Abstract
The origin of the red algae has remained an enigma. Historically the Rhodophyta were classified first as plants and later as the most ancient eukaryotic organisms. Recent molecular studies have indicated similarities between red and green plastids, which suggest that there was a single endosymbiotic origin for these organelles in a common ancestor of the rhodophytes and green plants. Previous efforts to confirm or reject this effort by analyses of nuclear DNA have been inconclusive; thus, additional molecular markers are needed to establish the relationship between the host cell lineages, independent of the evolutionary history of their plastids. To furnish such a data set we have sequenced the largest subunit of RNA polymerase II from two red algae, a green alga and a relatively derived amoeboid protist. Phylogenetic analyses provide strong statistical support for an early evolutionary emergence of the Rhodophyta that preceded the origin of the line that led to plants, animals, and fungi. These data, which are congruent with results from extensive analyses of nuclear rDNA, argue for a reexamination of current models of plastid evolution.
The origin of plastids as endosymbiotic cyanobacteria is well documented (1–4). Although some eukaryotic lineages (e.g., Chromophyta) apparently became photosynthetic secondarily by engulfing an already established eukaryotic alga, plastids of green plants, red algae, and cyanelles of glaucocystophytes are considered primary plastids, descended directly from free-living cyanobacteria (3). Despite their variation in pigment content and thylakoid membrane structure, molecular evidence suggests that all plastids evolved from a single cyanobacterial ancestor.
Most phylogenetic analyses of plastid-localized or plastid-derived genes (2, 3, 5, 6) and comparisons of genome organization (4, 7) have supported a monophyletic association of primary plastids with respect to extant cyanobacteria. These compelling but limited data have led to the contention that the endosymbiotic relationship was established in a single, common ancestor of red algae, green plants, and glaucocystophytes (3, 8, 9). The issue of how these organisms are related, as distinct from plastid interrelationships, must be established independently with characters indigenous to the host cells if the scenario suggested by plastid DNA evidence is to be confirmed.
Phylogenetic analyses of nuclear gene sequences, most often those encoding small-subunit ribosomal RNA (SSU rDNA), have helped to elucidate the early branches of eukaryotic evolution (10–12). The relationship between green plants and red algae, however, has not been fully delineated by rDNA evidence. Although SSU rDNA trees consistently show the Rhodophyta to have emerged before the common ancestor of plants, animals, and fungi (8, 13, 14), intervening branches are poorly supported statistically and are considered unreliable (3, 8). Other nuclear genes examined thus far also fail to provide robust support either for or against a monophyletic relationship of plants with red algae (6, 8, 15–18). Additional nuclear markers clearly are needed to resolve the relationship between the host components of these symbiotic associations.
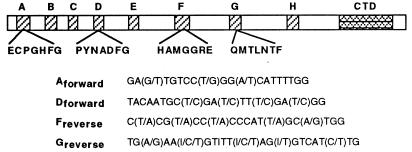
The largest subunits of DNA-dependent RNA polymerases have eight conserved core regions of amino acid similarity, designated A through H, that can be aligned among all eubacterial, archaebacterial, and eukaryotic genes (19). Several of these regions contain highly conserved sequence motifs on which to base PCR primers that can be used to recover sequences of the gene encoding the largest subunit of RNA polymerase II (RPB1) from widely divergent eukaryotic taxa (ref. 20 and Materials and Methods). RPB1 has shown promise for examining distant eukaryotic relationships because of its large size, a relatively constant G + C content, and the lack of paralogous copies (20, 21). To provide a large set of data from homologous genes, we have recovered and sequenced RPB1 from two red algae, a green alga, and the protist Acanthamoeba castellanii (see Table 1). Our phylogenetic analyses based on RPB1 sequences to our knowledge provide the first strong statistical support for an emergence of the Rhodophyta before the common ancestor of animals, fungi, and green plants.
Table 1.
Sequences used in phylogenetic analysis
| RPB1 | GenBank accession number |
|---|---|
| Acanthamoeba castellanii | ;;LEFT>U90211 |
| Arabidopsis thaliana | ;;LEFT>P31635 |
| Caenorhabditis elegans | ;;LEFT>P16356 |
| Drosophila melanogaster | ;;LEFT>P04052 |
| Giardia lamblia | ∗ |
| Homo sapiens | ;;LEFT>X63564 |
| Plasmodium falciparum | ;;LEFT>P14248 |
| Porphyra yezoensis | ;;LEFT>U90208 |
| Saccharomyces cerevisiae | ;;LEFT>P04050 |
| Schizosaccharomyces pombe | ;;LEFT>P36594 |
| Spirogyra sp. | ;;LEFT>U90210 |
| Bonnemaisonia hamifera | ;;LEFT>U90209 |
| Trichomonas vaginalis | ;;LEFT>U20501 |
| Trypanosoma brucei | ;;LEFT>P17545 |
| RPC1 S. cerevisiae | ;;LEFT>P04051 |
, Sequence from H.-P. Klenk.
MATERIALS AND METHODS
DNA Extraction, PCR Amplification, Cloning, and Sequencing.
