Abstract
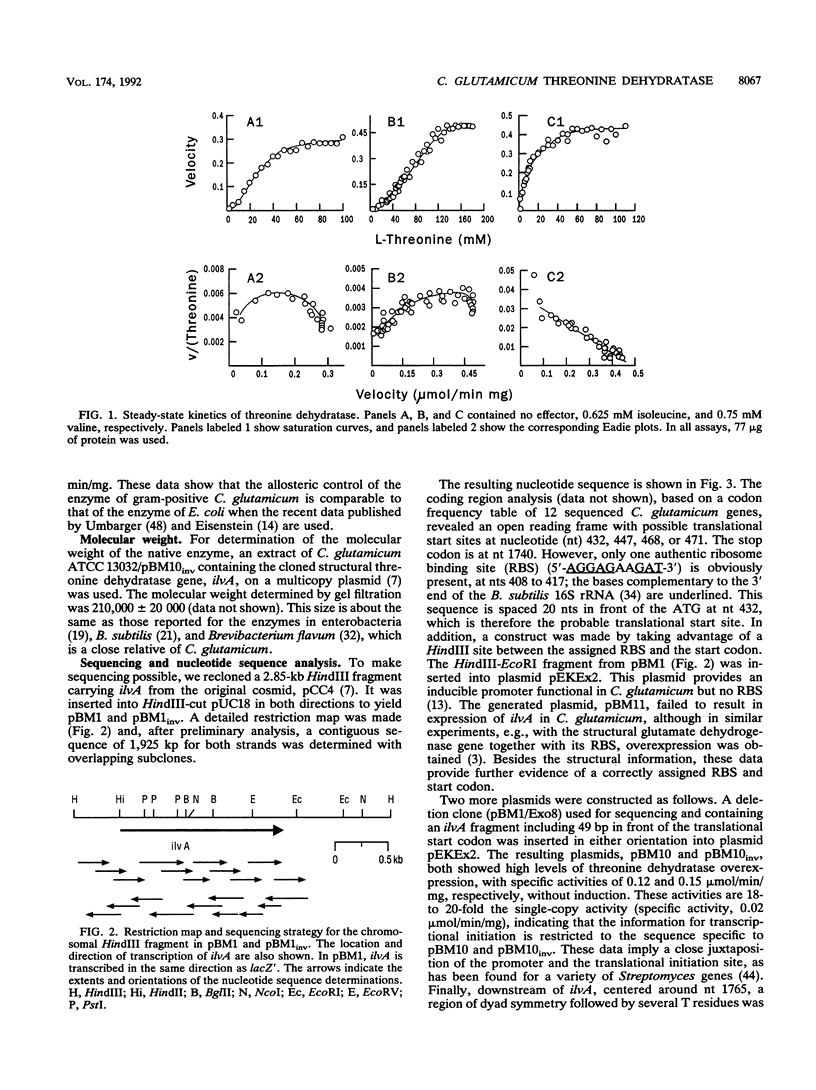
Threonine dehydratase activity is an important element in the flux control of isoleucine biosynthesis. The enzyme of Corynebacterium glutamicum demonstrates a marked sigmoidal dependence of initial velocity on the threonine concentration, a dependence that is consistent with substrate-promoted conversion of the enzyme from a low-activity to a high-activity conformation. In the presence of the negative allosteric effector isoleucine, the K0.5 increased from 21 to 78 mM and the cooperativity, as expressed by the Hill coefficient increased from 2.4 to 3.7. Valine promoted opposite effects: the K0.5 was reduced to 12 mM, and the enzyme exhibited almost no cooperativity. Sequence determination of the C. glutamicum gene for this enzyme revealed an open reading frame coding for a polypeptide of 436 amino acids. From this information and the molecular weight determination of the native enzyme, it follows that the dehydratase is a tetramer with a total mass of 186,396 daltons. Comparison of the deduced polypeptide sequence with the sequences of known threonine dehydratases revealed surprising differences from the C. glutamicum enzyme in the carboxy-terminal portion. This portion is greatly reduced in size, and a large gap of 95 amino acids must be introduced to achieve homology. Therefore, the C. glutamicum enzyme must be considered a small variant of threonine dehydratase that is typically controlled by isoleucine and valine but has an altered structure reflecting a topological difference in the portion of the protein most likely to be important for allosteric regulation.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börmann E. R., Eikmanns B. J., Sahm H. Molecular analysis of the Corynebacterium glutamicum gdh gene encoding glutamate dehydrogenase. Mol Microbiol. 1992 Feb;6(3):317–326. doi: 10.1111/j.1365-2958.1992.tb01474.x. [DOI] [PubMed] [Google Scholar]
- Calhoun D. H., Rimerman R. A., Hatfield G. W. Threonine deaminase from Escherichia coli. I. Purification and properties. J Biol Chem. 1973 May 25;248(10):3511–3516. [PubMed] [Google Scholar]
- Cordes C., Möckel B., Eggeling L., Sahm H. Cloning, organization and functional analysis of ilvA, ilvB and ilvC genes from Corynebacterium glutamicum. Gene. 1992 Mar 1;112(1):113–116. doi: 10.1016/0378-1119(92)90311-c. [DOI] [PubMed] [Google Scholar]
- Cox J. L., Cox B. J., Fidanza V., Calhoun D. H. The complete nucleotide sequence of the ilvGMEDA cluster of Escherichia coli K-12. Gene. 1987;56(2-3):185–198. doi: 10.1016/0378-1119(87)90136-3. [DOI] [PubMed] [Google Scholar]
- Dahlquist F. W. The meaning of Scatchard and Hill plots. Methods Enzymol. 1978;48:270–299. doi: 10.1016/s0076-6879(78)48015-2. [DOI] [PubMed] [Google Scholar]
- Datta P., Goss T. J., Omnaas J. R., Patil R. V. Covalent structure of biodegradative threonine dehydratase of Escherichia coli: homology with other dehydratases. Proc Natl Acad Sci U S A. 1987 Jan;84(2):393–397. doi: 10.1073/pnas.84.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decedue C. J., Hofler J. G., Burns R. O. Threonine deaminase from Salmonella typhimurium. Relationship between regulatory sites. J Biol Chem. 1975 Feb 25;250(4):1563–1570. [PubMed] [Google Scholar]
- Eikmanns B. J., Kleinertz E., Liebl W., Sahm H. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene. 1991 Jun 15;102(1):93–98. doi: 10.1016/0378-1119(91)90545-m. [DOI] [PubMed] [Google Scholar]
