Abstract
Cbl family proteins are evolutionarily conserved ubiquitin ligases that negatively regulate signaling from tyrosine kinase-coupled receptors. The mammalian cbl family consists of c-Cbl, Cbl-b, and the recently cloned Cbl-3 (also known as Cbl-c). In this study, we describe the detailed expression pattern of murine Cbl-3 and report the generation and characterization of Cbl-3-deficient mice. Cbl-3 exhibits an expression pattern distinct from those of c-Cbl and Cbl-b, with high levels of Cbl-3 expression in epithelial cells of the gastrointestinal tract and epidermis, as well as the respiratory, urinary, and reproductive systems. Cbl-3 expression was not detected in nonepithelial cells, but within epithelial tissues, the levels of Cbl-3 expression varied from undetectable in the alveoli of the lungs to very strong in the cecum and colon. Despite this restricted expression pattern, Cbl-3-deficient mice were viable, healthy, and fertile and displayed no histological abnormalities up to 18 months of age. Proliferation of epithelial cells in the epidermises and gastrointestinal tracts was unaffected by the loss of Cbl-3. Moreover, Cbl-3 was not required for attenuation of epidermal growth factor-stimulated Erk activation in primary keratinocytes. Thus, Cbl-3 is dispensable for normal epithelial development and function.
Signaling from receptor and cytoplasmic tyrosine kinases plays a major role in the control of fundamental cellular processes, including proliferation and differentiation (38, 48). The Cbl family of adapter proteins, which includes c-Cbl, Cbl-b, and Cbl-3/Cbl-c in mammals, as well as Sli-1 in Caenorhabditis elegans and D-Cbl in Drosophila, are negative regulators of tyrosine kinase signaling downstream of receptor tyrosine kinases and lymphocyte antigen receptors (reviewed in references 44 and 47). All cbl family members contain a conserved amino terminus with a novel phosphotyrosine-binding SH2-like domain termed a tyrosine kinase-binding (TKB) domain, a linker region, and a RING finger domain (28). The carboxy termini of different Cbl proteins are more divergent; however, they generally contain a proline-rich region.
Studies of Sli-1 and D-Cbl have established the role of Cbl proteins in C. elegans vulval development and Drosophila photoreceptor development and oogenesis as negative regulators of signaling from epidermal growth factor receptor (EGFR) homologs (16, 27, 34, 51). Similarly, in mammals, c-Cbl and Cbl-b are phosphorylated and recruited to the EGFR upon epidermal growth factor (EGF) stimulation and also interact with multiple intracellular signaling intermediates. Numerous overexpression studies have demonstrated that c-Cbl and Cbl-b are negative regulators of tyrosine kinase signaling (reviewed in references 44 and 47). Gene targeting studies of c-Cbl and Cbl-b have demonstrated that these proteins are important negative regulators of immunoreceptor signaling in thymocytes and mature T cells, respectively (1, 4, 31, 32). However, the roles of Cbl proteins appear to be complex, since c-Cbl or Cbl-b may also play positive regulatory roles in bone resorption, glucose transport, integrin-mediated adhesion, and T-cell receptor signaling (2, 8, 43, 52, 53).
An important breakthrough in our understanding of the mechanism by which Cbl proteins mediate their negative regulatory function came with the discovery that Cbl proteins act as ubiquitin ligases. Modification of receptor tyrosine kinases with ubiquitin terminates signaling by targeting these receptors for degradation (35, 50). Ubiquitination is a sequential process involving a ubiquitin-activating enzyme, or E1, a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). Cbl proteins have been shown to act as ubiquitin protein ligases by mediating the transfer of ubiquitin onto receptor tyrosine kinases, so enhancing their degradation (15, 23, 30). The conserved Cbl amino-terminal TKB, linker, and RING finger domains are sufficient for ubiquitin ligase activity (24).
Cbl-3 is the most recent mammalian cbl family member to be identified in humans (17, 20) and mice (9). Like other cbl's, Cbl-3 contains conserved amino-terminal TKB, linker, and RING finger regions. However, Cbl-3 possesses a much shorter proline-rich region than c-Cbl and Cbl-b and lacks their carboxy-terminal leucine zipper motif (17, 20). Nonetheless, the proline-rich region of Cbl-3 has been demonstrated to bind to various SH3 domain-containing proteins, including those of src family kinases such as Fyn (17, 20). An alternatively spliced form of human Cbl-3 termed Cbl-3S, which lacks part of the TKB domain, has also been reported in humans (17). Overexpression studies have shown that, like other cbl's, Cbl-3 is recruited to the EGFR upon EGF stimulation (17, 20). Cbl-3 functions as a negative regulator since overexpression of this protein attenuates EGF-stimulated mitogen-activated protein kinase activation and Elk-1 transactivation in 293T cells (17). Moreover, Cbl-3 appears to have ubiquitin ligase activity, since its overexpression results in enhanced endocytosis, ubiquitination, and degradation of the EGFR upon EGF stimulation (23). Cbl-3 has also been found to interact with the homologous-to-E6-associated-protein-C-terminus domain-containing E3 ligase Itch/AIP4 (5).
Cbl-3 has an expression pattern distinct from those of the other mammalian family members. While c-Cbl and Cbl-b are highly expressed in hematopoietic tissues and the testes (18, 22), Cbl-3 is not expressed in these organs and is instead expressed in the gastrointestinal tract, liver, kidney, pancreas, and prostate (17, 20). However, the physiological roles of Cbl-3 remain unknown. Here we report the detailed expression pattern of murine Cbl-3 and utilize gene-targeted deletion of Cbl-3 to characterize the requirements for this protein in vivo.
MATERIALS AND METHODS
Generation of Cbl-3-deficient mice.
A human CBL-3 cDNA probe was used to screen a mouse 129/J genomic phage library. Two overlapping clones that contained a region identical to exon 1 of mouse Cbl-3 (9) were isolated. A targeting vector was constructed in the plasmid pPNT containing a 5-kb long arm consisting of the Cbl-3 promoter region, cDNA encoding LacZ, PGK-Neor, a 650-bp short arm consisting of part of the first intron of Cbl-3, and a thymidine kinase cassette. The lacZ start codon was inserted in place of the Cbl-3 start codon by restriction digestion of the identical Kozak consensus sequences of Cbl-3 and lacZ with NcoI, followed by ligation of the long arm to lacZ.