Total cellular DNA was extracted as described previously (22) from an axenic conchocelis culture of Porphyra yezoensis (Rhodophyta), Bonnemaisonia hamifera (Rhodophyta), and an undetermined species of the green alga Spirogyra. Genomic DNA of Acanthamoeba castellanii was provided by Erik Bateman (Univ. of Vermont, Burlington). Plant, animal, and fungal RPB1 sequences available from GenBank were aligned and degenerate oligonucleotide primers designed based on strongly conserved gene domains (Fig. 1). RPB1 sequences were PCR-amplified from total DNA using primers from conserved regions D and F (Fig. 1); products were cloned into the pCR2.1 plasmid vector (Invitrogen) and sequenced in complementary directions. All clones large enough to contain a viable region D–F from RPB1 were sequenced, but only the dominant PCR product from each of the four taxa showed any homology to RPB1. Based on these sequences, taxon-specific primers were constructed and used in opposition to degenerate primers from regions A and G. The resulting PCR products were also cloned and sequenced.
Figure 1.
Degenerate oligonucleotide primers based on conserved amino acid motifs in RPB1 from plant, animal, and fungal sequences.
Phylogenetic Analysis.
The inferred amino acid sequences were aligned (available upon request) with published eukaryotic RPB1 sequences (Table 1) using Clustal V (23) and adjusted by eye. An initial alignment using RPB1 from plants, animals, fungi, and red algae resulted in 1,147 contiguous amino acid positions encompassing sequences from the region A primer through the region G primer (Fig. 1). As additional sequences were added to the alignment, gaps that could not be anchored reliably with conserved sequences on both ends were removed leaving a total of 941 aligned positions for phylogenetic analysis. Phylogenetic analyses were performed using parsimony (paup; ref. 24), neighbor-joining (phylip; ref. 25), and maximum likelihood (ML) algorithms (paml and molphy; refs. 26 and 27). To determine the strength of phylogenetic conclusions, 1,000 parsimony, 100 neighbor-joining, and 100 ML bootstrap replicates (28) and parsimony decay analysis (29) were performed. Paired-sites analyses (30, 31) were used to test both the most parsimonious and maximum-likelihood trees against alternative tree topologies.
RESULTS
Isolation of RBP1 Sequences.
The degenerate primers proved successful in amplifying regions D through F from all four taxa examined. In most other eukaryotes examined, RPB1 has been shown to occur as a single-copy gene (20, 21). The only exceptions are Trypanosoma brucei, in which two nearly identical copies of RPB1 are encoded at separate loci (32, 33), and soybean (Glycine max), which has multiple copies, presumably due to its polyploid ancestry (34). In both cases the additional copies are the result of recent, lineage-specific gene duplications. Thus, RPB1 does not appear to have been carried at paralogous loci over long periods of eukaryotic evolution. Sequencing of multiple-cloned PCR products gave no indication that more than one copy of RPB1 was present in any of the four taxa that we examined.
When used in opposition to sequence-specific primers, degenerate primers from regions A and G (Fig. 1) produced strong, single products in most cases. The sole exception was the A region primer, which, when used with the Spirogyra template, produced no products with sequence similarity to RPB1. As a result, only regions D through G were obtained from that isolate. A total of 3,051, 3,006, 3,773, and 2,412 contiguous base pairs were sequenced from Porphyra, Bonnemaisonia, Acanthamoeba, and Spirogyra, respectively.
No introns were found in either red algal sequence; however, a single intron has been reported near the 5′ end of the coding region for several other rhodophyte nuclear genes (15, 16). In each of our sequences, a small portion of the 5′ end is missing. Three introns are present between regions D and F in Spirogyra RPB1 in positions that correspond precisely to those in Arabidopsis (35). Nine introns occur between regions A and G in the sequence from Acanthamoeba. Only one, which matched an intron position reported in Schizosaccharomyces pombe (36), was coincident with any intron location from animal, plant, or fungal RPB1 genes (35, 36). The nearly complete absence of conserved intron locations in RPB1 from representatives of these different lineages (35) suggests that most introns were inserted subsequent to the divergence of the protist ancestors of those taxa. Consequently, the number and position of introns within RPB1 genes are not useful for determining relationships among major eukaryotic groups.
Phylogenetic Analyses.