- Eisenstein E. Cloning, expression, purification, and characterization of biosynthetic threonine deaminase from Escherichia coli. J Biol Chem. 1991 Mar 25;266(9):5801–5807. [PubMed] [Google Scholar]
- FLAVIN M., SLAUGHTER C. Threonine synthetase mechanism: studies with isotopic hydrogen. J Biol Chem. 1960 Apr;235:1112–1118. [PubMed] [Google Scholar]
- Feldner J., Grimminger H. Threonine deaminase from a nonsense mutant of Escherichia coli requiring isoleucine or pyridoxine: evidence for half-of-the-sites reactivity. J Bacteriol. 1976 Apr;126(1):100–107. doi: 10.1128/jb.126.1.100-107.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. S., Archer J. A., Sinskey A. J. The molecular structure of the Corynebacterium glutamicum threonine synthase gene. Mol Microbiol. 1990 Oct;4(10):1693–1702. doi: 10.1111/j.1365-2958.1990.tb00546.x. [DOI] [PubMed] [Google Scholar]
- Hatfield G. W., Burns R. O. Threonine deaminase from Salmonella typhimurium. 3. The intermediate substructure. J Biol Chem. 1970 Feb 25;245(4):787–791. [PubMed] [Google Scholar]
- Hatfield G. W., Umbarger H. E. Threonine deaminase from Bacillus subtilis. I. Purification of the enzyme. J Biol Chem. 1970 Apr 10;245(7):1736–1741. [PubMed] [Google Scholar]
- Hatfield G. W., Umbarger H. E. Threonine deaminase from Bacillus subtilis. II. The steady state kinetic properties. J Biol Chem. 1970 Apr 10;245(7):1742–1747. [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hofler J. G., Burns R. O. Threonine deaminase from Salmonella typhimurium. Effect of regulatory ligands on the binding of substrates and substrate analogues to the active sites and the differentiation of the activator and inhibitor sites from the active sites. J Biol Chem. 1978 Feb 25;253(4):1245–1251. [PubMed] [Google Scholar]
- Hyde C. C., Ahmed S. A., Padlan E. A., Miles E. W., Davies D. R. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988 Nov 25;263(33):17857–17871. [PubMed] [Google Scholar]
- Kalinowski J., Cremer J., Bachmann B., Eggeling L., Sahm H., Pühler A. Genetic and biochemical analysis of the aspartokinase from Corynebacterium glutamicum. Mol Microbiol. 1991 May;5(5):1197–1204. doi: 10.1111/j.1365-2958.1991.tb01893.x. [DOI] [PubMed] [Google Scholar]
- Lawther R. P., Wek R. C., Lopes J. M., Pereira R., Taillon B. E., Hatfield G. W. The complete nucleotide sequence of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 1987 Mar 11;15(5):2137–2155. doi: 10.1093/nar/15.5.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl W., Bayerl A., Schein B., Stillner U., Schleifer K. H. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol Lett. 1989 Dec;53(3):299–303. doi: 10.1016/0378-1097(89)90234-6. [DOI] [PubMed] [Google Scholar]
- MACGEE J., DOUDOROFF M. A new phosphorylated intermediate in glucose oxidation. J Biol Chem. 1954 Oct;210(2):617–626. [PubMed] [Google Scholar]
- MONOD J., WYMAN J., CHANGEUX J. P. ON THE NATURE OF ALLOSTERIC TRANSITIONS: A PLAUSIBLE MODEL. J Mol Biol. 1965 May;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Marceau M., McFall E., Lewis S. D., Shafer J. A. D-serine dehydratase from Escherichia coli. DNA sequence and identification of catalytically inactive glycine to aspartic acid variants. J Biol Chem. 1988 Nov 15;263(32):16926–16933. [PubMed] [Google Scholar]
