Abstract
The retinoblastoma tumor suppressor protein (RB) is targeted for inactivation in the majority of human tumors, underscoring its critical role in attenuating cellular proliferation. RB inhibits proliferation by repressing the transcription of genes that are essential for cell cycle progression. To repress transcription, RB assembles multiprotein complexes containing chromatin-modifying enzymes, including histone deacetylases (HDACs). However, the extent to which HDACs participate in transcriptional repression and are required for RB-mediated repression has not been established. Here, we investigated the role of HDACs in RB-dependent cell cycle inhibition and transcriptional repression. We find that active RB mediates histone deacetylation on cyclin A, Cdc2, topoisomerase IIα, and thymidylate synthase promoters. We also demonstrate that this deacetylation is HDAC dependent, since the HDAC inhibitor trichostatin A (TSA) prevented histone deacetylation at each promoter. However, TSA treatment blocked RB repression of only a specific subset of genes, thereby demonstrating that the requirement of HDACs for RB-mediated transcriptional repression is promoter specific. The HDAC-independent repression was not associated with DNA methylation or gene silencing but was readily reversible. We show that this form of repression resulted in altered chromatin structure and was dependent on SWI/SNF chromatin remodeling activity. Importantly, we find that cell cycle inhibitory action of RB is not intrinsically dependent on the ability to recruit HDAC activity. Thus, while HDACs do play a major role in RB-mediated repression, they are dispensable for the repression of critical targets leading to cell cycle arrest.
The retinoblastoma tumor suppressor, RB, functions as a negative regulator of cell cycle progression that is frequently inactivated in human cancers (10, 22, 75). In G0 and early G1 cells, RB is hypophosphorylated and inhibits the transition into the S phase of the cell cycle. Mitogenic signaling cascades activate CDK4/cyclin D1 complexes that initiate the phosphorylation of RB on a subset of serine and threonine residues (65). Subsequent phosphorylation catalyzed by CDK2/cyclin E leads to RB hyperphosphorylation (23). These combined events serve to functionally inactivate RB and thereby facilitate progression through the S phase (2, 23). In contrast with mitogenic signaling pathways, antimitogens (e.g., transforming growth factor β or DNA damage) serve to inhibit RB phosphorylation and prevent progression through the cell cycle (28). Thus, RB integrates multiple signaling cascades to modify proliferation. In cancer, RB is inactivated through the activity of several disparate mechanisms. These modes of inactivation include the biallelic inactivation of the RB gene, binding by oncoproteins of DNA tumor viruses, and aberrant phosphorylation (2, 3, 30, 59, 62, 76). Through these distinct mechanisms of RB inactivation, tumors are able to evade cell cycle regulation and proliferate uncontrollably.
RB inhibits cellular proliferation by assembling complexes involved in transcriptional repression. Biochemical analyses have shown that RB interacts with a plethora (>100) of different cellular proteins (46). The significance of most of these interactions remains elusive. However, the E2F family of transcription factors represents critical targets of RB (8, 16, 49). E2F complexes exist in vivo as heterodimers composed of subunits from E2F (E2F1 to E2F6) and DP (DP1 and DP2) gene families. E2F-DP heterodimers bind to specific DNA sequences and function as transcriptional activators. E2F-responsive genes include cell cycle regulators, such as cyclin E, cyclin A, cdc2, and cdk2 (5, 12, 18, 20, 29, 54, 61, 73), as well as factors important for DNA synthesis, including DNA polymerase α, thymidine kinase, and dihydrofolate reductase (31, 53, 57, 69). Recently, E2F proteins have been shown to directly interact with the promoters of many of these genes (72, 78).
Genetic and biochemical analyses have shown RB to functionally antagonize E2F activity (27, 64). In addition, we have recently shown that RB potently represses a significant number of E2F-regulated genes that are requisite for cell cycle progression (42). Currently, there are two models which describe how RB impinges upon E2F-directed transcription: (i) RB binds to the E2F family of transcription factors, thus blocking their transactivation capacity (17, 27), and (ii) RB assembles large multiprotein complexes at E2F-regulated promoters that actively repress transcription (7, 63, 77). A number of functional studies demonstrate that E2F-dependent repression is required for RB to inhibit proliferation. For example, E2F alleles which displace E2F/RB complexes from DNA inhibit RB-dependent cell cycle control (82).
To facilitate transcriptional repression, RB interacts not only with E2F but also with multiple corepressor molecules. Many of these interactions are mediated by LXCXE motifs that are present in the corepressor and which interact with the A/B pocket domain of RB (6, 15, 68). For example, RB interacts with histone deacetylases (HDAC) (6, 11, 36, 39, 40), ATP-dependent chromatin-modifying enzymes BRG-1 and BRM (15, 68), and histone methyltransferases (52) which contain LXCXE motifs. It is believed that these enzymes play critical roles in RB-mediated transcriptional repression. Such an idea is supported by the observation that the combined loss of BRG-1 and BRM compromise RB-mediated transcriptional repression and cell cycle inhibition (71). In contrast, the discrete role of HDACs in cell cycle control is less clear.