The targeting vector was linearized with NotI and electroporated into E14K embryonic stem (ES) cells. The ES cells were selected in medium containing 300 μg of G418/ml and 2 μM ganciclovir. Four hundred G418- and ganciclovir-resistant ES cell colonies were picked and screened for homologous recombination by using PCR with two sets of primers, one of which was specific for the neomycin resistance cassette (5′-CCA GCT CAT TCC TCC CAC TC-3′ and 5′-GGA AGG ATT GGA GCT ACG GGG GTG-3′) and the other of which comprised flanking primers specific for a region of intron 1 (5′-GCC AGA CCC AAA TCA CCA GTT TAG-3′ and 5′-AGT TTA GAC CTT GCT CTG GGT TCC-3′). Positive colonies were further analyzed by digestion of their genomic DNA with SpeI, followed by Southern blotting with a 250-bp external random hexamer 32P-labeled DNA probe located 5′ of the long arm to confirm that homologous recombination had occurred. To confirm that the ES cells had undergone a single recombination event, genomic DNA was digested with NcoI and hybridized with a Neor gene-specific probe.
Six correctly targeted ES cell clones were identified, and four were microinjected into 3.5-day-old C57BL/6J blastocysts. Backcrossing of chimeric males with C57BL/6 females resulted in establishment of three independent Cbl-3−/− mouse strains as verified by Southern blotting. For PCR genotyping, the mutant locus was detected as described above and the wild-type locus was detected with a primer specific for Cbl-3 exon 1 (5′-CAG CTA CTT GGA GAG GTG GCA AAG-3′) and the flanking primers described above. No differences in phenotypes were detected among Cbl-3-deficient mice derived from the three different ES cell clones. Mice were maintained at the animal facilities of the Ontario Cancer Institute under specific-pathogen-free conditions according to institutional guidelines.
Northern blot analysis.
To confirm loss of Cbl-3 expression in gene-targeted mice, total RNA was isolated from various mouse tissues by using TRIZOL reagent (Gibco BRL) according to the manufacturer's instructions. For Northern analysis, total RNA (30 μg/lane) was electrophoresed through 1% agarose-formaldehyde gels. Northern blots were hybridized with Cbl-3- or β-actin (Clontech)-specific [α-32P]dCTP-labeled cDNA probes. Two Cbl-3-specific probes were used that recognized either exons 2 to 7 or exons 4 to 8 (both of which are 3′ of the targeted region), with comparable results.
Detection of β-galactosidase expression.
Immediately after sacrifice, mice were perfused with ice-cold Ca2+- and Mg2+-free phosphate-buffered saline (PBS) followed by 1% glutaraldehyde. Organs were removed and further fixed for 1 h at room temperature in fixative solution containing 0.2% glutaraldehyde, 2% formaldehyde, 2 mM MgCl2, and 5 mM EGTA in 0.1 M phosphate buffer. After being washed in wash buffer containing 2 mM MgCl2, 0.02% NP-40, and 0.01% sodium deoxycholate in 0.1 M phosphate buffer, organs were incubated for 1 h in 15% sucrose in PBS at 4°C and then overnight in 30% sucrose in PBS at 4°C. Organs were then embedded in Tissue Tek OCT compound (Sakura) and frozen on dry ice. Cryosections (10 and 40 μm) were fixed again in fixative solution minus formaldehyde for 10 min, washed three times in wash buffer, and stained overnight at 37°C in wash buffer containing 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 1 mg of 4-chromo-5-bromo-3-indolyl-β-d-galactosidase (X-Gal; Gibco BRL)/ml. X-Gal is hydrolyzed by β-galactosidase to form a blue precipitate. Slides were counterstained with nuclear fast red, dehydrated, and mounted in Entellan mounting medium.
Analysis of Cbl-3 expression by in situ hybridization.
Mice were sacrificed by cervical dislocation and immediately perfused with ice-cold Ca2+- and Mg2+-free PBS for 3 min followed by 4% paraformaldehyde (PFA) in Ca2+- and Mg2+-free PBS for 10 min. Organs were removed and fixed further in 4% PFA overnight at 4°C, dehydrated, and embedded in paraffin blocks. In situ hybridization of paraffin sections was performed as described previously (14).
Histology and immunohistochemistry.
For hematoxylin and eosin (H&E) staining, tissues were fixed with 4% PFA as described above and 3-μm-thick paraffin sections were stained with H&E. For immunohistochemistry, heat-induced epitope retrieval was carried out in 10 mM citrate buffer, pH 6.0. Sections were incubated with monoclonal rat anti-mouse Ki67 (DAKO) at a 1/150 dilution for 1 h at room temperature, followed by biotinylated rabbit anti-rat immunoglobulin G, streptavidin-horseradish peroxidase, and diaminobenzidine tetrahydrochloride reagent.
Isolation and stimulation of primary keratinocytes.
Keratinocytes were isolated from 1- to 5-day-old littermate mice as previously described (13), with the following modifications: (i) skins were floated overnight in Dispase II (Roche) to separate dermises from epidermises, and (ii) keratinocytes were cultured on collagen I- or IV-coated plates (BD Biocoat cellware) in KGM-2 medium (Clonetics). Once keratinocytes were almost confluent, cells were starved overnight in KGM (Clonetics) containing only antibiotics and 0.1% fatty acid-free bovine serum albumin (Sigma). Keratinocytes were stimulated in starvation medium containing 10 ng of EGF (Peprotech)/ml and lysed in radioimmunoprecipitation assay buffer. Western blots were probed with anti-phospho-Erk1,2 (Cell Signaling), stripped, and reprobed with antibody that detects total Erk1 and Erk2 proteins (Cell Signaling). To detect protein expression levels, lysates of unstimulated keratinocytes were probed with rabbit anti-c-Cbl (Santa Cruz), mouse anti-Cbl-b (Santa Cruz), or rabbit anti-keratin 1 (Cedarlane).
RESULTS
Targeting of the murine Cbl-3 gene.
