Abstract
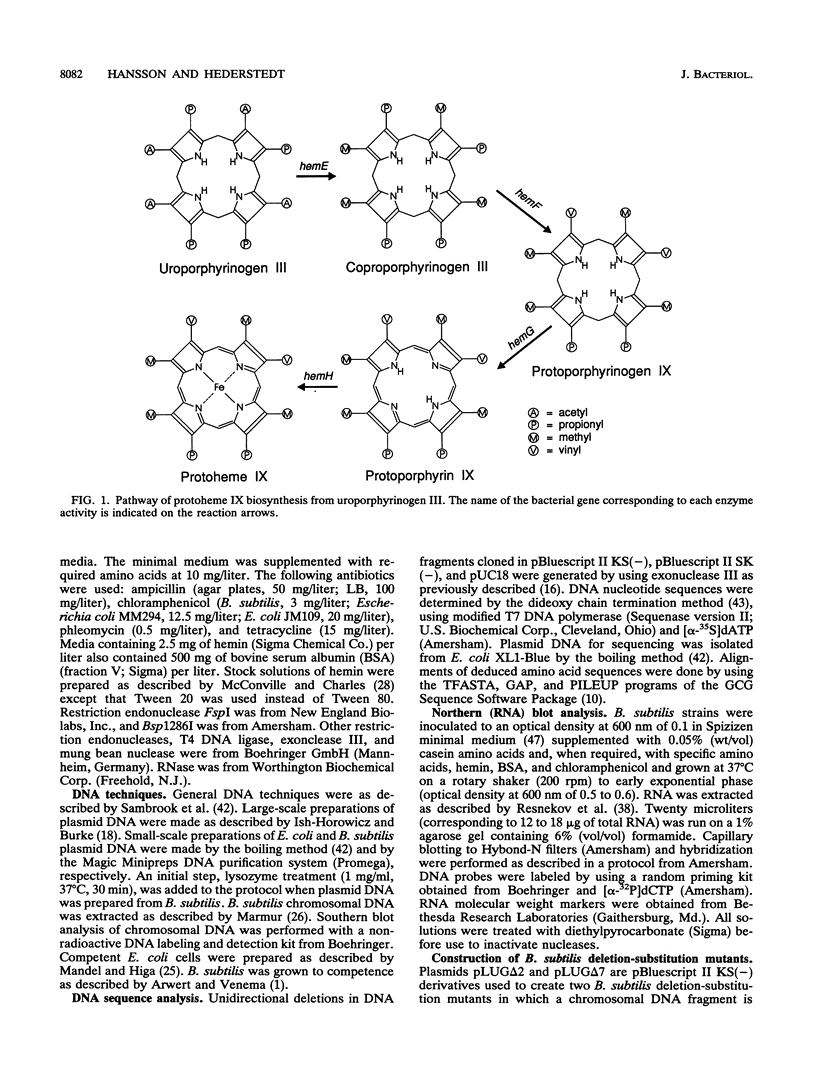
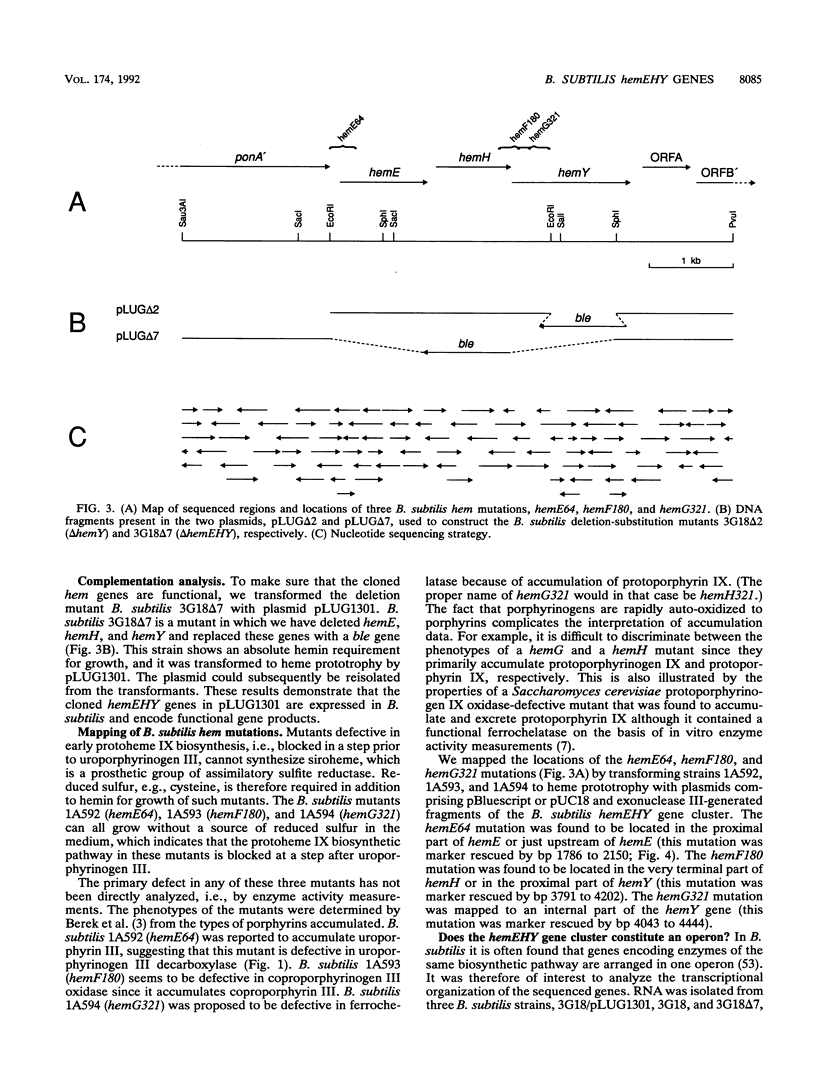
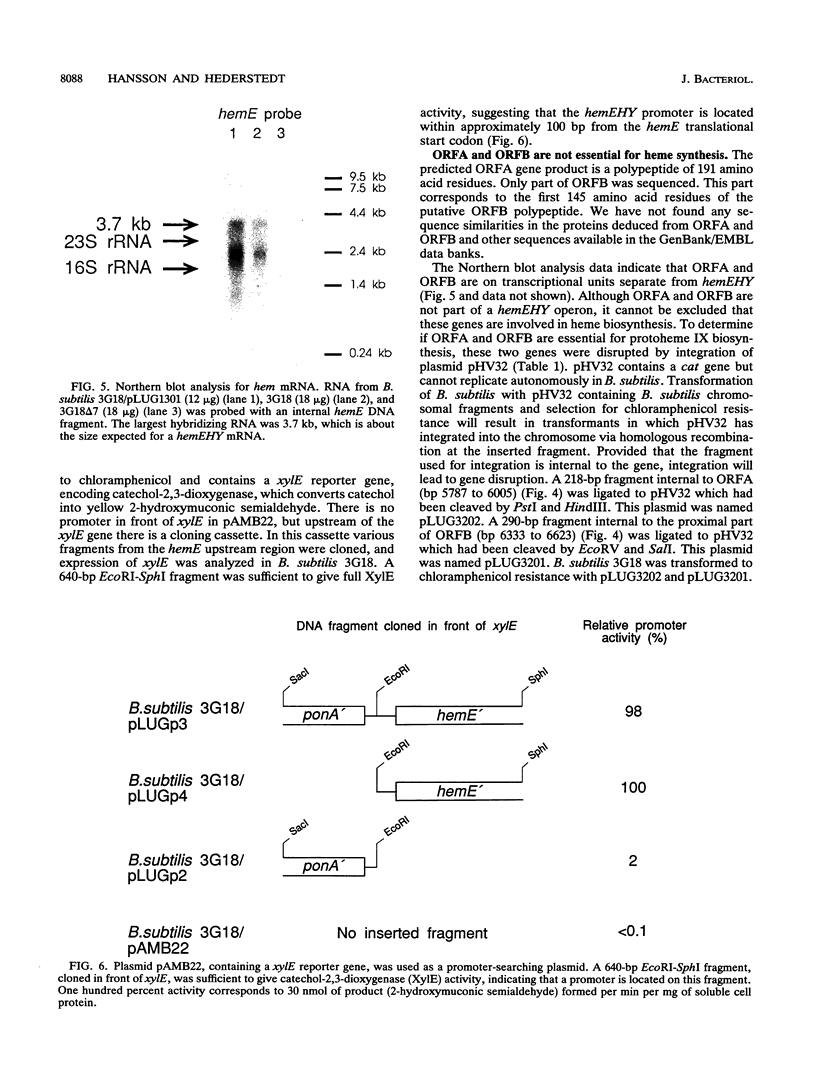
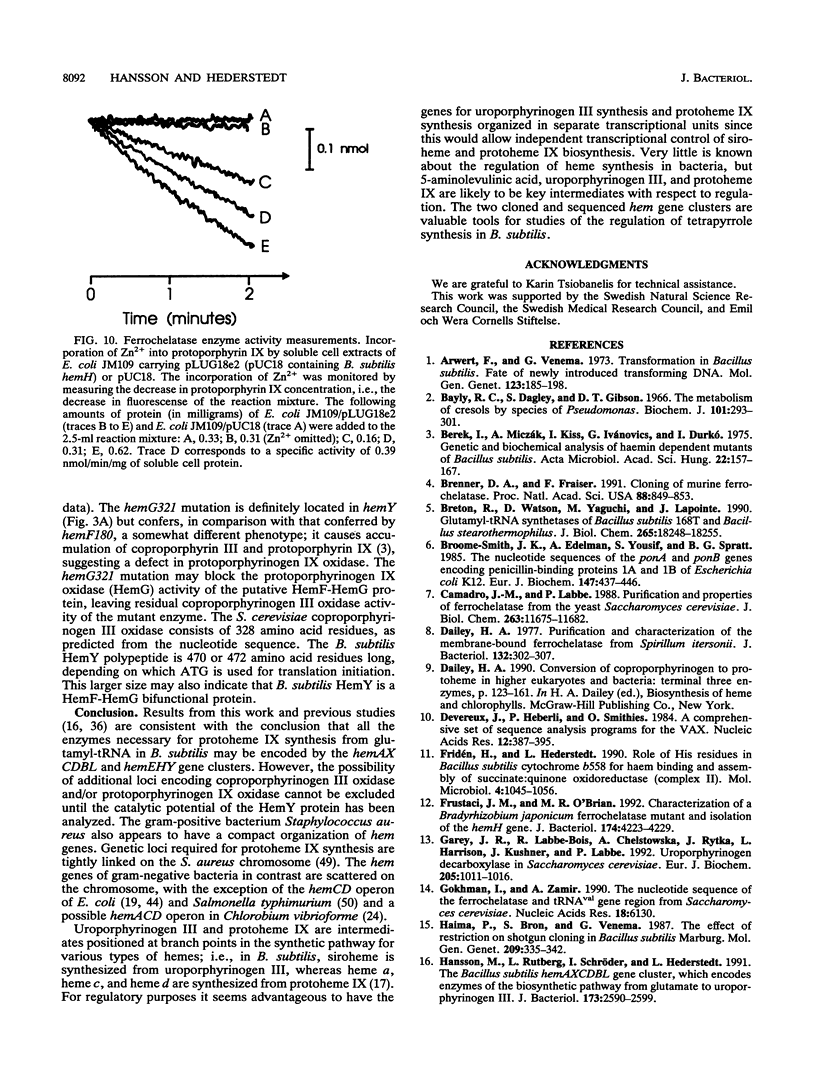
Mutations that cause a block in a late step of the protoheme IX biosynthetic pathway, i.e., in a step after uroporphyrinogen III, map at 94 degrees on the Bacillus subtilis chromosomal genetic map. We have cloned and sequenced the hem genes at this location. The sequenced region contains six open reading frames: ponA, hemE, hemH, hemY, ORFA, and ORFB. The ponA gene product shows over 30% sequence identity to penicillin-binding proteins 1A of Escherichia coli, Streptococcus pneumoniae, and Streptococcus oralis and probably has a role in cell wall metabolism. The hemE gene was identified from amino acid sequence comparisons as encoding uroporphyrinogen III decarboxylase. The hemH gene was identified by enzyme activity analysis of the HemH protein expressed in E. coli. It encodes a water-soluble ferrochelatase which catalyzes the final step in protoheme IX synthesis, the insertion of ferrous iron into protoporphyrin IX. The function of the hemY gene product was not elucidated, but mutation analysis shows that it is required for a late step in protoheme IX synthesis. The hemY gene probably encodes an enzyme with coproporphyrinogen III oxidase or protoporphyrinogen IX oxidase activity or both of these activities. Inactivation of the ORFA and ORFB genes did not block protoheme IX synthesis. Preliminary evidence for a hemEHY mRNA was obtained, and a promoter region located in front of hemE was identified. From these combined results we conclude that the hemEHY gene cluster encodes enzymes for the synthesis of protoheme IX from uroporphyrinogen III and probably constitutes an operon.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arwert F., Venema G. Transformation in Bacillus subtilis. Fate of newly introduced transforming DNA. Mol Gen Genet. 1973;123(2):185–198. doi: 10.1007/BF00267334. [DOI] [PubMed] [Google Scholar]