Acetylation and deacetylation of core histones is a key mechanism by which transcription can be altered (21). In general, transcriptional activators recruit histone acetyltransferases (HATs), which add acetyl groups to the lysine residues of histone tails. This acetylation neutralizes the positive charge on lysine, resulting in loosening of the chromatin structure. Such disruption is believed to unfold DNA, allowing basal transcription machinery to gain access to the promoter regions of genes that are to be activated. In contrast, HDACs remove acetyl groups from lysine residues of core histones, thereby preventing basal transcription machinery access to the promoter. Indeed, transcriptional repressors recruit HDACs and make use of their transcription suppression function (21, 24-26, 35, 51). HDACs belong to a family of enzymes which is divided into three different classes, encompassing >10 distinct proteins (43). Three of these HDACs (HDAC1 to HDAC3) have been shown to interact with RB (6, 9, 11, 36, 37, 39, 40). It was initially demonstrated that RB could recruit HDAC1 and mediate histone deacetylation at a synthetic promoter (39). Further studies have shown that several endogenous RB/E2F-regulated promoters exhibit changes in promoter histone acetylation as a function of cell cycle position (47, 58). However, no direct evidence has elucidated a role for HDACs in cell cycle inhibition and transcriptional repression mediated by RB.
Here we focused on delineating the role of HDACs in RB function. We found that RB-mediated repression occurs in concert with the deacetylation of histones. These deacetylation events are catalyzed through the action of HDACs, as addition of the HDAC inhibitor trichostatin A (TSA) prevents RB-mediated histone deacetylation. The functional requirement of HDACs exhibited promoter specificity, as only a subset of RB targets remained repressed when HDAC activity was inhibited. The HDAC-independent repression was shown to be readily reversible and independent of DNA methylation, and such repression required functional SWI/SNF activity. Importantly, inhibition of HDAC activity with TSA was not sufficient to overcome RB-mediated cell cycle arrest. Together, these data provide critical insights into the action of RB in transcriptional repression and the relative role of HDACs in cell cycle control.
MATERIALS AND METHODS
Cell culture.
The A5-1 cell line, harboring conditional expression of a phosphorylation site mutant RB (PSM-RB) (1), was cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-denatured fetal bovine serum, glutamine, penicillin-streptomycin, G418 (400 μg/ml), hygromycin B (200 μg/ml), and doxycycline (Dox; 1 μg/ml). To induce expression of PSM-RB, cells were washed with phosphate-buffered saline (PBS) and maintained in media lacking Dox for times as indicated. TSA (Sigma, St. Louis, Mo.) was added to the culture media in concentrations as described. Freshly prepared medium containing 5 μM 5-aza-2-deoxycytidine (Sigma) was added every 24 h for 96 h, and cells were subsequently maintained in the presence or absence of Dox for another 24 h.
Western blotting.
Immunoblotting was performed by following standard biochemical techniques. To detect histone H4 and acetylated histone H4, proteins were isolated by acid extraction. Antibodies against the following proteins were used: RB 851 (gift from Jean Wang), cyclin A (sc-751; Santa Cruz), Cdc2 (sc-747; Santa Cruz), topoisomerase IIα (topoIIα; TopoGen, Inc.), thymidylate synthase (TS; gift from Masakazu Fukushima), RNRII (sc-10848; Santa Cruz), BRG-1 (sc-17796; Santa Cruz), β-tubulin (sc-5274; Santa Cruz), FLAG M2 (F-3165; Sigma), histone H4 (07-108; Upstate Biotechnology), acetylated histone H4 (06-866; Upstate Biotechnology), HDAC1 (sc-7872; Santa Cruz), and HDAC3 (sc-11417; Santa Cruz).
Plasmids.
Primers were used to amplify from rat genomic DNA the following gene promoter regions: topoIIα, bases −217 to −19 (79); TS, bases −124 to −5 (38); Cdc2, bases −193 to −1 (66); cyclin A, bases −135 to +33 (67); and ribonucleotide reductase II, bases −310 to −26. These promoter regions were cloned into the firefly luciferase expression vector pGL2-Basic (Promega, Madison, Wis.) and then confirmed by DNA sequencing. The pTS-dnBRG-1 construct has been described previously (13, 14).
Generation of stable cell lines and transcriptional reporter assays.
The reporter constructs along with the empty vector pGL2-B were integrated into the A5-1 cell line, and three clones were isolated for each construct. Each clone was cultured in the presence or absence of Dox for 18 h. Cells were then harvested, and luciferase activity was quantified by using the Promega luciferase assay kit. Luciferase activity was normalized to total protein concentration by using the DC protein assay (Bio-Rad, Hercules, Calif.). Reporter assays were also performed on cells treated with a 100 nM concentration of the HDAC inhibitor TSA (Sigma) for 18 h in the presence or absence of Dox. The pTS-dnBRG-1 plasmid was transfected into A5-1 cells and selected for the stable inducible expression of dnBRG-1 by FLAG immunoblot.