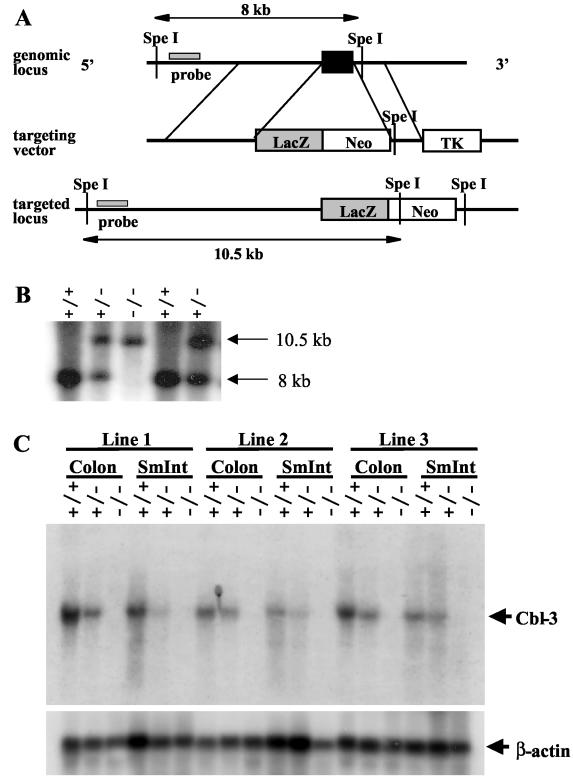
To investigate the in vivo functions of Cbl-3, we generated gene-targeted Cbl-3-deficient mice. The murine Cbl-3 gene was disrupted in ES cells by using a targeting vector that removed exon 1 of Cbl-3 and replaced it with the neomycin resistance cassette (Fig. 1A). Exon 1 encodes part of the TKB domain, including residues that have been shown in structure-function studies of c-Cbl to be essential for TKB domain integrity and thus c-Cbl function (28). To allow investigation of the expression pattern of Cbl-3, we inserted the lacZ gene, which codes for β-galactosidase, at the Cbl-3 start codon. As a result, β-galactosidase should be expressed in place of Cbl-3 in mice with the targeted allele. Four independent Cbl-3+/− ES cell clones that had undergone homologous recombination were injected into C57BL/6 blastocysts, and three of these clones gave rise to germ line transmission. Heterozygous mice were intercrossed to produce homozygous mutant offspring. Loss of both copies of the wild-type Cbl-3 allele was confirmed by Southern blotting (Fig. 1B).
FIG. 1.
Gene targeting of Cbl-3. (A) Exon 1 of Cbl-3 is shown as a black box. The flanking probe and the expected fragment sizes after SpeI digestion of wild-type (8 kb) and mutant (10.5 kb) genomic DNA are indicated. Neo, neomycin resistance cassette; TK, thymidine kinase. (B) Southern blot analysis of genomic DNA from tails of offspring from Cbl-3 heterozygote intercrosses showing germ line transmission of the targeted allele. Genomic DNA was digested with SpeI and hybridized to the 5′ flanking probe shown in panel A. Sizes of bands representing the wild-type and mutant Cbl-3 alleles are indicated. (C) Northern blot analysis of RNA isolated from the colons and small intestines (SmInt) of Cbl-3+/+, Cbl-3+/−, and Cbl-3−/− mice. The blot was hybridized with a probe specific for Cbl-3 exons 2 to 7 (outside the targeted region) and then stripped and reprobed for β-actin as a loading control. Loss of Cbl-3 mRNA expression can be seen in tissues of Cbl-3−/− mice from all three cell lines that underwent germ line transmission.
Since an antibody that recognizes the carboxy terminus of murine Cbl-3 is not available, we verified that gene targeting had produced a null mutation by using Northern blot analysis. It has previously been shown by Northern blotting that human CBL-3 is highly expressed in the colon and small intestine (17, 20). RNA was extracted from Cbl-3+/+, Cbl-3+/−, and Cbl-3−/− mice, and Cbl-3 expression was analyzed by Northern blotting with a probe outside of the targeted region. Although Cbl-3 expression was seen in colons and small intestines from Cbl-3+/+ mice, Cbl-3+/− mice exhibited a gene dose-dependent decrease in expression levels, and no Cbl-3 transcript was detected in tissues from Cbl-3−/− mice derived from all three ES cell clones, indicating that gene targeting had been successful (Fig. 1C).
Cbl-3 is not required for viability.
To determine whether Cbl-3 is required during development, offspring from Cbl-3+/− intercrosses were genotyped by PCR at 3 weeks of age. Normal Mendelian ratios of Cbl-3+/+, Cbl-3+/−, and Cbl-3−/− offspring were observed, indicating that Cbl-3 is not required for embryogenesis or postnatal survival (Table 1). Breeding of male and female Cbl-3−/− mice resulted in healthy offspring, indicating that Cbl-3 is not necessary for fertility. Cbl-3−/− mice did not exhibit any increases in mortality or disease compared to Cbl-3+/− or Cbl-3+/+ control littermates up to 18 months of age. Thus, Cbl-3 is dispensable for development and is not required for viability or fertility.
TABLE 1.
Offspring from Cbl-3+/− intercrosses are born at Mendelian frequencies
| Cbl-3 genotype | No. of mice with genotype from line:
|
% of mice with genotype from line:
|
||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | |
| +/+ | 55 | 67 | 9 | 24 | 26 | 28 |
| +/− | 123 | 131 | 15 | 54 | 52 | 47 |
| −/− | 48 | 56 | 8 | 21 | 22 | 25 |
Analysis of Cbl-3 expression.
Although Cbl family members are critical regulators of tyrosine kinase signaling, nothing was known about their expression patterns beyond results of whole-organ Northern blot analysis of adult tissues. Nonetheless, previous studies indicate that different Cbl proteins are differentially expressed in various tissues, suggesting that these proteins may play distinct biological roles. For example, although c-Cbl and Cbl-b are widely expressed, highest expression levels are seen in hematopoietic tissues and testes (18, 21). In contrast, whole-organ Northern blot analysis of human Cbl-3 shows highest expression levels in the small intestine and colon, with expression also seen in the pancreas, prostate, liver, kidney, and trachea (17, 20). To confirm that mouse Cbl-3 has a similar expression pattern, we analyzed Cbl-3 expression in various murine tissues by Northern blotting. In addition to that in the mouse small intestine and colon, Cbl-3 expression was seen in mouse kidney, liver, and trachea but not in mouse thymus or spleen or in murine embryonic fibroblasts (data not shown). Cbl-3 expression is observed only late in embryogenesis, starting at embryonic day 15. Thus, the expression pattern of Cbl-3 is conserved between mice and humans.