- Bayly R. C., Dagley S., Gibson D. T. The metabolism of cresols by species of Pseudomonas. Biochem J. 1966 Nov;101(2):293–301. doi: 10.1042/bj1010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berek I., Miczák A., Kiss I., Ivánovics G., Durkó I. Genetic and biochemical analysis of haemin dependent mutants of Bacillus subtilis. Acta Microbiol Acad Sci Hung. 1975;22(2):157–167. [PubMed] [Google Scholar]
- Brenner D. A., Frasier F. Cloning of murine ferrochelatase. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):849–853. doi: 10.1073/pnas.88.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton R., Watson D., Yaguchi M., Lapointe J. Glutamyl-tRNA synthetases of Bacillus subtilis 168T and of Bacillus stearothermophilus. Cloning and sequencing of the gltX genes and comparison with other aminoacyl-tRNA synthetases. J Biol Chem. 1990 Oct 25;265(30):18248–18255. [PubMed] [Google Scholar]
- Broome-Smith J. K., Edelman A., Yousif S., Spratt B. G. The nucleotide sequences of the ponA and ponB genes encoding penicillin-binding protein 1A and 1B of Escherichia coli K12. Eur J Biochem. 1985 Mar 1;147(2):437–446. doi: 10.1111/j.1432-1033.1985.tb08768.x. [DOI] [PubMed] [Google Scholar]
- Camadro J. M., Labbe P. Purification and properties of ferrochelatase from the yeast Saccharomyces cerevisiae. Evidence for a precursor form of the protein. J Biol Chem. 1988 Aug 25;263(24):11675–11682. [PubMed] [Google Scholar]
- Dailey H. A., Jr Purification and characterization of the membrane-bound ferrochelatase from Spirillum itersonii. J Bacteriol. 1977 Oct;132(1):302–307. doi: 10.1128/jb.132.1.302-307.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridén H., Hederstedt L. Role of His residues in Bacillus subtilis cytochrome b558 for haem binding and assembly of succinate: quinone oxidoreductase (complex II). Mol Microbiol. 1990 Jun;4(6):1045–1056. doi: 10.1111/j.1365-2958.1990.tb00677.x. [DOI] [PubMed] [Google Scholar]