RT-PCR.
A5-1 cells were maintained in the presence or absence of Dox for 18 h prior to RNA extraction by using TRIzol (Invitrogen, Carlsbad, Calif.). Reverse transcription (RT) of purified RNA was performed using oligo(dT) priming and Superscript II RT (Invitrogen). cDNA was then amplified for 24 to 28 cycles by using the following primer pairs: for topoIIα, 5′-TGCCCAGTTAGCTGGGTCAGTG-3′ and 5′-TGAGCATTGTAAAGATGTACCT-3′ (200 bp); for TS, 5′-TTTTATGTGGTGAATGGGGAGC-3′ and 5′-TGGGAAAGGTCTTGGTTCTCGC-3′ (231 bp); for Cdc2, 5′-GGATTGTGTTTTGTCACTCCCG-3′ and 5′-CCTATGCTCCAGATGTCAACCG-3′ (229 bp); for cyclin A, 5′-GAGAATGTCAACCCCGAAAAAG-3′ and 5′-TGGTGAAGGCAGGCTGTTTAC-3′ (205 bp); for RNRII, 5′-CTTCAACGCCATTGAGACAA-3′ and 5′-TCACAGTGCAGACCCTCATC-3′ (234 bp); for β-actin, 5′-ATGGATGACGATATCGCTGC-3′ and 5′-CTTCTGACCCATACCCACCA-3′ (150 bp); and for BRG1-FLAG, 5′-GCCCGTGGACTTCAAG-3′ and 5′-CGTCGTCCTTGTAGTCG-3′ (450 bp). PCR products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. RT-PCR was also performed on RNA extracted from A5-1 cells that had been treated with 100 nM TSA for 18 h in the presence or absence of Dox.
BrdU incorporation and flow cytometry.
A5-1 cells were cultured in the presence or absence of Dox and 100 nM TSA for 16 h. To detect progression through S phase, cells were pulse-labeled with bromodeoxyuridine (BrdU; Amersham). Following 8 h of labeling, cells were fixed and BrdU incorporation was detected by indirect immunofluorescence (anti-BrdU; Accurate Scientific). Results are representative of three independent experiments. Flow cytometry was performed as previously described (32).
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (78) with a few modifications. A5-1 cells were cultured in 15-cm culture plates with or without Dox and TSA for 24 h. Formaldehyde (Fisher Scientific) was added directly into the culture medium to a final concentration of 1% and fixed for 15 min at room temperature with mild shaking. To stop the fixation reaction, glycine was added to a final concentration of 0.125 M. Cells were then washed with ice-cold PBS and harvested by trypsinization (20% in PBS). Cells were lysed in cell lysis buffer (5 mM PIPES [pH 8.0], 85 mM KCl, 0.5% NP-40, 0.5 mM phenylmethylsulfonyl fluoride, 100 ng of leupeptin and aprotinin per ml) and then incubated on ice for 10 min. Nuclei were collected by microcentrifugation (3,000 × g for 5 min) and resuspended in nucleus lysis buffer (50 mM Tris-Cl [pH 8.1], 10 mM EDTA, 1% sodium dodecyl sulfate [SDS], 0.5 mM phenylmethylsulfonyl fluoride, 100 ng of leupeptin and aprotinin per ml). After incubation on ice for 10 min, nuclei were sonicated seven times with 10-s pulses and then centrifuged. Chromatin solution was precleared with Staphylococcus aureus protein A-positive cells for 15 min at 4°C. Prior to use, these cells were blocked with sheared herring sperm DNA and bovine serum albumin for at least 3 h at 4°C. Precleared chromatin from approximately 107 cells was incubated with 1 μg of the indicated antibodies and without an antibody for at least 3 h at 4°C. Mock IP buffer contained nuclear lysis buffer only. Antibodies against the following proteins were used: acetylated histone H4 (06-866; Upstate Biotechnology), dimethyl-K9 histone H3 (07-212; Upstate Biotechnology), E2F4 (sc-1082X; Santa Cruz), Dbf-4 (sc-11354; Santa Cruz), and HDAC1 (sc-7872; Santa Cruz). Staph A cells were then added and incubated at room temperature for 15 min. Staph A-immune complexes were washed twice in dialysis buffer (2 mM EDTA, 50 mM Tris-Cl [pH 8.0], 0.2% N-lauryl sarcosine), five times in IP wash buffer (100 mM Tris-Cl [pH 9.0], 500 mM LiCl, 1% NP-40, 1% deoxycholic acid) and twice in Tris-EDTA (10 mM Tris [pH 8.0], 1 mM EDTA) buffer. After washing, immune complexes were eluted by adding elution buffer (50 mM NaHCO3, 1% SDS). Inputs were processed from 1% of the total chromatin used in IPs. Cross-links were reversed by the addition of NaCl to a final concentration of 300 mM, and RNA was removed by the addition of 10 μg of RNase per sample followed by incubation at 65°C for 6 h. DNA was purified using the QIAquick PCR Purification Kit by following the manufacturer's protocol (QIAGEN, Valencia, Calif.). Promoter regions for the genes encoding the indicated proteins were then amplified with the following primer pairs: for cyclin A, 5′-CGACCGGCGCTCCTGGTGACGTC-3′ and 5′-TGGCGGCCGACGCACGGAGCA-3′; for Cdc2, 5′-TGAGCTCAAGAGTCAGTTGGCGCC-3′ and 5′-CGGCACAGCAGTTTCAAACTCAC-3′; for topoIIα, 5′-GACCGTCTGCGATTGATTGC-3′ and 5′-TGACCGTCCTGAAGGGGCTC-3′; for TS, 5′-GGGTCTGTCAATTTCGG-3′ and 5′-GAGCAGTCTGGTGGCAGTGTAGTC-3′; for myogenin, 5′-AGAGGGAAGGGGAATCACAT-3′ and 5′-TCCATCAGGTCGGAAAAGAC-3′; and for hypoxan- thine-guanine phosphoribosyltransferase, 5′-CAGGCCCAACTTGTCAGAAC-3′ and 5′-TGCACAACACCTCAGAGACG-3′. PCR was performed in a 50-μl volume containing 3 μl of purified DNA, 50 ng of each primer set, 0.25 U of Taq DNA polymerase (Promega), and 5 μCi of [α-32 P]dCTP. PCR parameters were 94°C for 4 min; 27 cycles of 94°C for 30 s, annealing for 30 s, 72°C for 30 s, and a final extension at 72°C for 10 min. PCR products were separated on a 6% polyacrylamide gel and visualized with a phosphorimager (Molecular Dynamics, Sunnyvale, Calif.).
Restriction enzyme accessibility assay.
The restriction enzyme accessibility assay was used to investigate nucleosome position at the cyclin A promoter. This assay was performed as previously described (4, 56). Nuclei were harvested from A5-1 cells that were grown in the presence and absence of Dox for 18 h. Nuclei permeabilized in 0.5% NP-40 were digested with EagI for 2 h at 37°C followed by digestion in 1 mg of proteinase K (Sigma) per ml overnight. Protein-free genomic DNA was extracted and subsequently digested overnight with KpnI and EcoRI. Digested DNA was then resolved by agarose gel electrophoresis and transferred onto an Immobilon-Ny nylon membrane (Millipore). Enzyme accessibility was visualized by radioactive Southern blot with a probe generated by PCR against bases −135 to +33 of the cyclin A gene promoter.
RESULTS
Active RB represses cell cycle genes.
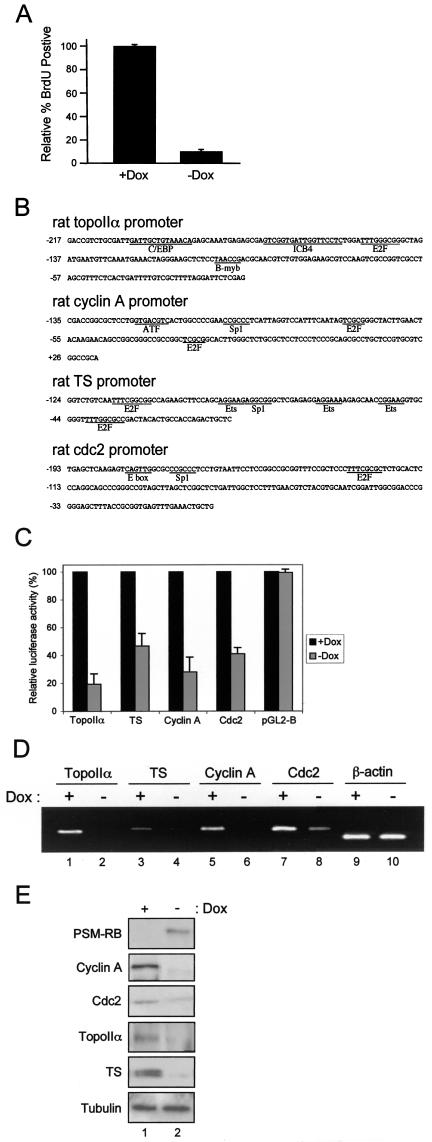
Studying the function of RB in mediating transcriptional repression is hampered, as wild-type RB is readily phosphorylated and inactivated by CDK/cyclin complexes in most cell types. To circumvent this problem, we utilized a PSM-RB that is refractory to phosphorylation and is therefore constitutively active. In this study, a Rat-1-derived cell line with tetracycline-regulated PSM-RB expression (A5-1) was utilized to specifically analyze the downstream action of RB. In order to confirm the cell cycle inhibitory potential of PSM-RB in this setting, we examined the BrdU incorporation of A5-1 cells cultured in the presence or absence of the tetracycline analogue Dox. Consistent with previously reported data, we found that the induction of PSM-RB upon removal of Dox dramatically reduced the number of BrdU-positive cells (Fig. 1A). Therefore, A5-1 cells represent an effective model for the study of RB activity.