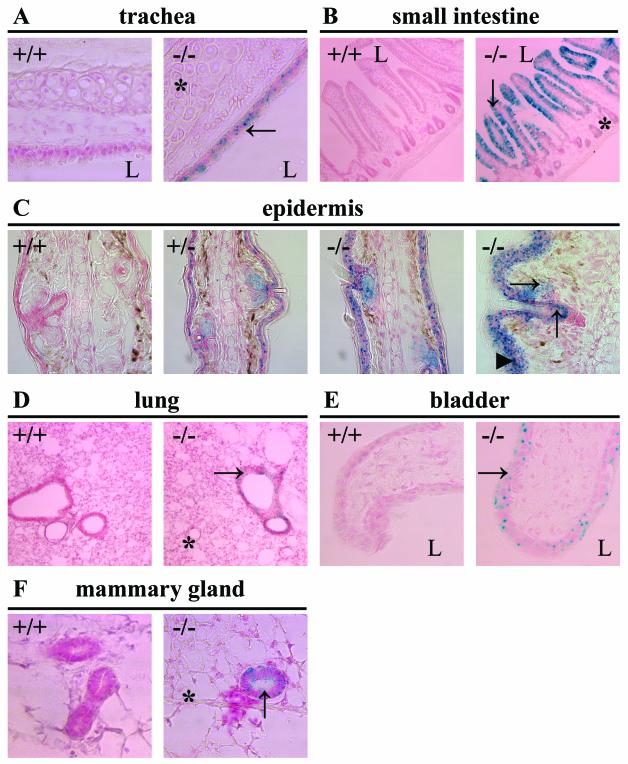
Since organs where Cbl-3 expression is detected contain multiple tissue and cell types—for example, smooth muscle, connective tissue, and epithelial cells in the intestine—Northern blotting could not reveal the cell type specificity of Cbl-3 expression. Thus, we made use of the lacZ gene, which was inserted in place of Cbl-3 exon 1 by our gene targeting strategy, to facilitate the analysis of Cbl-3 expression. X-Gal staining for β-galactosidase activity was performed on various tissues from Cbl-3+/+, Cbl-3+/−, and Cbl-3−/− mice. In agreement with previous Northern blotting results (17, 20), we observed X-Gal staining in the tracheas, small intestines, kidney tubules, and livers of Cbl-3+/− and Cbl-3−/− mice (Fig. 2 and data not shown). Interestingly, in all these tissues, X-Gal staining was found only in epithelial cells. In the small intestines, X-Gal staining was detected in epithelial cells of the villi but not in those of the crypts. The stratified squamous epithelia of the rectums and biliary epithelial cells of the livers were also positive for β-galactosidase activity. However, X-Gal staining was not detected in the pancreases or prostates, which do show Cbl-3 expression in humans as detected by Northern blot analysis (17, 20). X-Gal staining also revealed novel Cbl-3 expression in the epidermises, bronchi and bronchioles of the lungs, transitional epithelia of the bladders, mammary epithelia, uterine endometria, vaginas, epithelial linings of the epididymides, esophagi, and stomachs (Fig. 2 and data not shown). Consistent with results of Northern analysis, organs such as the lymph nodes, thymuses, hearts, and skeletal muscles showed no increase in X-Gal staining in Cbl-3+/− and Cbl-3−/− mice. Additionally, no X-Gal staining was seen in the bones or the eyes.
FIG. 2.
Analysis of Cbl-3 expression by X-Gal staining. (A) Tracheas. Cbl-3 expression is seen in the epithelium (→) but not the cartilage (*). (B) Small intestines. Cbl-3 is expressed in the villi (↓) but not the crypts (*). (C) Ears. Cbl-3 expression is seen in the suprabasal layers of the epidermis (▴), the hair follicle (↑), and the sebaceous gland (→). (D) Lungs. Cbl-3 is expressed in the bronchioles (→) but not the alveoli (*). (E) Bladders. Cbl-3 is expressed in the transitional epithelium (→). (F) Mammary glands. Cbl-3 is expressed in the epithelium (↑) but not the fat pad (*). Cryosections from Cbl-3+/+ (+/+), Cbl-3+/− (+/−), and Cbl-3−/− (−/−) mice were stained with X-Gal and counterstained with nuclear fast red. Since Cbl-3−/− mice express lacZ under the control of the Cbl-3 promoter, cells that would normally express Cbl-3 express β-galactosidase instead, resulting in hydrolysis of X-Gal to form a blue precipitate. Tissues from Cbl-3+/− mice showed an X-Gal staining pattern similar to, but fainter than, that of Cbl-3−/− tissues. L, lumen.
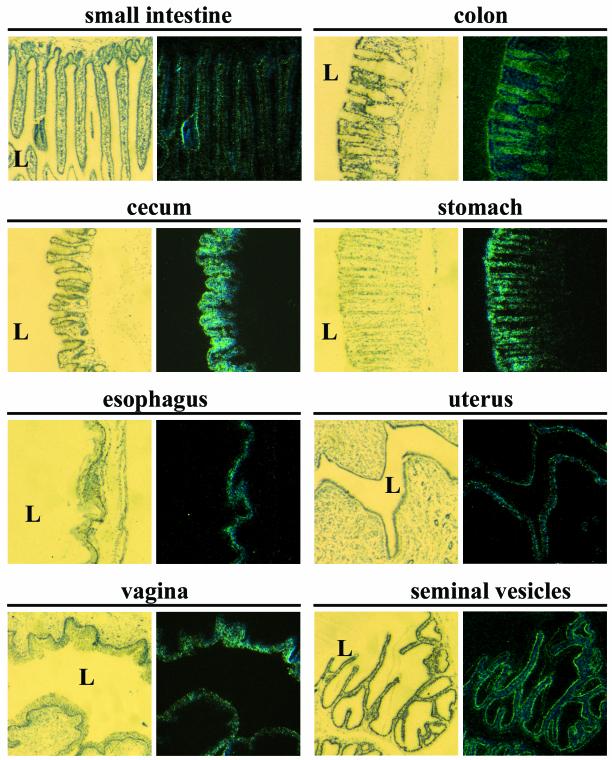
To confirm that β-galactosidase expression truly reflects the endogenous expression pattern of Cbl-3, in situ hybridization was performed (Fig. 3). In the small intestines, Cbl-3 expression was stronger in the villi than it was in the crypts. Highest expression levels of Cbl-3 were observed in the gastrointestinal tracts, especially in the colons, ceca, and gastric mucosae, with expression also seen in the esophagi (Fig. 3). In confirmation of the X-Gal staining pattern, Cbl-3 expression was seen in the epidermises, large airways (but not alveoli) of the lungs, and the respiratory epithelia of the tracheas (data not shown). The kidneys and bladders showed Cbl-3 expression in the tubules and transitional epithelia, respectively (data not shown). In the reproductive organs, Cbl-3 was strongly expressed in the endometria in the uteruses, the stratified squamous epithelia of the vaginas, and the epithelial linings of the seminal vesicles (Fig. 3). Cbl-3 was expressed at lower levels in the mammary gland epithelia, whereas little or no Cbl-3 expression was seen in the testes (data not shown). Thus, while Cbl-3 is expressed in numerous organs, its expression is restricted to epithelial cells.
FIG. 3.
In situ hybridization analysis of Cbl-3 expression. Tissue sections from wild-type mice were hybridized with a 33P-labeled RNA probe spanning Cbl-3 exons 2 to 8 (3′ of targeted region). The left column shows microscope images taken under bright field illumination to visualize the toluidine blue counterstain. The right column shows the corresponding dark field images for visualization of Cbl-3 expression. No hybridization was seen with a sense probe. L, lumen.