- Frustaci J. M., O'Brian M. R. Characterization of a Bradyrhizobium japonicum ferrochelatase mutant and isolation of the hemH gene. J Bacteriol. 1992 Jul;174(13):4223–4229. doi: 10.1128/jb.174.13.4223-4229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garey J. R., Labbe-Bois R., Chelstowska A., Rytka J., Harrison L., Kushner J., Labbe P. Uroporphyrinogen decarboxylase in Saccharomyces cerevisiae. HEM12 gene sequence and evidence for two conserved glycines essential for enzymatic activity. Eur J Biochem. 1992 May 1;205(3):1011–1016. doi: 10.1111/j.1432-1033.1992.tb16868.x. [DOI] [PubMed] [Google Scholar]
- Gokhman I., Zamir A. The nucleotide sequence of the ferrochelatase and tRNA(val) gene region from Saccharomyces cerevisiae. Nucleic Acids Res. 1990 Oct 25;18(20):6130–6130. doi: 10.1093/nar/18.20.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haima P., Bron S., Venema G. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol Gen Genet. 1987 Sep;209(2):335–342. doi: 10.1007/BF00329663. [DOI] [PubMed] [Google Scholar]
- Hansson M., Rutberg L., Schröder I., Hederstedt L. The Bacillus subtilis hemAXCDBL gene cluster, which encodes enzymes of the biosynthetic pathway from glutamate to uroporphyrinogen III. J Bacteriol. 1991 Apr;173(8):2590–2599. doi: 10.1128/jb.173.8.2590-2599.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. M., Mgbeje B. I., Thomas S. D., Alwan A. F. Nucleotide sequence for the hemD gene of Escherichia coli encoding uroporphyrinogen III synthase and initial evidence for a hem operon. Biochem J. 1988 Jan 15;249(2):613–616. doi: 10.1042/bj2490613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel J. A., Boels J. M., Beldman G., Venema G. Nucleotide sequence of the Synechococcus sp. PCC7942 branching enzyme gene (glgB): expression in Bacillus subtilis. Gene. 1990 Apr 30;89(1):77–84. doi: 10.1016/0378-1119(90)90208-9. [DOI] [PubMed] [Google Scholar]