FIG. 1.
PSM-RB (active RB) represses a set of cell cycle genes and induces cell cycle arrest. (A) A5-1 cells were cultured in the presence or absence of Dox for 16 h prior to BrdU labeling for an additional 8 h. The percentage of BrdU-positive cells was determined from three independent experiments. (B) The cloned rat regulatory regions ofdifferent RB target genes analyzed in the present study. (C) The A5-1 parental cell line was stably transfected with rat promoter constructs for the indicated genes or empty vector (pGL2-B) driving expression of the firefly luciferase gene. Three clones were selected for each construct, maintained in the presence or absence of Dox for 18 h, and then assayed for luciferase activity. (D) A5-1 cells were cultured in the presence or absence of Dox for 18 h. Total RNA was then extracted and reverse transcribed into cDNA. This cDNA was subjected to linear PCR amplification with specific primers for the indicated genes. (E) A5-1 cells were cultured in the presence (lane 1) or absence (lane 2) of Dox for 24 h. Total protein was isolated, resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and immunoblotted with antibodies as indicated.
We have recently identified and validated a large number of RB-repressed target genes by using microarray analysis (42). To initially investigate the promoter activity of representative genes, we cloned regions of the genes encoding the cyclin A, Cdc2, topoIIα, and TS promoters into the luciferase reporter vector pGL2-B (Fig. 1B). These cloned promoter fragments represent less than two nucleosomes of DNA and are thus useful for investigating the regulatory action of RB on a relatively small region of chromatin. The reporter constructs and the vector (pGL2-B) were then integrated into A5-1 cells to provide an effective means for studying promoter activity in the context of chromatin. Three independent clones for each construct were selected and maintained in medium with or without Dox for 18 h. We found that the activity of cyclin A, Cdc2, topoIIα, and TS promoters was reduced three- to fivefold upon induction of PSM-RB expression (Fig. 1C, compare +Dox to −Dox). In contrast, PSM-RB had no effect on the basal transcription of pGL2-B. These data demonstrate that active RB represses the promoter activity of these genes in chromatin.
Next we investigated the correlation between promoter activity and endogenous RNA levels (Fig. 1D). We utilized RT-PCR to determine the levels of endogenous RNA in A5-1 cells that were cultured in the presence or absence of Dox. Induction of active RB attenuated the expression of cyclin A, Cdc2, topoIIα, and TS RNA (Fig. 1D, compare +Dox to −Dox). In contrast, induction of PSM-RB had no effect on the expression of the β-actin gene. Consistent with these observations, endogenous protein levels of cyclin A, Cdc2, topoIIα, and TS were also attenuated when Dox was removed from the medium (Fig. 1E, compare +Dox to −Dox). The attenuations are due to the presence of PSM-RB and not merely to differences in loading, as revealed by immunoblotting for β-tubulin. Immunoblotting with a PSM-RB-specific antibody clearly demonstrated that PSM-RB is expressed only in the absence of Dox (Fig. 1E). Taken together, these results confirm that active RB inhibits the expression of the genes encoding cyclin A, Cdc2, topoIIα, and TS by repressing promoter activity.
Active RB induces histone deacetylation at promoters of specific cell cycle genes.
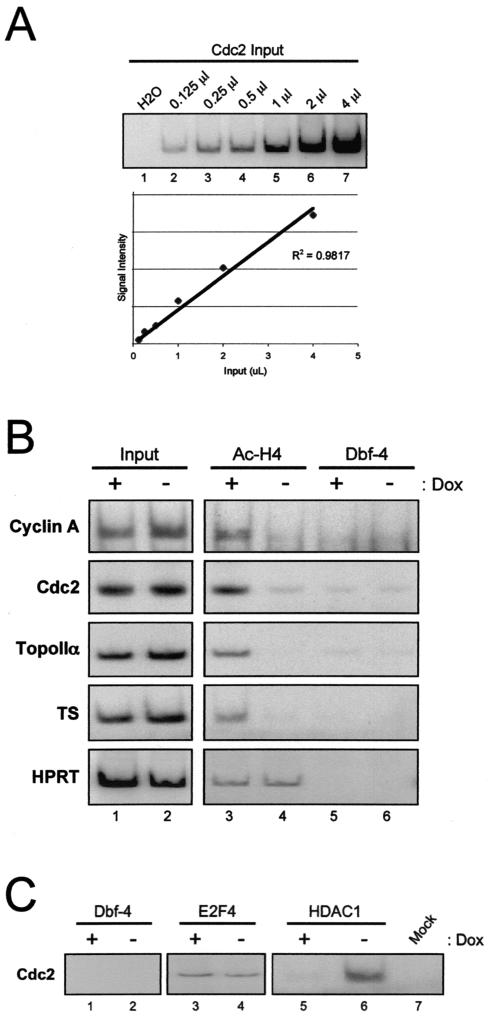