Normal epithelial tissue development in the absence of Cbl-3.
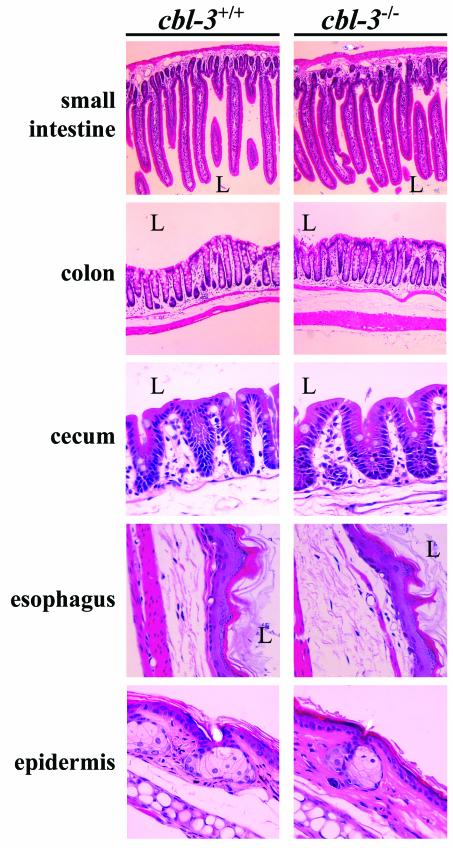
EGFR signaling is essential for the development of multiple epithelial tissues, and EGFR−/− mice show impaired development of the lungs and gastrointestinal tracts, as well as defects in epidermal and hair follicle differentiation (29, 41, 45). Since our analysis of the expression pattern of Cbl-3 revealed that this adapter is specifically expressed in the epithelial tissues of multiple organs, we analyzed these organs for any developmental defects due to the absence of Cbl-3. Tissues were isolated from Cbl-3+/+, Cbl-3+/−, and Cbl-3−/− mice and stained with H&E to allow identification of any gross histological defects. However, examination of multiple tissues including the small intestines, colons, ceca, stomachs, esophagi, epidermises, tracheas, lungs, kidneys, bladders, mammary glands, uteruses, and epididymides revealed no discernible differences between Cbl-3-deficient and control tissues (Fig. 4 and data not shown). H&E staining of tissues from 15- to 18-month-old mice did not reveal any histological defects, indicating that Cbl-3 deficiency did not lead to epithelial defects in older animals. Even areas that showed highest levels of Cbl-3 expression, such as the gastrointestinal tracts, displayed no defects in Cbl-3−/− mice. Comparison of carmine-stained mammary gland whole mounts from virgin (days 30, 40, 50, 60, 70, and 80), pregnant (day 12 of pregnancy), and involuting (days 4 and 7 after weaning) mice did not reveal any obvious differences between Cbl-3+/+, Cbl-3 +/−, and Cbl-3−/− mammary glands (data not shown). Furthermore, no functional defects were seen in Cbl-3-deficient mammary tissue since Cbl-3−/− mice were able to support healthy litters. Thus, Cbl-3 is not required for the development of epithelial tissues in vivo.
FIG. 4.
Tissue morphology is unaffected by Cbl-3 deficiency. Tissues from Cbl-3+/+ and Cbl-3−/− mice were stained with H&E. Tissues from five young mice per genotype (three males and two females, ages 10 to 12 weeks), and four old mice per genotype (two males and two females, ages 15 to 18 months) derived from two different Cbl-3+/− ES cell lines were analyzed. Pictures of tissues from young mice are shown. L, lumen.
Cbl-3 is not required for regulation of epithelial cell proliferation in the epidermis or gastrointestinal tract.
The EGFR is a critical regulator of epithelial cell proliferation (11, 12, 46). Since Cbl-3 has been shown to negatively regulate EGFR signaling (17), we evaluated whether loss of Cbl-3 expression resulted in changes in epithelial proliferation. Tissues were isolated from Cbl-3+/+ and Cbl-3−/− mice, and sections were stained with anti-Ki67 antibody to visualize proliferating cells. Ki67 is a nuclear antigen that is expressed in the G1, S, G2, and M phases of the cell cycle but is absent in resting (G0) cells (39). In the small intestine, colon, and cecum, proliferating cells are normally localized in the crypts, whereas in the stratified squamous epithelium of the esophagus and epidermis, proliferation occurs only in the basal layer (3, 42, 46). Since Cbl-3 is expressed in the villi but not the crypts of the small intestine, we hypothesized that Cbl-3 may negatively regulate proliferation in the villus epithelium. However, in both Cbl-3+/+ and Cbl-3−/− small intestines, proliferation was limited to the crypts (Fig. 5). Also, similar levels of proliferating cells were observed in the stomachs, the crypts of the colons and ceca, and the basal layers of the esophagi and epidermises of both Cbl-3+/+ and Cbl-3−/− mice. Thus, basal levels of proliferation in the epidermis and gastrointestinal tract appear to be unaffected by Cbl-3 deficiency.
FIG. 5.
Normal levels of basal proliferation in the absence of Cbl-3. Tissues sections from Cbl-3+/+ and Cbl-3−/− mice were stained with anti-Ki67 to visualize proliferating cells. We analyzed tissues from four different 10- to 12-week-old mice per genotype (two males and two females with littermate controls derived from two different Cbl-3+/− ES cell lines). L, lumen.
EGF-stimulated Erk activation in primary keratinocytes.
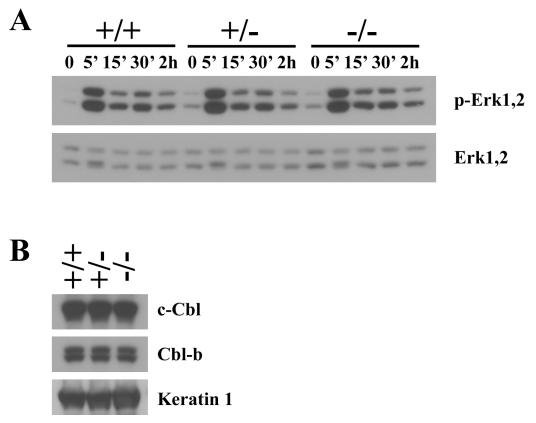
It has been previously shown that Cbl-3 overexpression in cell lines inhibits EGF-induced Erk activation by reducing the duration of Erk2 phosphorylation (17). To investigate whether loss of Cbl-3 would result in enhanced Erk activation in primary cells, we examined EGF signaling in keratinocytes, which were found by X-Gal staining (Fig. 2) and in situ hybridization (data not shown) to express Cbl-3. We isolated primary keratinocytes from Cbl-3+/+, Cbl-3+/−, and Cbl-3−/− littermate mice and assessed levels of Erk1 and Erk2 phosphorylation in response to EGF stimulation (Fig. 6A). In contrast to the findings of the overexpression studies, loss of Cbl-3 did not alter the level or kinetics of Erk activation or dephosphorylation. To assess whether overexpression of c-Cbl or Cbl-b may be compensating for the lack of Cbl-3 in these cells, we measured c-Cbl and Cbl-b protein levels by Western blotting. There was no change in c-Cbl or Cbl-b protein levels in Cbl-3-deficient keratinocytes compared to those in control keratinocytes (Fig. 6B). However, primary keratinocytes do express significant levels of c-Cbl and Cbl-b, indicating that endogenous levels of other cbl family members may be able to compensate for the absence of Cbl-3.