- Kleppe G., Strominger J. L. Studies of the high molecular weight penicillin-binding proteins of Bacillus subtilis. J Biol Chem. 1979 Jun 10;254(11):4856–4862. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Labbe-Bois R. The ferrochelatase from Saccharomyces cerevisiae. Sequence, disruption, and expression of its structural gene HEM15. J Biol Chem. 1990 May 5;265(13):7278–7283. [PubMed] [Google Scholar]
- Majumdar D., Avissar Y. J., Wyche J. H., Beale S. I. Structure and expression of the Chlorobium vibrioforme hemA gene. Arch Microbiol. 1991;156(4):281–289. doi: 10.1007/BF00262999. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Martin C., Briese T., Hakenbeck R. Nucleotide sequences of genes encoding penicillin-binding proteins from Streptococcus pneumoniae and Streptococcus oralis with high homology to Escherichia coli penicillin-binding proteins 1a and 1b. J Bacteriol. 1992 Jul;174(13):4517–4523. doi: 10.1128/jb.174.13.4517-4523.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville M. L., Charles H. P. Isolation of haemin-requiring mutants of Escherichia coli K12. J Gen Microbiol. 1979 Jul;113(1):155–164. doi: 10.1099/00221287-113-1-155. [DOI] [PubMed] [Google Scholar]
- Miczák A., Berek I., Ivanovics G. Mapping the uroporphyrinogen decarboxylase, coproporphyrinogen oxidase and ferrochelatase loci in Bacillus subtilis. Mol Gen Genet. 1976 Jul 5;146(1):85–87. doi: 10.1007/BF00267986. [DOI] [PubMed] [Google Scholar]
- Miyamoto K., Nakahigashi K., Nishimura K., Inokuchi H. Isolation and characterization of visible light-sensitive mutants of Escherichia coli K12. J Mol Biol. 1991 Jun 5;219(3):393–398. doi: 10.1016/0022-2836(91)90180-e. [DOI] [PubMed] [Google Scholar]
- Nakahashi Y., Taketani S., Okuda M., Inoue K., Tokunaga R. Molecular cloning and sequence analysis of cDNA encoding human ferrochelatase. Biochem Biophys Res Commun. 1990 Dec 14;173(2):748–755. doi: 10.1016/s0006-291x(05)80099-3. [DOI] [PubMed] [Google Scholar]