RB has been shown to recruit HDACs to the promoters of target genes (81). This event is believed to be responsible for transcriptional repression and the subsequent antiproliferative action of RB. However, the extent to which HDACs are required for RB to act as a transcriptional repressor and tumor suppressor is unclear. To assess the action of HDAC enzymatic activity in RB-mediated transcriptional repression, we utilized in vivo formaldehyde cross-linking of DNA-protein complexes followed by ChIP assay. Initially, we validated the linearity of PCR amplification over a wide range of template concentrations. Increasing amounts of input chromatin were amplified with primers specific for the Cdc2 promoter gene in the presence of [α-32P]dCTP for quantitation. As shown in Fig. 2A, the PCR was linear throughout the titration, indicating that changes in chromatin levels could be accurately observed by radioactive PCR. Using this approach, we then investigated the promoters of the cyclin A, Cdc2, topoIIα, and TS genes for changes in histone H4 acetylation, as these targets were repressed by PSM-RB (Fig. 2B). Specifically, A5-1 cells were cultured in the presence or absence of Dox for 24 h and were formaldehyde cross-linked, and ChIP assays were performed by utilizing antibody to acetylated histone H4. For each target promoter, input lanes (nonimmunoprecipitated DNA) confirmed that equal amounts of chromatin were used in all ChIP assays (Fig. 2B, lanes 1 and 2). The antibody to Dbf-4 was utilized as a negative control, since the replication factor Dbf-4 is not expected to occupy these promoters (Fig. 2A, lanes 5 and 6). We found that, in the presence of PSM-RB, acetylated histone H4 association with the promoters of cyclin A, Cdc2, topoIIα, and TS was nearly abolished (Fig. 2B, compare lanes 3 and 4). This effect is specific to RB-repressed genes, as no change in histone acetylation was observed on the hypoxanthine-guanine phosphoribosyltransferase promoter (Fig. 2B). In contrast with acetylated histone H4 occupancy, which was diminished in the presence of PSM-RB, we found that E2F4 occupancy was unaffected by the induction of active RB (Fig. 2C, compare lanes 3 and 4). Moreover, we detected HDAC1 preferentially bound to Cdc2 promoter in the presence of PSM-RB (Fig. 2C, compare lanes 5 and 6). Therefore, the decrease in acetylated histone H4 on these promoters is a consequence of PSM-RB induction, and not all protein-promoter interactions were similarly affected. The reduction in acetylated histone H4 at these promoters strongly suggests that these promoters are actively deacetylated. This observation is consistent with RB/HDAC complexes acting on these promoters.
FIG. 2.
Active RB induces histone deacetylation at promoters of specific cell cycle genes. (A) Total chromatin was isolated from A5-1 cells cultured in the presence of Dox, and increasing amounts of chromatin (0 to 4 μl) were subjected to PCR in the presence of [α-32P]dCTP and primers specific for the cdc2 promoter. Production of PCR product was quantified by using a phosphorimager. (B) A5-1 cells were cultured in the presence (lanes 1, 3, and 5) or absence (lanes 2, 4, and 6) of Dox for 24 h and cross-linked with formaldehyde, and ChIP assays were performed as described in Materials and Methods. Residency of acetylated histone H4 at the indicated gene promoters was determined by carrying out the ChIP assay with antibodies specific to acetylated histone H4 (lanes 3 and 4). Input (lanes 1 and 2) refers to PCR containing 1% of the total chromatin used in IP. IP with Dbf-4 (lanes 5 and 6) is a negative control. PCR products were detected by autoradiography. HPRT, hypoxanthine-guanine phosphoribosyltransferase. (C) Cells were cultured as described for panel B, except that immunoprecipitation was performed with antibodies specific for Dbf-4 (lanes 1 and 2), E2F4 (lanes 3 and 4), and HDAC1 (lanes 5 and 6). The mock represents a ChIP assay that was performed without the inclusion of chromatin substrate. PCR products were detected by autoradiography.
RB-induced histone deacetylation at specific promoters is mediated by HDACs.