FIG. 6.
EGF-induced Erk phosphorylation in primary keratinocytes. (A) Primary keratinocytes isolated from Cbl-3+/+, Cbl-3+/−, and Cbl-3−/− mice were stimulated with 10 ng of EGF/ml for the indicated time periods. Active Erk1 and Erk2 were detected by Western blotting by using phospho-specific antibody. Levels of total Erk1 and Erk2 protein are shown in the lower panel. Results are representative of those from five independent experiments. (B) c-Cbl and Cbl-b expression. Primary keratinocytes isolated from Cbl-3+/+, Cbl-3+/−, and Cbl-3−/− mice were lysed, and levels of c-Cbl, Cbl-b, and keratin 1 protein expression were assessed by Western blotting.
DISCUSSION
While all three mammalian cbl family members can attenuate EGFR signaling and mediate ubiquitination of the EGFR upon overexpression in cell lines, the properties unique to each family member are less clear. Gene targeting studies have demonstrated that different cbl family members have distinct physiological requirements. While c-Cbl regulates T-cell receptor signaling in thymocytes in vivo, the closely related Cbl-b is required for regulation of mature T-cell signaling instead (1, 4, 31, 32). The physiological roles of Cbl family members in nonhematopoietic tissues are not well defined. Although c-Cbl and Cbl-b are widely expressed, the generation of gene-targeted mice has shown that loss of either family member does not lead to defects in development, with the exception of increased ductal branching and density seen in the mammary glands of c-Cbl−/− mice (31). The cloning of the third mammalian Cbl family member, Cbl-3, in humans, along with the discovery that this protein exhibits structural dissimilarities and a distinct expression pattern compared with c-Cbl and Cbl-b, suggested that Cbl-3 may play a distinct role in vivo (17, 20). By analyzing the expression pattern of Cbl-3 in mice in detail, we demonstrate that Cbl-3 is expressed specifically in epithelial tissues, suggesting that it would play an important role in these tissues. However, by utilizing gene targeting to generate mice lacking Cbl-3 expression, we show that Cbl-3 is in fact not required for normal development or function of epithelial tissues in vivo.
Through a combined approach of X-Gal staining and in situ hybridization, we demonstrated that Cbl-3 expression is restricted to epithelial cells. Cbl-3 expression was not observed in nonepithelial tissues. Within the epithelial lineage, Cbl-3 expression levels varied significantly between different organs and different regions within an organ. For example, in the lungs, Cbl-3 expression was seen in the bronchi and bronchioles but not in the surrounding alveoli. Since other cbl family members have been shown to negatively regulate antigen receptor-stimulated proliferation in lymphocytes and EGFR signaling in cell lines, we hypothesized that Cbl-3 deficiency would lead to defects in development and/or increased proliferation due to enhanced signaling from growth factor receptors. However, surprisingly, no developmental abnormalities were seen in Cbl-3−/− mice. Proliferation in the small intestine is restricted to the crypts of Lieberkühn, whereas the villi are composed of differentiated enterocytes, goblet cells, and enteroendocrine cells (3, 33, 42). Since Cbl-3 expression is enriched in nonproliferative, differentiated cells of the small intestine, we hypothesized that Cbl-3 may negatively regulate the proliferative potential of these cells. However, basal levels of proliferation in the gastrointestinal tract and epidermis were unaffected by Cbl-3 deficiency. Moreover, the kinetics of EGF-induced mitogen-activated protein kinase activation in primary keratinocytes were unchanged by Cbl-3 deficiency.
The reasons for the lack of requirement for Cbl-3 for epithelial development and function in vivo are unclear, but this finding may be due to compensation between Cbl family members. Whereas C. elegans possesses only one Cbl gene, humans and mice have three different Cbl family members. The presence of multiple Cbl proteins in mammals, all with conserved TKB and RING finger domains, suggests that there may be functional compensation between Cbl family members. Indeed, while mice deficient in c-Cbl or Cbl-b display mild defects that are for the most part limited to the immune system, mice deficient in both c-Cbl and Cbl-b have an early embryonic lethal phenotype (25, 36). Although highest expression levels of c-Cbl and Cbl-b are found in hematopoietic tissues and the testes, these two proteins are also expressed in epithelial cells, including keratinocytes (10, 18, 26). To address the issue of functional redundancy, Cbl-3-deficient mice will be intercrossed with c-Cbl- and Cbl-b-deficient mice to generate Cbl-3/c-Cbl and Cbl-3/Cbl-b double-deficient animals.
Epithelial tissues undergo continuous regeneration, and most tumors arise from epithelial tissues (Ontario Cancer Registry, 2000). Alterations in EGFR signaling can lead to epithelial tumor formation. For example, human squamous cell carcinomas have a high frequency of EGFR gene amplifications, rearrangements, and overexpression (6, 37). Moreover, transgenic expression of transforming growth factor α or ErbB2 in the epidermis induces the development of papillomas and/or squamous cell carcinomas (7, 19, 49) and EGFR signaling has been found to be essential for Sos-dependent skin tumor formation (40). Since Cbl-3 has been shown to negatively regulate EGFR signaling in cell lines, the crossing of Cbl-3-, Cbl-3/c-Cbl-, and Cbl-3/Cbl-b-deficient mice with transgenic mice that are susceptible to epithelial tumors may help to reveal roles for this protein in tumor formation in vivo.
Acknowledgments
We thank B. Song, M. Zhao, S. Arya, A. Oliveira-dos-Santos, I. Kozieradzki, T. Wada, M. Crackower, N. Joza, M. Cheng, K. Bachmaier, R. Sarao, M. Nghem, and H. Jones for comments and assistance.