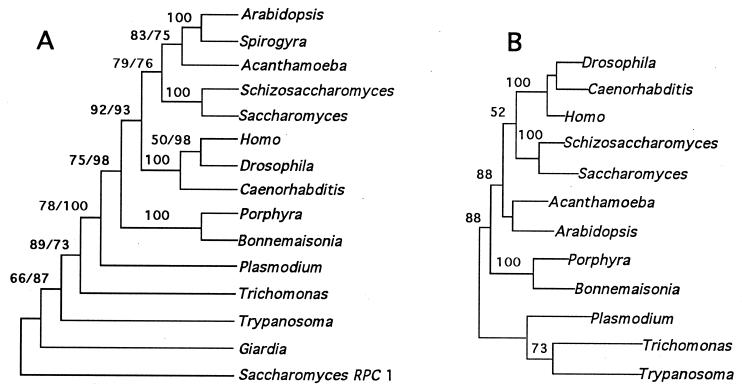
Trees produced from RPB1-encoded amino acid sequences using parsimony, neighbor-joining, and maximum likelihood algorithms (Fig. 2) all exclude a sister relationship between plants and red algae. In agreement with SSU rDNA studies (13, 37), these RPB1-based trees support a monophyletic association of plants, animals, fungi, and certain protists as a late-evolving group of so-called “crown” taxa (38), but indicate that red algae originated before the common ancestor of these lineages. Robust bootstrap support from parsimony (92%), distance (93%), and ML (88%) analyses exclude red algae from this crown group (Fig. 2). More significantly, the red algal and plant sequences form a clade only 3 times in 1,000 and 100 bootstrapped parsimony and ML replicates, respectively, and not once in 100 neighbor-joining replicates. Parsimony decay analysis indicates that the red algae are excluded from the crown clade in all trees up to 11 steps longer than the most parsimonious. With the exception of the unambiguously monophyletic clades of the animal, plant, fungal, and rhodophyte sequences themselves, this is the best supported of all nodes on the tree. The tree produced by ML analysis using paml, which considered rate variations among sites, also excludes red algae from the crown taxa (Fig. 2).
Figure 2.
Phylogenetic analyses based on RPB1 sequences. (A) Congruent bootstrapped parsimony (paup 3.1.1; ref. 24) and neighbor-joining (protdist, phylip3.53; ref. 25) trees. Trees are unrooted, but the largest subunit of polymerase III RPC1 from S. cerevisiae was used as the outgroup (21). Distances were estimated using the Dayhoff PAM matrix for amino acid substitutions. Percent appearances in 1,000 parsimony/100 distance bootstrap replicates are given above each node. Only one value is given when bootstrap values agree. (B) Tree with the maximum likelihood (codeml, paml 1.1; ref. 26) using the Jones, Taylor, Thornton matrix for amino acid substitutions. Branch lengths are approximately proportional to the estimated number of substitutions leading to the subsequent node. Six categories were used to estimate the γ distribution for rate variation, and likelihood estimates made with α = 1 and α = 0.5 produced the same tree topology. Because paml disregards any position in an alignment in which a gap occurs in any one of the aligned sequences, truncated sequences (Giardia, RPC1, Spirogyra) were deleted from the analysis to prevent loss of large portions of the data set.
The significance of this early divergence of the rhodophyte sequences was tested further by statistical comparison of the most parsimonious and maximum-likelihood trees with tree topologies in which the red algal and green plant sequences were constrained to form a monophyletic group. Using both methods, removing plants from the crown or placing red algae within the crown was rejected at P = 0.06 or better (Table 2).
Table 2.
Statistical tests of alternative phylogenetic hypotheses constraining red algae and plants to be closely related
| Hypothesis | Different steps/ln L | Standard error | P |
|---|---|---|---|
| Parsimony | |||
| (Pl,(R,(A,(F,(Ac,P))))) | Best | ||
| (Pl,((R,P),(A,(F,Ac)))) | 39 | 12.05 | <0.001 |
| (Pl,((A,F),(Ac,(P,R)))) | 25 | 13.5 | 0.06 |
| Likelihood | |||
| (Pl,(R,((Ac,P),(F,A)))) | Best | ||
| (Pl,(((R,P),Ac),(F,A))) | −37.4 | 17.3 | 0.03 |
| (Pl,((R,(P,Ac)),(F,A))) | −32.0 | 11.8 | 0.006 |
Tests were performed by supplying user trees in parsimony (protpars-phylip) and likelihood; (protml-molphy) analyses. All taxa were included in the analyses, but for clarity, deeper branching sequences are not shown in the hypothetical trees shown. ln L, natural log likelihood. A, animals; Ac, Acanthamoeba, F, fungi; P, plants; Pl, Plasmodium; R, Rhodophytes. The P value given is the confidence limit for rejection of the alternative tree topology.
DISCUSSION
Results from RPB1 analyses (Fig. 2) generally are consistent with views of eukaryotic evolution developed in earlier studies of nuclear genes (10–13). The amitochondrial flagellates Giardia and Trichomonas, and the kinetoplastid Trypanosoma, represent the most basal lineages; Plasmodium branches off next, followed by the red algae and, finally, by the crown taxa, which group together with strong support. The position of Plasmodium and other apicomplexans is somewhat problematic in rDNA trees. In a number of studies that include sequences of basal eukaryotes (13, 14, 39, 40), apicomplexans emerge before the Rhodophyta in agreement with our results. Depending on the sequences examined and methods employed, however, apicomplexans often nest well within the crown taxa (3, 41). The RPB1 gene of P. falciparum contains several long insertions (42) that make alignment with eukaryotic homologues uncertain in these regions and may affect the rate of evolution in other parts of the gene. Although it is possible that the position of Plasmodium in our analyses is artifactual, removal of the sequence from phylogenetic analyses does not otherwise change tree topologies or affect bootstrap values significantly.
