Abstract
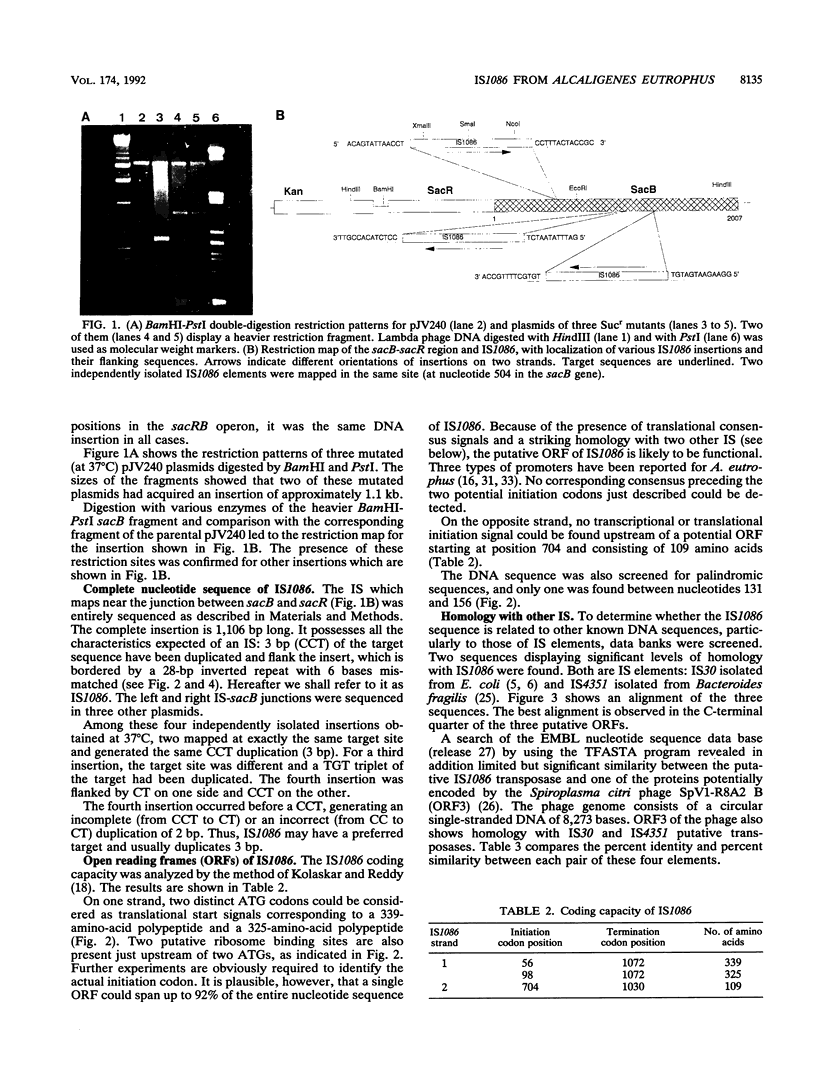
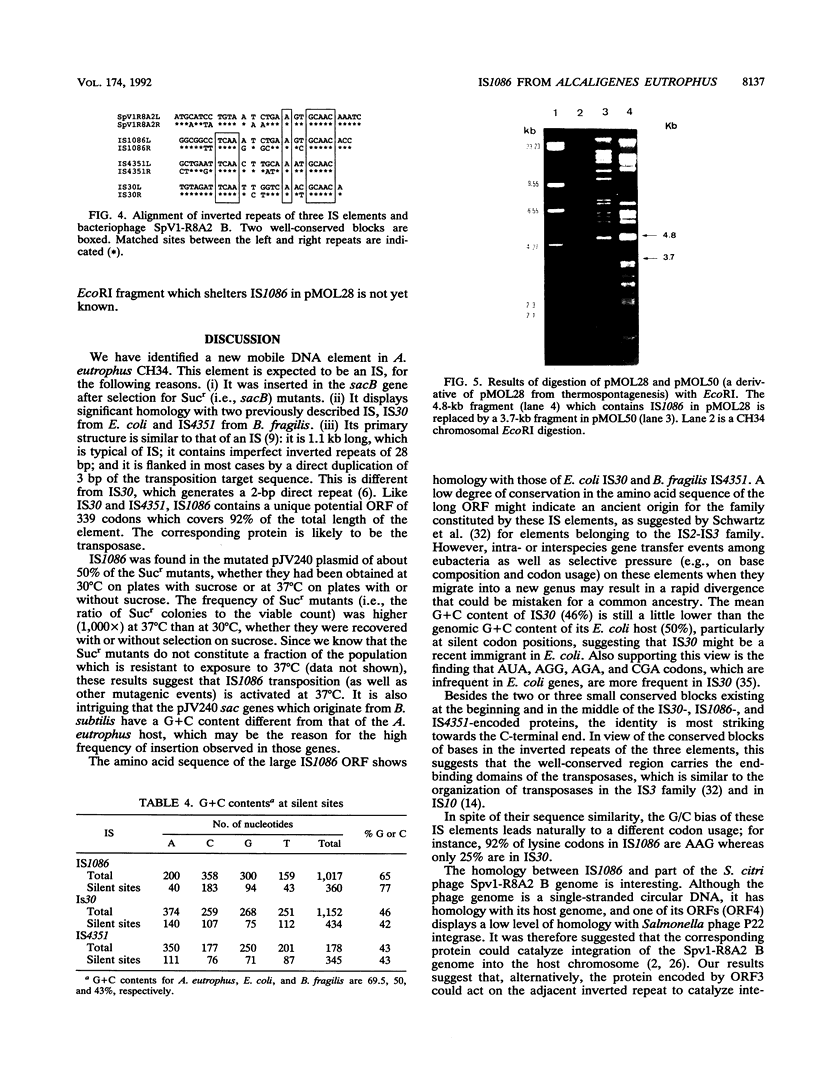
A new insertion sequence (IS), designated IS1086, was isolated from Alcaligenes eutrophus CH34 by being trapped in plasmid pJV240, which contains the Bacillus subtilis sacB and sacR genes. The 1,106-bp IS1086 element contains partially matched (22 of 28 bp) terminal-inverted repeats and a long open reading frame. Hybridization data suggest the presence of one copy of IS1086 in the strain CH34 heavy-metal resistance plasmid pMOL28 and at least two copies in its chromosome. Analysis of the IS1086 nucleotide sequence revealed striking homology with two other IS elements, IS30 and IS4351, suggesting that they are three close members in a family of phylogenetically related insertion sequences. One open reading frame of the Spiroplasma citri phage SpV1-R8A2 B was also found to be related to this IS family but to a lesser extent. Comparison of the G+C contents of IS30 and IS1086 revealed that they conform to their respective hosts (46 versus 50% for IS30 and Escherichia coli and 64.5% for IS1086 and A. eutrophus). The pressure on the AT/GC ratio led to a very different codon usage in these two closely related IS elements. Results suggesting that IS1086 transposition might be activated by some forms of stress are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M. M., Amann E., Lurz R., Rückert B., Bagdasarian M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983 Dec;26(2-3):273–282. doi: 10.1016/0378-1119(83)90197-x. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Caspers P., Dalrymple B., Iida S., Arber W. IS30, a new insertion sequence of Escherichia coli K12. Mol Gen Genet. 1984;196(1):68–73. doi: 10.1007/BF00334094. [DOI] [PubMed] [Google Scholar]
- Dalrymple B., Caspers P., Arber W. Nucleotide sequence of the prokaryotic mobile genetic element IS30. EMBO J. 1984 Sep;3(9):2145–2149. doi: 10.1002/j.1460-2075.1984.tb02104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972 Sep;72(2):349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C. I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985 Nov;164(2):918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Ferrari E., Hoch J. A. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J Bacteriol. 1983 Mar;153(3):1424–1431. doi: 10.1128/jb.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Huisman O., Errada P. R., Signon L., Kleckner N. Mutational analysis of IS10's outside end. EMBO J. 1989 Jul;8(7):2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrossek D., Krüger N., Steinbüchel A. Characterization of alcohol dehydrogenase genes of derepressible wild-type Alcaligenes eutrophus H16 and constitutive mutants. J Bacteriol. 1990 Sep;172(9):4844–4851. doi: 10.1128/jb.172.9.4844-4851.