- Nakahigashi K., Nishimura K., Miyamoto K., Inokuchi H. Photosensitivity of a protoporphyrin-accumulating, light-sensitive mutant (visA) of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10520–10524. doi: 10.1073/pnas.88.23.10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville D. M., Jr Molecular weight determination of protein-dodecyl sulfate complexes by gel electrophoresis in a discontinuous buffer system. J Biol Chem. 1971 Oct 25;246(20):6328–6334. [PubMed] [Google Scholar]
- Niaudet B., Goze A., Ehrlich S. D. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene. 1982 Oct;19(3):277–284. doi: 10.1016/0378-1119(82)90017-8. [DOI] [PubMed] [Google Scholar]
- O'Neill G. P., Chen M. W., Söll D. delta-Aminolevulinic acid biosynthesis in Escherichia coli and Bacillus subtilis involves formation of glutamyl-tRNA. FEMS Microbiol Lett. 1989 Aug;51(3):255–259. doi: 10.1016/0378-1097(89)90406-0. [DOI] [PubMed] [Google Scholar]
- Petricek M., Rutberg L., Schröder I., Hederstedt L. Cloning and characterization of the hemA region of the Bacillus subtilis chromosome. J Bacteriol. 1990 May;172(5):2250–2258. doi: 10.1128/jb.172.5.2250-2258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
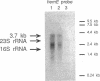
- Resnekov O., Rutberg L., von Gabain A. Changes in the stability of specific mRNA species in response to growth stage in Bacillus subtilis. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8355–8359. doi: 10.1073/pnas.87.21.8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romana M., Le Boulch P., Roméo P. H. Rat uroporphyrinogen decarboxylase cDNA: nucleotide sequence and comparison to human uroporphyrinogen decarboxylase. Nucleic Acids Res. 1987 Sep 11;15(17):7211–7211. doi: 10.1093/nar/15.17.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roméo P. H., Raich N., Dubart A., Beaupain D., Pryor M., Kushner J., Cohen-Solal M., Goossens M. Molecular cloning and nucleotide sequence of a complete human uroporphyrinogen decarboxylase cDNA. J Biol Chem. 1986 Jul 25;261(21):9825–9831. [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasarman A., Nepveu A., Echelard Y., Dymetryszyn J., Drolet M., Goyer C. Molecular cloning and sequencing of the hemD gene of Escherichia coli K-12 and preliminary data on the Uro operon. J Bacteriol. 