Analyses based on rDNA indicate that Trichomonas branches before the kinetoplastids, whereas RPB1 phylogenies place Trypanosoma in the more basal position. Statistical tests of the relative branching order among ancient protist groups have suggested that rDNA data may not be adequate to resolve the pattern of early eukaryotic evolution (13), and there is growing evidence that Trichomonas is derived from an ancestor that contained mitochondria (43). Additional RPB1 sequences from putatively basal protists are needed to determine whether RPB1 and rDNA consistently support different evolutionary histories among early eukaryotes.
As shown previously (21), parsimony analysis of RPB1 weakly supports a plant/fungi relationship. Maximum likelihood, however, which is less sensitive to extreme rate variations in sequence divergence (44), groups animals and fungi together in agreement with rDNA trees (13, 37) and the majority of protein sequences examined (12, 45).
Although the preponderance of molecular evidence supports a sister relationship between animals and fungi, a sizable number of genes have supported alternative arrangements that group either animals or fungi together with plants (45). It has been argued that these contradictory tree topologies based on different molecular characters reflect very close divergence times among the ancestral protists that gave rise to the plant, animal, and fungal lineages (45). Consistent with this argument, the poor bootstrap support of branches within the crown and the different branching orders among crown taxa deduced from likelihood as compared with parsimony and distance algorithms suggest that relatively little phylogenetic signal is present in RPB1 sequences for these nodes of the tree. It is all the more significant, therefore, that the red algae are strongly excluded from the crown group when each of the tree-building methods is used. This suggests that the rhodophyte branch diverged appreciably earlier than the nearly simultaneous radiation that apparently produced the rest of the crown taxa.
When Did the Red Algae Originate?
The origin of red algae and their relationship to other eukaryotic organisms, chlorophytic algae and plants in particular, have been disputed for over a century. Although the Rhodophyta were classified originally as plants in the Linnaean system, as early as the mid-19th century it was argued that this arrangement was unnatural (see ref. 8 for review). Because they lack flagella and basal bodies and their plastids resemble cyanobacteria in structure and pigment content, red algae were thought to represent the earliest diverging eukaryotes. Subsequent phylogenetic analysis using cytological characters placed the red algae in the position of ancestor to all eukaryotic organisms (46). Comparative sequence data from a growing number of nuclear genes suggest that the red algae are not the most primitive eukaryotic lineage, but rather that they diverged just before the common ancestor of plants, animals, and fungi (13, 17, 18, 47). Still another position for red algae in the eukaryotic hierarchy has been suggested because of similarities between rhodophyte chloroplast and mitochondrial genomes and those of green plants.
Are Red Algae Plants?
The similarities in gene content and DNA sequences between their plastids (see Introduction) have led to the suggestion that rhodophytes and chlorophytes are sister taxa that emanated from a common photosynthetic ancestor (4, 8). A close relationship between red algae and plants also has been argued on the basis of mitochondrial gene content (48). The mitochondrial genome of Chondrus crispus (Rhodophyta) shares with plant mitochondria several open reading frames that are absent in animals and fungi. If these genes were present in the mitochondria of the common ancestor of all four lineages, however, their current distribution suggests a shared loss in animals and fungi, rather than a link between red algae and plants. Moreover, two succinate dehydrogenase genes (SDHB and SDHC) that are still present in the Chondrus mitochondrial genome (48) have been transferred to the nucleus in animals, plants, and fungi, suggesting a common ancestor for the latter three groups. As has been noted previously, because of extreme variation in composition and evolutionary rates among mitochondrial genomes, caution must be exercised when interpreting similarities in gene content or arrangement as shared ancestral or derived characters (49).
Combined phylogenetic analyses of multiple mitochondrial genes have strongly supported a common origin for red algae and plants (50), but outgroup rooting has been problematic. Inclusion of a number of additional protist lineages in the analyses disrupts this monophyletic relationship (50). As more protists are examined it will help to clarify whether molecular phylogenies of mitochondrial genes consistently link red algae with plants. As pointed out in a recent review (51), however, some mitochondria may have been transferred secondarily between eukaryotic hosts, a possibility that has been largely ignored in most analyses of mitochondrial evolution. The abundant evidence for numerous cases of secondary plastid endosymbiosis (3) suggests that an analogous secondary transfer of mitochondria should be taken seriously when mitochondrial- and nuclear-based evolutionary histories are in clear conflict.
Evidence from Nuclear Genes.
Despite similarities in their organellar genomes, a relationship between red algae and green plants is strongly rejected in all of our phylogenetic analyses of RPB1 sequences (Fig. 2; Table 2). Phylogenetic studies of other nuclear genes are generally in agreement with this result but have failed to provide adequate statistical support. For example, SSU rDNA phylogenetic treatments, including those accounting for rate variation among sites and different G + C content among taxa, consistently exclude the Rhodophyta from the plant/animal/fungi clade (8, 13, 14).
Phylogenetic studies based on large-subunit rDNA (18) and β-tubulin sequences (16) do not support a close relationship between plants and red algae. Analyses of actin genes from two different rhodophytes (17, 47) place the red algae as the immediate outgroup to the plant/animal/fungal clade, in agreement with our results. Only trees based on genes encoding cytosolic glyceraldehyde-3-phosphate dehydrogenase (GapC) (6, 15) have suggested a relationship between the red algal and plant sequences; however, statistical support is lacking for this relationship (an equally parsimonious tree does not include the rhodophyte–plant clade) (6). Moreover, anomalous branching patterns due to suspected paralogous gene copies (6) complicate GapC analyses.