E.K.G. is supported by a scholarship from the Natural Sciences and Engineering Research Council (NSERC) of Canada. J.M.P. is supported by grants from the Canadian Institute for Health Research (CIHR), the National Cancer Institute of Canada (NCIC), and the Institute for Molecular Biotechnology of the Austrian Academy of Sciences (IMBA). J.M.P. holds a Canada research chair in cell biology.
REFERENCES
- 1.Bachmaier, K., C. Krawczyk, I. Kozieradzki, Y. Y. Kong, T. Sasaki, A. Oliveira-dos-Santos, S. Mariathasan, D. Bouchard, A. Wakeham, A. Itie, J. Le, P. S. Ohashi, I. Sarosi, H. Nishina, S. Lipkowitz, and J. M. Penninger. 2000. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature 403:211-216. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, C. A., V. Ribon, M. Kanzaki, D. C. Thurmond, S. Mora, S. Shigematsu, P. E. Bickel, J. E. Pessin, and A. R. Saltiel. 2000. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature 407:202-207. [DOI] [PubMed] [Google Scholar]
- 3.Bergman, R. A., A. K. Afifi, and P. M. Heidger. 1996. Histology. W. B. Saunders Company, Orlando, Fla.
- 4.Chiang, Y. J., H. K. Kole, K. Brown, M. Naramura, S. Fukuhara, R. J. Hu, I. K. Jang, J. S. Gutkind, E. Shevach, and H. Gu. 2000. Cbl-b regulates the CD28 dependence of T-cell activation. Nature 403:216-220. [DOI] [PubMed] [Google Scholar]
- 5.Courbard, J. R., F. Fiore, J. Adelaide, J. P. Borg, D. Birnbaum, and V. Ollendorff. 2002. Interaction between two ubiquitin-protein isopeptide ligases of different classes, CBLC and AIP4/ITCH. J. Biol. Chem. 277:45267-45275. [DOI] [PubMed] [Google Scholar]
- 6.Derynck, R. 1992. The physiology of transforming growth factor-alpha. Adv. Cancer Res. 58:27-52. [DOI] [PubMed] [Google Scholar]
- 7.Dominey, A. M., X. J. Wang, L. E. King, Jr., L. B. Nanney, T. A. Gagne, K. Sellheyer, D. S. Bundman, M. A. Longley, J. A. Rothnagel, and D. A. Greenhalgh. 1993. Targeted overexpression of transforming growth factor alpha in the epidermis of transgenic mice elicits hyperplasia, hyperkeratosis, and spontaneous, squamous papillomas. Cell Growth Differ. 4:1071-1082. [PubMed] [Google Scholar]
- 8.Feshchenko, E. A., S. K. Shore, and A. Y. Tsygankov. 1999. Tyrosine phosphorylation of C-Cbl facilitates adhesion and spreading while suppressing anchorage-independent growth of V-Abl-transformed NIH3T3 fibroblasts. Oncogene 18:3703-3715. [DOI] [PubMed] [Google Scholar]
- 9.Fiore, F., V. Ollendorff, and D. Birnbaum. 2001. Characterization of the mouse Cblc/Cbl3 gene. Biochem. Biophys. Res. Commun. 280:182-187. [DOI] [PubMed] [Google Scholar]
- 10.Fukazawa, T., S. Miyake, V. Band, and H. Band. 1996. Tyrosine phosphorylation of Cbl upon epidermal growth factor (EGF) stimulation and its association with EGF receptor and downstream signaling proteins. J. Biol. Chem. 271:14554-14559. [DOI] [PubMed] [Google Scholar]
- 11.Giraud, A. S. 2000. X. Trefoil peptide and EGF receptor/ligand transgenic mice. Am. J. Physiol. Gastrointest. Liver Physiol. 278:G501-G506. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto, K. 2000. Regulation of keratinocyte function by growth factors. J. Dermatol. Sci. 24(Suppl. 1):S46-S50. [DOI] [PubMed] [Google Scholar]
- 13.Hennings, H. 1994. Primary culture of keratinocytes from newborn mouse epidermis in medium with lowered levels of Ca2+, p. 21-23. In I. Leigh and F. Watt (ed.), Keratinocyte methods. Cambridge University Press, Cambridge, United Kingdom.
- 14.Hui, C. C., and A. L. Joyner. 1993. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet. 3:241-246. [DOI] [PubMed] [Google Scholar]
- 15.Joazeiro, C. A., S. S. Wing, H. Huang, J. D. Leverson, T. Hunter, and Y. C. Liu. 1999. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286:309-312. [DOI] [PubMed] [Google Scholar]
- 16.Jongeward, G. D., T. R. Clandinin, and P. W. Sternberg. 1995. Sli-1, a negative regulator of let-23-mediated signaling in C. elegans. Genetics 139:1553-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keane, M. M., S. A. Ettenberg, M. M. Nau, P. Banerjee, M. Cuello, J. Penninger, and S. Lipkowitz. 1999. Cbl-3: a new mammalian cbl family protein. Oncogene 18:3365-3375. [DOI] [PubMed] [Google Scholar]
- 18.Keane, M. M., O. M. Rivero-Lezcano, J. A. Mitchell, K. C. Robbins, and S. Lipkowitz. 1995. Cloning and characterization of cbl-b: a SH3 binding protein with homology to the c-cbl proto-oncogene. Oncogene 10:2367-2377. [PubMed] [Google Scholar]
- 19.Kiguchi, K., D. Bol, S. Carbajal, L. Beltran, S. Moats, K. Chan, J. Jorcano, and J. DiGiovanni. 2000. Constitutive expression of erbB2 in epidermis of transgenic mice results in epidermal hyperproliferation and spontaneous skin tumor development. Oncogene 19:4243-4254. [DOI] [PubMed] [Google Scholar]
- 20.Kim, M., T. Tezuka, Y. Suziki, S. Sugano, M. Hirai, and T. Yamamoto. 1999. Molecular cloning and characterization of a novel cbl-family gene, cbl-c. Gene 239:145-154. [DOI] [PubMed] [Google Scholar]
- 21.Langdon, W. Y., J. W. Hartley, S. P. Klinken, S. K. Ruscetti, and H. C. Morse III. 1989. v-cbl, an oncogene from a dual-recombinant murine retrovirus that induces early B-lineage lymphomas. Proc. Natl. Acad. Sci. USA 86:1168-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langdon, W. Y., C. D. Hyland, R. J. Grumont, and H. C. Morse III. 1989. The c-cbl proto-oncogene is preferentially expressed in thymus and testis tissue and encodes a nuclear protein. J. Virol. 63:5420-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levkowitz, G., H. Waterman, S. A. Ettenberg, M. Katz, A. Y. Tsygankov, I. Alroy, S. Lavi, K. Iwai, Y. Reiss, A. Ciechanover, S. Lipkowitz, and Y. Yarden. 1999. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 4:1029-1040. [DOI] [PubMed] [Google Scholar]