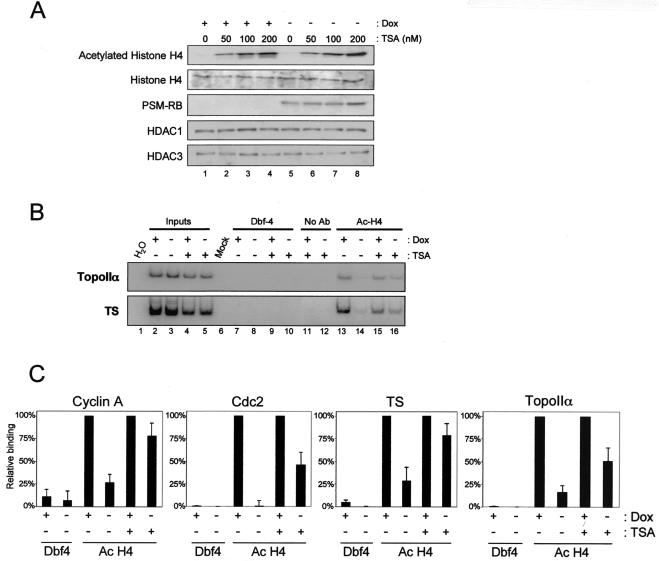
Histone deacetylation at the promoters of cyclin A, Cdc2, topoIIα, and TS in the presence of active RB (shown in Fig. 2) supports a critical role for HDAC in transcriptional repression. To determine the explicit requirement of HDACs for this transcriptional repression, we used TSA, a pharmacological inhibitor of HDAC activity. Initially, we determined the effect of TSA on bulk histone acetylation (Fig. 3A). A5-1 cells were cultured in the presence and absence of Dox and TSA for 24 h. Cells were then harvested, and the levels of acetylated histone H4 and total histone H4 were evaluated by immunoblotting (Fig. 3A). We found that TSA treatment resulted in the marked accumulation of acetylated histone H4 in a dose-dependent manner (Fig. 3A, top panel, compare lanes 1, 2, 3, and 4). Importantly, analysis of total histone H4 levels showed that TSA inhibited deacetylation and did not merely augment the expression of histone H4. Furthermore, TSA did not affect the expression of PSM-RB, HDAC1, or HDAC3. While performing these studies, we found that 100 nM TSA had a minimal effect on cell viability (data not shown). Therefore, we employed 100 nM TSA to study the requirement of HDAC activity for RB-mediated transcriptional repression.
FIG. 3.
RB-induced deacetylation at specific promoters is mediated by HDACs. (A) A5-1 cells were maintained in media with (lanes 1 to 4) or without (lanes 5 to 8) Dox and the HDAC inhibitor TSA (0, 50, 100, and 200 nM) for 24 h. Proteins were resolved by SDS-PAGE and immunoblotted with antibodies as indicated. (B) A5-1 cells were cultured in the presence or absence of Dox and TSA (100 nM). Cells were cross-linked after 24 h, and the ChIP assay was performed with an acetylated histone H4-specific antibody (lanes 13 to 16). Inputs (lanes 2 to 5) represent PCR with 1% of chromatin utilized in immunoprecipitations. Controls include H2 O (PCR control, lane 1), Mock (IP without chromatin, lane 6), Dbf-4 (nonspecific antibody, lanes 7 to 10), and No Ab (IP without antibody, lanes 11 to 12). (C) PCR products from Fig. 3B were quantified by using the phosphorimager, and data were normalized to account for variation in inputs.
To investigate the requirement of HDAC activity for RB-mediated histone deacetylation, we used TSA to inhibit HDAC activity and then monitored promoter histone acetylation. For this analysis, A5-1 cells were treated with or without Dox and TSA for 24 h. These cells were then used as substrates for ChIP assays with antibodies specific for acetylated histone H4 (Fig. 3B). Nonimmunoprecipitated chromatin (Inputs) showed the relative amounts of chromatin used in each IP (Fig. 3B, lanes 2 to 5). IP with nonspecific antibody to Dbf4 (Fig. 3B, lanes 7 to 10), without an antibody (Fig. 3B, lanes 11 to 12), and without chromatin (Fig. 3B, lane 6) did not result in appreciable PCR product and thus provided evidence of specificity for the ChIP assay. Consistent with data described above, we observed histone H4 deacetylation on the promoters analyzed (Fig. 3B, compare lanes 13 and 14). Surprisingly, TSA by itself did not augment the histone H4 acetylation observed on any promoter in the presence of Dox (Fig. 3B, compare lanes 13 and 15), indicating that HDAC activity was likely absent from the promoter in normal asynchronously proliferating cells. However, inhibition of HDAC activity by TSA resulted in marked acetylation of histone H4 on the promoters of topoIIα and TS in the presence of PSM-RB (Fig. 3B, compare lane 14 to lane 16). Similar results were obtained for cyclin A and Cdc2 promoters. The use of [α-32P]dCTP in PCR enabled the quantitative analysis of product signal intensity. PCR product bands from acetylated histone H4 IPs were normalized to their corresponding inputs to account for variations in chromatin used in the ChIP assay. PCR-amplified products that were quantified by using phosphorimaging clearly showed that TSA blocks deacetylation of these promoters (Fig. 3C). Collectively, these data demonstrate that RB-induced deacetylation of cyclin A, Cdc2, topoIIα, and TS promoters requires HDAC activity. Since histone deacetylation is a critical mechanism of transcriptional repression, these results suggest that TSA may reverse RB-mediated cell cycle inhibition.
HDAC activity is dispensable for RB-mediated cell cycle arrest.