- Menkel E., Thierbach G., Eggeling L., Sahm H. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl Environ Microbiol. 1989 Mar;55(3):684–688. doi: 10.1128/aem.55.3.684-688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima R., Shiio I. Regulation of aspartate family amino acid biosynthesis in Brevibacterium flavum. VI. Effects of isoleucine and valine on threonine dehydratase activity and its formation. J Biochem. 1972 Jun;71(6):951–960. doi: 10.1093/oxfordjournals.jbchem.a129866. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Myers E. W., Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988 Mar;4(1):11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Gomi T., Konishi K., Date T., Nakashima H., Nose K., Matsuda Y., Peraino C., Pitot H. C., Fujioka M. Human liver serine dehydratase. cDNA cloning and sequence homology with hydroxyamino acid dehydratases from other sources. J Biol Chem. 1989 Sep 25;264(27):15818–15823. [PubMed] [Google Scholar]
- Parsot C. Evolution of biosynthetic pathways: a common ancestor for threonine synthase, threonine dehydratase and D-serine dehydratase. EMBO J. 1986 Nov;5(11):3013–3019. doi: 10.1002/j.1460-2075.1986.tb04600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A. T., Wood W. A. The mechanism of action of 5'-adenylic acid-activated threonine dehydrase. J Biol Chem. 1965 Dec;240(12):4703–4709. [PubMed] [Google Scholar]
- Reinscheid D. J., Eikmanns B. J., Sahm H. Analysis of a Corynebacterium glutamicum hom gene coding for a feedback-resistant homoserine dehydrogenase. J Bacteriol. 1991 May;173(10):3228–3230. doi: 10.1128/jb.173.10.3228-3230.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Samach A., Hareven D., Gutfinger T., Ken-Dror S., Lifschitz E. Biosynthetic threonine deaminase gene of tomato: isolation, structure, and upregulation in floral organs. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2678–2682. doi: 10.1073/pnas.88.7.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrumpf B., Schwarzer A., Kalinowski J., Pühler A., Eggeling L., Sahm H. A functionally split pathway for lysine synthesis in Corynebacterium glutamicium. J Bacteriol. 1991 Jul;173(14):4510–4516. doi: 10.1128/jb.173.14.4510-4516.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohl W. R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992 Mar 11;20(5):961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon B. E., Little R., Lawther R. P. Analysis of the functional domains of biosynthetic threonine deaminase by comparison of the amino acid sequences of three wild-type alleles to the amino acid sequence of biodegradative threonine deaminase. Gene. 1988 Mar 31;63(2):245–252. doi: 10.1016/0378-1119(88)90528-8. [DOI] [PubMed] [Google Scholar]
- Tanizawa K., Asano S., Masu Y., Kuramitsu S., Kagamiyama H., Tanaka H., Soda K. The primary structure of thermostable D-amino acid aminotransferase from a thermophilic Bacillus species and its correlation with L-amino acid aminotransferases. J Biol Chem. 1989 Feb 15;264(5):2450–2454. [PubMed] [Google Scholar]
- UMBARGER H. E. Evidence for a negative-feedback mechanism in the biosynthesis of isoleucine. Science. 1956 May 11;123(3202):848–848. doi: 10.1126/science.123.3202.848. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Williams A. L., Jr, Tinoco I., Jr A dynamic programming algorithm for finding alternative RNA secondary structures. Nucleic Acids Res. 1986 Jan 10;14(1):299–315. doi: 10.1093/nar/14.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]