Taken as a whole, most of the evidence from nuclear genes supports an emergence of the Rhodophyta before the common ancestor of plants, animals, and fungi. Phylogenetic analyses of RPB1 sequences provide the first statistically robust rejection of a sister relationship between the red and green host cell lineages. These data from nuclear genes indicate that a shared presence of primary plastids, by itself, does not establish that red algae and plants are closely related.
Other Protists Contain Primary Plastids.
Red algae and plants are not the only photosynthetic eukaryotes containing plastids that are regarded as primary in origin. Although the cyanelles of the Glaucocystophyta appear to be monophyletic with red and green plastids based on gene sequences (52), analysis of nuclear SSU rDNA (53) significantly rejects a close relationship between their host cells and those of either plants or red algae. The rhizopod amoeba Paulinella chromatophora contains a cyanelle similar in appearance to the glaucocystophytes but, based on all available evidence, the host cell is derived from a nonphotosynthetic protist line that is unrelated to any other group containing primary plastids (54). An analogous situation appears to be true for species of the dinoflagellate genus Dinophysis (55).†
Although molecular analyses have suggested a single, primary origin for plastids (refs. 4 and 9; but see ref. 56 for an opposing view), the vast majority of evidence from nuclear genes suggests that the host cell lines are not related. How can the apparent monophyletic relationship of plastids that are now classified as primary (i.e., those thought to be descended directly from an endosymbiotic cyanobacterium) be reconciled with the polyphyletic relationships of their host cells?
Are Primary Plastids Monophyletic?
Although most molecular studies of plastid genes indicate a common ancestor for all plastids, a polyphyletic origin of rhodoplasts and chloroplasts has been argued based primarily on differences in gene content (56) and sequence analysis of the large subunit of ribulose-1,5-bisphospate carboxylase (rbcL) (57). It is possible to reconcile these incongruities if widely divergent host taxa independently engulfed different, but genetically related, Cyanobacteria leading to the appearance of monophyletic plastids (2). The different evolutionary histories indicated by nuclear and plastid genes of rhodophytes, chlorophytes, and glaucocystophytes suggest that this is more than a formal possibility; a close examination of plastid phylogenies and of the development of more recent endosymbiotic relationships make it a very likely one.
Phylogenetic analysis of a broad range of Cyanobacteria (5) suggests that plastids may appear to be monophyletic because most extant Cyanobacteria arose after the divergence of the line that led to plastids. If most of the cyanobacterial taxa now available for phylogenetic sampling evolved after plastid–host cell relationships were established, they must by definition be monophyletic, with plastids excluded from that clade.
Moreover, the ancestors of primary plastids may not have been adopted as endosymbionts in a random manner. Numerous examples of modern symbiotic relationships (58–61) provide evidence that certain lineages are adopted preferentially as endosymbionts by widely divergent hosts. For example, members of the green algal genus Trebouxia, the most common algal phycobiont in lichens, have been acquired independently by a wide variety of both ascomycete and basidiomycete fungal taxa (58). In these cases, phylogenetic analysis of the symbiotic Trebouxia clearly would result in an incorrect assessment of the relationships among their fungal hosts. The strong support for independent origins of the rhodophyte and chlorophyte nuclear genomes suggests that plastid DNA evidence may have been misinterpreted in just this way. If plastids descended from an ancient cyanobacterial lineage that was adept at endosymbiotic associations, one that became rare or extinct during the subsequent radiation of other cyanobacterial groups, then the monophyletic relationship of plastids and the polyphyletic origins of host cells need not be mutually inconsistent.
Loss of Plastids?
Because of the complexity of plastid–host cell interactions, in particular the mechanisms for transport of proteins encoded by nuclear genes into the plastid, it has been argued that primary plastids originated only once, relatively early in eukaryotic evolution, but were subsequently lost from many extant lineages (62). There is only fragmentary molecular evidence for this scenario. One possible example, a heme oxygenase in birds and mammals, shares approximately 35% amino acid sequence identity with a plastid heme oxygenase from red algae (5). A homologous protein, however, has not been reported in invertebrate animals, fungi, or protists. It is therefore unclear whether this protein is derived from the plastid of a photosynthetic ancestor or has been acquired more recently, only by vertebrate animals (5). Perhaps additional evidence will be found as the genomes of crown eukaryotes are examined further. Given the paucity of current evidence and its largely anecdotal character, wholesale loss of plastids by so many different lineages is not well supported at this time.
Are Primary Plastids Really Primary?
