Res. 1995;5:42–52. doi: 10.1101/gr.5.1.42. [DOI] [PubMed] [Google Scholar]
- 28.Stenico M, Nigro L, Bertorelle G, Calafell F, Capitanio M, Corrain C, Barbujani G. Am J Hum Genet. 1996;59:1363–1375. [PMC free article] [PubMed] [Google Scholar]
- 29.Excoffier L, Poloni E S, Santachiara-Benerecetti A S, Semino O, Langaney A. In: Molecular Biology and Human Diversity. Boyce A J, Mascie-Taylor C G N, editors. Cambridge: Cambridge Univ. Press; 1996. pp. 141–155. [Google Scholar]