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kolaskar A. S., Reddy B. V. A method to locate protein coding sequences in DNA of prokaryotic systems. Nucleic Acids Res. 1985 Jan 11;13(1):185–194. doi: 10.1093/nar/13.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M., Houba C., Gerits J. Extrachromosomal inheritance controlling resistance to cadmium, cobalt, copper and zinc ions: evidence from curing in a Pseudomonas [proceedings]. Arch Int Physiol Biochim. 1978 May;86(2):440–442. [PubMed] [Google Scholar]
- Mergeay M., Nies D., Schlegel H. G., Gerits J., Charles P., Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985 Apr;162(1):328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987 Jan;84(1):166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R. K., Schilperoort R. A., Nuti M. P. Large plasmids of fast-growing rhizobia: homology studies and location of structural nitrogen fixation (nif) genes. J Bacteriol. 1981 Mar;145(3):1129–1136. doi: 10.1128/jb.145.3.1129-1136.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen J. L., Odelson D. A., Macrina F. L. Complete nucleotide sequence of insertion element IS4351 from Bacteroides fragilis. J Bacteriol. 1987 Aug;169(8):3573–3580. doi: 10.1128/jb.169.8.3573-3580.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudin J., Aullo P., Vignault J. C., Bové J. M. Complete nucleotide sequence of the genome of Spiroplasma citri virus SpV1-R8A2 B. Nucleic Acids Res. 1990 Mar 11;18(5):1293–1293. doi: 10.1093/nar/18.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouch D. A., Skurray R. A. IS257 from Staphylococcus aureus: member of an insertion sequence superfamily prevalent among gram-positive and gram-negative bacteria. Gene. 1989;76(2):195–205. doi: 10.1016/0378-1119(89)90160-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert P., Krüger N., Steinbüchel A. Molecular analysis of the Alcaligenes eutrophus poly(3-hydroxybutyrate) biosynthetic operon: identification of the N terminus of poly(3-hydroxybutyrate) synthase and identification of the promoter. J Bacteriol. 1991 Jan;173(1):168–175. doi: 10.1128/jb.173.1.168-175.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E., Kröger M., Rak B. IS150: distribution, nucleotide sequence and phylogenetic relationships of a new E. coli insertion element. Nucleic Acids Res. 1988 Jul 25;16(14B):6789–6802. doi: 10.1093/nar/16.14.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Betcke A., Warnecke U., Böcker C., Zaborosch C., Friedrich B. Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1990 Jun;172(6):2920–2929. doi: 10.1128/jb.172.6.2920-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K., Wada Y., Doi H., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):1981–1986. doi: 10.1093/nar/19.suppl.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]