As discussed above, the establishment of a complex protein transport system that allowed the transformation of an endosymbiotic cyanobacterium into a full-fledged plastid may have been a rare or even unique event (62). Once this protein import mechanism was in place, however, its transfer to a secondary host was less difficult as evidenced by the number of eukaryotic algae that have been adopted as endosymbionts (3). Primary plastids are defined by the presence of two outer membranes; the inner is thought to be derived from the original cyanobacterial cell membrane and the outer remains from the phagocytotic vacuole of the host cell. The additional membranes present in secondary plastids are explained as vestiges of a later endosymbiotic engulfment involving a eukaryotic alga, rather than a cyanobacterium (63, 64).
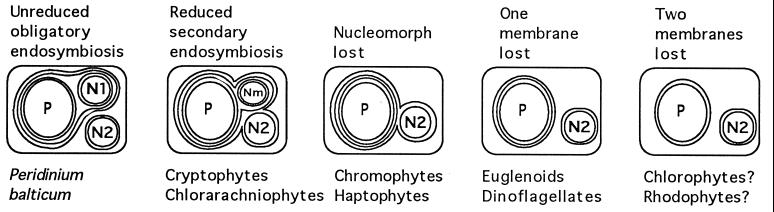
Although this model is consistent with the number of membranes surrounding the plastids of certain algal groups, there is a great deal of variation in the ensemble of interactions between the secondary host and its eukaryotic algal endosymbiont, particularly in the extent of reduction of the primary host cell. In some secondary relationships the primary host cell nucleus has been retained in a reduced form, which is called the nucleomorph. In the case of the Cryptophyta, analysis of nucleomorph SSU rDNA has established that the eukaryotic endosymbiont was a red alga (39). In chromophytes, all of the genes needed for the development and functioning of plastids were transferred from the primary to the secondary host cell nucleus, and the primary cell nucleus and cytoplasm were lost completely (see Fig. 3).
Figure 3.
Hypothetical evolutionary reduction of secondary plastids to give the appearance of primary plastids. Organisms representative of the condition are given below each diagram. Conditions 1 (65) through 4 are widely accepted (3, 4, 8, 9, 54, 55, 63, 64) to explain membrane variability in the plastids of photosynthetic eukaryotes. N1, primary host cell nucleus; N2, secondary host cell nucleus; Nm, nucleomorph (vestigial endosymbiont nucleus); P, plastid.
Euglenoids and many dinoflagellates also have lost the primary host nucleus but possess only three membranes rather than the four predicted for a secondary endosymbiotic relationship. If the eukaryotic endosymbiont could be reduced to this great an extent, one can readily imagine that the other additional membrane was lost in some secondarily photosynthetic lineages. Viewed in this way, a continuum may exist in secondary plastids (Fig. 3), wherein the most reduced forms are indistinguishable from primary plastids.
Secondary plastids that have masqueraded as primary would explain the different evolutionary histories implied by nucleus- and plastid-based characters. If a single cyanobacterium was the endosymbiotic ancestor of all plastids, perhaps in a glaucocystophyte-like protist, then it is possible that in all other photosynthetic eukaryotes plastids were acquired secondarily. Perhaps the observed similarity in mitochondrial genomes of red algae and plants, and the GapC data that are discordant with analyses of other nuclear genes, are evidence for reduced secondary plastid endosymbiosis in one or both groups. If this proposal is true, additional evidence of a lost primary endosymbiont should exist in the nuclei of the putative secondary host(s). As more is known about the nuclear genomes of glaucocystophytes and red algae, a clearer picture of the evolutionary history of plastids should emerge.
Conclusion.
Our analysis of RPB1 sequences, combined with most evidence from other nonplastid characters, indicates that red algae and plants do not share a common host cell ancestor. It remains unclear whether primary plastids were established independently in several different host cells, whether they originated once and were subsequently lost from many lineages, or whether plastid membrane evolution has been misinterpreted in some groups. What is clear is that the plastid and nuclear symbiotic components, now present in the cells of photosynthetic eukaryotes, need not have followed congruent evolutionary pathways. Molecular evidence from each of the symbiotic components must be viewed in a common framework to gain a complete understanding of the evolutionary histories of the diverse and complex interrelationships that make up photosynthetic eukaryotic cells.
Acknowledgments
We thank H.-P. Klenk for providing the RPB1 sequence from G. lamblia, E. Bateman for Acanthamoeba DNA, J. R. Waaland and E. Duffield for culture material of P. yezoensis, T. hamifera, and Spirogyra, A. L. Denton for collaboration on primer design and technical assistance, and B. F. Lang and J. D. Palmer for constructive input.
ABBREVIATIONS
- RPB1
RNA polymerase II largest subunit
- SSU rDNA
gene encoding small-subunit ribosomal RNA
- ML
maximum likelihood
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. U90208–U90211U90208U90209U90210U90211).
To our knowledge, the endosymbionts of Paulinella and Dinophysis have not been characterized at the molecular level and they may turn out to be descended from cyanobacterial ancestors that are clearly distinct from rhodoplasts, chloroplasts, and cyanelles.