1987 Sep;169(9):4257–4262. doi: 10.1128/jb.169.9.4257-4262.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder I., Hederstedt L., Kannangara C. G., Gough P. Glutamyl-tRNA reductase activity in Bacillus subtilis is dependent on the hemA gene product. Biochem J. 1992 Feb 1;281(Pt 3):843–850. doi: 10.1042/bj2810843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketani S., Nakahashi Y., Osumi T., Tokunaga R. Molecular cloning, sequencing, and expression of mouse ferrochelatase. J Biol Chem. 1990 Nov 15;265(32):19377–19380. [PubMed] [Google Scholar]
- Tien W., White D. C. Linear sequential arrangement of genes for the biosynthetic pathway of protoheme in Staphylococcus aureus. Proc Natl Acad Sci U S A. 1968 Dec;61(4):1392–1398. doi: 10.1073/pnas.61.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Delling J., Elliott T. The genes required for heme synthesis in Salmonella typhimurium include those encoding alternative functions for aerobic and anaerobic coproporphyrinogen oxidation. J Bacteriol. 1992 Jun;174(12):3953–3963. doi: 10.1128/jb.174.12.3953-3963.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zagorec M., Buhler J. M., Treich I., Keng T., Guarente L., Labbe-Bois R. Isolation, sequence, and regulation by oxygen of the yeast HEM13 gene coding for coproporphyrinogen oxidase. J Biol Chem. 1988 Jul 15;263(20):9718–9724. [PubMed] [Google Scholar]
- Zalkin H., Ebbole D. J. Organization and regulation of genes encoding biosynthetic enzymes in Bacillus subtilis. J Biol Chem. 1988 Feb 5;263(4):1595–1598. [PubMed] [Google Scholar]
- Zukowski M. M., Miller L. Hyperproduction of an intracellular heterologous protein in a sacUh mutant of Bacillus subtilis. Gene. 1986;46(2-3):247–255. doi: 10.1016/0378-1119(86)90409-9. [DOI] [PubMed] [Google Scholar]