- 24.Lill, N. L., P. Douillard, R. A. Awwad, S. Ota, M. L. Lupher, Jr., S. Miyake, N. Meissner-Lula, V. W. Hsu, and H. Band. 2000. The evolutionarily conserved N-terminal region of Cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J. Biol. Chem. 275:367-377. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Y. C., and H. Gu. 2002. Cbl and Cbl-b in T-cell regulation. Trends Immunol. 23:140-143. [DOI] [PubMed] [Google Scholar]
- 26.Meisner, H., and M. P. Czech. 1995. Coupling of the proto-oncogene product c-Cbl to the epidermal growth factor receptor. J. Biol. Chem. 270:25332-25335. [DOI] [PubMed] [Google Scholar]
- 27.Meisner, H., A. Daga, J. Buxton, B. Fernandez, A. Chawla, U. Banerjee, and M. P. Czech. 1997. Interactions of Drosophila Cbl with epidermal growth factor receptors and role of Cbl in R7 photoreceptor cell development. Mol. Cell. Biol. 17:2217-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng, W., S. Sawasdikosol, S. J. Burakoff, and M. J. Eck. 1999. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature 398:84-90. [DOI] [PubMed] [Google Scholar]
- 29.Miettinen, P. J., J. E. Berger, J. Meneses, Y. Phung, R. A. Pedersen, Z. Werb, and R. Derynck. 1995. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature 376:337-341. [DOI] [PubMed] [Google Scholar]
- 30.Miyake, S., M. L. Lupher, Jr., B. Druker, and H. Band. 1998. The tyrosine kinase regulator Cbl enhances the ubiquitination and degradation of the platelet-derived growth factor receptor alpha. Proc. Natl. Acad. Sci. USA 95:7927-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy, M. A., R. G. Schnall, D. J. Venter, L. Barnett, I. Bertoncello, C. B. Thien, W. Y. Langdon, and D. D. Bowtell. 1998. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol. Cell. Biol. 18:4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naramura, M., H. K. Kole, R. J. Hu, and H. Gu. 1998. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc. Natl. Acad. Sci. USA 95:15547-15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pacha, J. 2000. Development of intestinal transport function in mammals. Physiol. Rev. 80:1633-1667.11015621 [Google Scholar]
- 34.Pai, L. M., G. Barcelo, and T. Schupbach. 2000. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell 103:51-61. [DOI] [PubMed] [Google Scholar]
- 35.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 36.Rao, N., I. Dodge, and H. Band. 2002. The Cbl family of ubiquitin ligases: critical negative regulators of tyrosine kinase signaling in the immune system. J. Leukoc. Biol. 71:753-763. [PubMed] [Google Scholar]
- 37.Reichmann, E. 1994. Oncogenes and epithelial cell transformation. Semin. Cancer Biol. 5:157-165.8061331 [Google Scholar]
- 38.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211-225. [DOI] [PubMed] [Google Scholar]
- 39.Scholzen, T., and J. Gerdes. 2000. The Ki-67 protein: from the known and the unknown. J. Cell. Physiol. 182:311-322. [DOI] [PubMed] [Google Scholar]
- 40.Sibilia, M., A. Fleischmann, A. Behrens, L. Stingl, J. Carroll, F. M. Watt, J. Schlessinger, and E. F. Wagner. 2000. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell 102:211-220. [DOI] [PubMed] [Google Scholar]
- 41.Sibilia, M., and E. F. Wagner. 1995. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science 269:234-238. [DOI] [PubMed] [Google Scholar]
- 42.Stappenbeck, T. S., M. H. Wong, J. R. Saam, I. U. Mysorekar, and J. I. Gordon. 1998. Notes from some crypt watchers: regulation of renewal in the mouse intestinal epithelium. Curr. Opin. Cell Biol. 10:702-709. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka, S., M. Amling, L. Neff, A. Peyman, E. Uhlmann, J. B. Levy, and R. Baron. 1996. c-Cbl is downstream of c-Src in a signalling pathway necessary for bone resorption. Nature 383:528-531. [DOI] [PubMed] [Google Scholar]
- 44.Thien, C. B., and W. Y. Langdon. 2001. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell. Biol. 2:294-307. [DOI] [PubMed] [Google Scholar]
- 45.Threadgill, D. W., A. A. Dlugosz, L. A. Hansen, T. Tennenbaum, U. Lichti, D. Yee, C. LaMantia, T. Mourton, K. Herrup, R. C. Harris, J. A. Barnard, S. H. Yuspa, R. J. Coffey, and T. Magnuson. 1995. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269:230-234. [DOI] [PubMed] [Google Scholar]
- 46.Tomic-Canic, M., M. Komine, I. M. Freedberg, and M. Blumenberg. 1998. Epidermal signal transduction and transcription factor activation in activated keratinocytes. J. Dermatol. Sci. 17:167-181. [DOI] [PubMed] [Google Scholar]
- 47.Tsygankov, A. Y., A. M. Teckchandani, E. A. Feshchenko, and G. Swaminathan. 2001. Beyond the RING: CBL proteins as multivalent adapters. Oncogene 20:6382-6402. [DOI] [PubMed] [Google Scholar]
- 48.van der Geer, P., T. Hunter, and R. A. Lindberg. 1994. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu. Rev. Cell Biol. 10:251-337. [DOI] [PubMed] [Google Scholar]
- 49.Vassar, R., and E. Fuchs. 1991. Transgenic mice provide new insights into the role of TGF-alpha during epidermal development and differentiation. Genes Dev. 5:714-727. [DOI] [PubMed] [Google Scholar]
- 50.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell. Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 51.Yoon, C. H., J. Lee, G. D. Jongeward, and P. W. Sternberg. 1995. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c-cbl. Science 269:1102-1105. [DOI] [PubMed] [Google Scholar]
- 52.Zell, T., C. S. Warden, A. S. Chan, M. E. Cook, C. L. Dell, S. W. Hunt III, and Y. Shimizu. 1998. Regulation of beta 1-integrin-mediated cell adhesion by the Cbl adaptor protein. Curr. Biol. 8:814-822. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, Z., C. Elly, L. Qiu, A. Altman, and Y. C. Liu. 1999. A direct interaction between the adaptor protein Cbl-b and the kinase zap-70 induces a positive signal in T cells. Curr. Biol. 9:203-206. [DOI] [PubMed] [Google Scholar]