References
- 1.Gray M W, Doolittle W F. Microbiol Rev. 1982;46:1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delwiche C F, Kuhsel M, Palmer J D. Mol Phylogenet Evol. 1995;4:110–128. doi: 10.1006/mpev.1995.1012. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya D, Medlin L. J Phycol. 1995;31:489–498. [Google Scholar]
- 4.Reith M. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:549–575. [Google Scholar]
- 5.Nelissen B, Van de Peer Y, Wilmotte A, De Wachter R. Mol Biol Evol. 1995;12:1166–1173. doi: 10.1093/oxfordjournals.molbev.a040289. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y-H, Ragan M. Curr Genet. 1995;28:324–332. doi: 10.1007/BF00326430. [DOI] [PubMed] [Google Scholar]
- 7.Reith M E, Munholland J. Plant Cell. 1993;5:465–475. doi: 10.1105/tpc.5.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ragan M A, Gutell R A. Bot J Linn Soc. 1995;118:81–105. [Google Scholar]
- 9.Palmer J D. Nature (London) 1993;364:762–763. [Google Scholar]
- 10.Kamaishi T, Hashimoto T, Nakamura Y, Nakamura F, Murata S, Okada N, Okamoto K-i, Shimizu M, Hasegawa M. J Mol Evol. 1996;42:257–263. doi: 10.1007/BF02198852. [DOI] [PubMed] [Google Scholar]
- 11.Sogin M L, Gunderson J H, Elwood H J, Alonso R A, Peattie D A. Science. 1989;243:75–77. doi: 10.1126/science.2911720. [DOI] [PubMed] [Google Scholar]
- 12.Baldauf S L, Palmer J D. Proc Natl Acad Sci USA. 1993;90:11558–11562. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S, Rzhetsky A. J Mol Evol. 1996;42:183–193. doi: 10.1007/BF02198844. [DOI] [PubMed] [Google Scholar]
- 14.Van de Peer Y, Rensing S A, Maier U-G, De Wachter R. Proc Natl Acad Sci USA. 1996;93:7732–7736. doi: 10.1073/pnas.93.15.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liaud M-F, Valentin C, Martin W, Bouget F-Y, Kloareg B, Cerff R. J Mol Evol. 1994;38:319–327. doi: 10.1007/BF00163149. [DOI] [PubMed] [Google Scholar]
- 16.Liaud M-F, Brandt U, Cerff R. Plant Mol Evol. 1995;28:313–325. doi: 10.1007/BF00020250. [DOI] [PubMed] [Google Scholar]
- 17.Bouget F Y, Kerbourc’h C, Laiud M-F, Loiseaux de Goër S, Quatrano R S, Cerff R, Kloareg B. Curr Genet. 1995;28:164–172. doi: 10.1007/BF00315783. [DOI] [PubMed] [Google Scholar]
- 18.Perraso R, Baroin A, Liang H Q, Bachellerie J P, Adoutte A. Nature (London) 1989;339:142–144. doi: 10.1038/339142a0. [DOI] [PubMed] [Google Scholar]
- 19.Jokerst R S, Weeks J R, Zehring W A, Greenleaf A L. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 20.Sidow A, Thomas W K. Curr Biol. 1994;4:596–603. doi: 10.1016/s0960-9822(00)00131-7. [DOI] [PubMed] [Google Scholar]
- 21.Klenk H-P, Zillig W, Lanzendörfer M, Brampp B, Palm P. Arch Protistenkd. 1995;145:221–230. [Google Scholar]
- 22.Stiller J W, Waaland J R. J Phycol. 1993;29:506–517. [Google Scholar]
- 23.Higgens D G, Bleasby A J, Fuchs R. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 24.Swofford D L. paup. Champaign, IL: Ill. Natl. Hist. Surv.; 1993. , Version 3.1.1. [Google Scholar]
- 25.Felsenstein J. Cladistics. 1989;5:164–165. [Google Scholar]
- 26.Yang Z. paml. Pennsylvania State University, University Park, PA: Inst. Mol. Evol. Genet.; 1995. , Version 1.1. [Google Scholar]
- 27.Adachi J, Hasagawa M. Computer Science Monograph. Vol. 27. Tokyo: Institute of Statistical Mathematics; 1993. [Google Scholar]
- 28.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 29.Bremer K. Evolution. 1988;42:795–803. doi: 10.1111/j.1558-5646.1988.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 30.Kishino H, Hasagawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. Syst Zool. 1985;34:152–161. [Google Scholar]
- 32.Evers R, Hammer A, Köck J, Jess W, Borst P, Mémet S, Cornelissen C A. Cell. 1989;56:585–597. doi: 10.1016/0092-8674(89)90581-3. [DOI] [PubMed] [Google Scholar]
- 33.Smith J L, Levin J R, Ingles C J, Agabian N. Cell. 1989;56:815–827. doi: 10.1016/0092-8674(89)90686-7. [DOI] [PubMed] [Google Scholar]
- 34.Dietrich M A, Prenger J P, Guilfoyle T J. Plant Mol Biol. 1990;15:207–223. doi: 10.1007/BF00036908. [DOI] [PubMed] [Google Scholar]
- 35.Nawrath C, Schell J, Koncz C. Mol Gen Genet. 1990;223:65–75. doi: 10.1007/BF00315798. [DOI] [PubMed] [Google Scholar]
- 36.Asuma Y, Yamagishi M, Ueshima R, Ishihama A. Nucleic Acids Res. 1991;19:461–468. doi: 10.1093/nar/19.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wainwright P O, Hinkle G, Sogin M L, Stickel S K. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 38.Knoll A H. Science. 1992;256:622–627. doi: 10.1126/science.1585174. [DOI] [PubMed] [Google Scholar]
- 39.Douglas S E, Murphy C A, Spencer D F, Gray M W. Nature (London) 1991;350:148–151. doi: 10.1038/350148a0. [DOI] [PubMed] [Google Scholar]
- 40.Cavalier-Smith T, Chao E E. J Mol Evol. 1996;43:551–562. doi: 10.1007/BF02202103. [DOI] [PubMed] [Google Scholar]
- 41.Hinkle G, Leipe D D, Nerad T A, Sogin M L. Nucleic Acids Res. 1994;22:465–469. doi: 10.1093/nar/22.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W-B, Bzik D J, Gu H, Tanaka M, Fox B A, Inselburg J. Nucleic Acids Res. 1989;17:9621–9636. doi: 10.1093/nar/17.23.9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bui E T N, Bradley P J, Johnson P J. Proc Natl Acad Sci USA. 1996;93:9651–9656. doi: 10.1073/pnas.93.18.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felsenstein J. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 45.Nikoh N, Hayase N, Iwabe N, Kuma K-i, Miyata T. Mol Biol Evol. 1994;11:762–768. doi: 10.1093/oxfordjournals.molbev.a040156. [DOI] [PubMed] [Google Scholar]
- 46.Liscomb D L. In: The Hierarchy of Life. Fernholm B, Bremer K, Jörnvall H, editors. Amsterdam: Elsevier; 1989. pp. 161–178. [Google Scholar]
- 47.Takahashi H, Takano H, Yokoyama A, Hara Y, Kawano S, Toh e-A, Kuroiwa T. Curr Genet. 1995;28:484–490. doi: 10.1007/BF00310820. [DOI] [PubMed] [Google Scholar]
- 48.Leblanc C, Boyen C, Richard O, Bonnard G, Grienenberger J-M, Kloareg B. J Mol Biol. 1995;250:484–495. doi: 10.1006/jmbi.1995.0392. [DOI] [PubMed] [Google Scholar]
- 49.Burger G, Plante I, Lonergan K, Gray M W. J Mol Biol. 1995;245:522–537. doi: 10.1006/jmbi.1994.0043. [DOI] [PubMed] [Google Scholar]
- 50.Paquin, B., Laforest, M.-J., Forget, L., Roewer, I., Wang, Z., Longcore, J. & Lang, B. F. (1997) Curr. Gen., in press. [DOI] [PubMed]
- 51.Gray M W. Curr Opin Genet Dev. 1993;3:884–890. doi: 10.1016/0959-437x(93)90009-e. [DOI] [PubMed] [Google Scholar]
- 52.Helmchen T A, Bhattacharya D, Melkonian M. J Mol Evol. 1995;41:203–210. doi: 10.1007/BF00170674. [DOI] [PubMed] [Google Scholar]
- 53.Bhattacharya D, Helmchen T, Bibeau C, Melkonian M. Mol Biol Evol. 1995;12:415–420. doi: 10.1093/oxfordjournals.molbev.a040216. [DOI] [PubMed] [Google Scholar]
- 54.Bhattacharya D, Helmchen T, Melkonian M. J Eukaryotic Microbiol. 1995;42:65–69. doi: 10.1111/j.1550-7408.1995.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 55.Schnepf E. In: Origin of Plastids. Lewin R A, editor. London: Chapman & Hall; 1993. pp. 53–76. [Google Scholar]
- 56.Loiseaux-de Goër S. Prog Phycol Res. 1994;10:137–177. [Google Scholar]
- 57.Valentin K, Cattolico R A, Zetsche K. In: Origin of Plastids. Lewin R A, editor. London: Chapman & Hall; 1993. pp. 193–221. [Google Scholar]
- 58.Ahmadjian N. The Lichen Symbiosis. New York: Wiley; 1993. [Google Scholar]
- 59.Rowan R, Knowlton N. Proc Natl Acad Sci USA. 1995;92:2850–2853. doi: 10.1073/pnas.92.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johanowicz D L, Hoy M A. Ann Entomol Soc Am. 1996;89:435–441. [Google Scholar]
- 61.Gast R J, Caron D A. Mol Biol Evol. 1996;13:1192–1197. doi: 10.1093/oxfordjournals.molbev.a025684. [DOI] [PubMed] [Google Scholar]
- 62.Cavalier-Smith T. In: Origin of Plastids. Lewin R A, editor. London: Chapman & Hall; 1993. pp. 291–348. [Google Scholar]
- 63.Gibbs S P. Ann NY Acad Sci. 1981;361:193–207. doi: 10.1111/j.1749-6632.1981.tb46519.x. [DOI] [PubMed] [Google Scholar]
- 64.Whatley J M, Whatley F R. New Phytol. 1981;87:223–247. [Google Scholar]
- 65.Tomas R W, Cox E R. J Phycol. 1973;9:304–323. [Google Scholar]