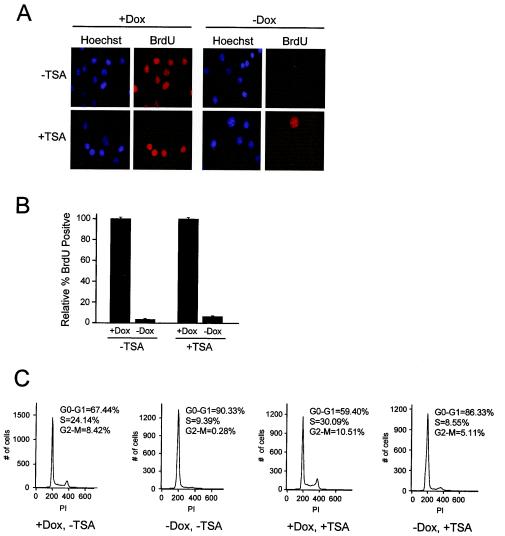
To monitor the influence of HDAC inhibition on cell cycle progression, A5-1 cells were cultured in the presence or absence of Dox and TSA for 16 h before being pulsed with BrdU for 8 h. Surprisingly, cells cultured in the absence of Dox remained inhibited for BrdU incorporation in the presence of TSA (Fig. 4A and B). To examine cell cycle distribution, we performed flow cytometric analysis of cells treated with or without Dox and TSA. As shown in Fig. 4C, we found that treatment with or without TSA, concurrent with PSM-RB (−Dox) expression, did not significantly alter cell cycle position. Taken together, these data demonstrate that RB is able to maintain cell cycle arrest even in the absence of HDAC activity.
FIG. 4.
HDAC activity is dispensable for RB-mediated cell cycle arrest. (A) A5-1 cells were cultured in the presence or absence of Dox and TSA for 16 h prior to BrdU labeling for an additional 8 h. Cells were fixed and stained with anti-BrdU antibody and Hoechst to identify proliferating cells and nuclei, respectively. (B) The percentage of BrdU-positive cells was determined from three independent experiments. (C) A5-1 cells were cultured in the presence or absence of Dox and TSA for 24 h. Cells were harvested, fixed with ethanol, and stained with propidium iodide (PI). Cell cycle distribution was then determined by flow cytometry. DNA content (PI intensity) is plotted against cell number. The percentage of cells in G0-G1, S, and G2-M phases was determined by ModFit software.
HDAC requirement for repression is promoter specific.
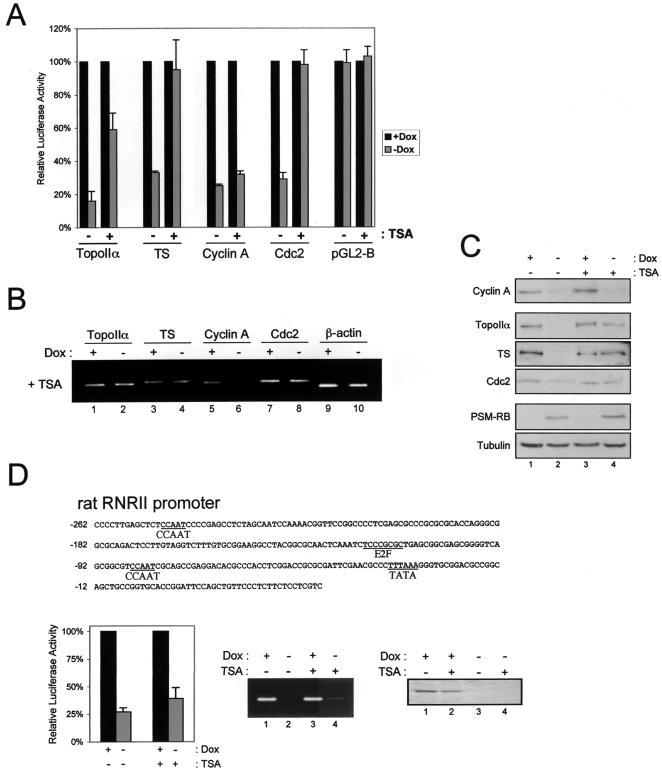
The failure of TSA to rescue cells from RB-mediated cycle arrest suggested that inhibition of HDAC activity was not sufficient to alleviate transcriptional repression. Having established that histone deacetylation at cyclin A, Cdc2, topoIIα, and TS promoters by RB is the result of HDAC activity (Fig. 3B), we sought to elucidate the functional significance of this event (Fig. 5). A5-1 cells with integrated luciferase reporters of cyclin A, Cdc2, topoIIα, TS, and vector pGL2-B were cultured in the presence or absence of Dox and TSA for 18 h. Consistent with the failure of TSA treatment to augment promoter histone acetylation in the absence of PSM-RB (+Dox), TSA did not significantly stimulate basal promoter activity (not shown). Analysis of RB-mediated repression showed that TSA significantly alleviated RB-mediated promoter repression of Cdc2, topoIIα, and TS (Fig. 5A). However, the promoter activity of cyclin A was not recovered in the presence of TSA (Fig. 5A).
FIG. 5.
HDAC-dependent deacetylation is required by RB to repress specific promoters. (A) A5-1-integrated reporter cell lines described in the legend of Fig. 1C were cultured in the presence or absence of Dox and 100 nM TSA for 18 h. Cells were harvested and assayed for luciferase activity. (B) A5-1 cells were cultured in 100 nM TSA in the presence or absence of Dox for 18 h. Total RNA was then extracted and reverse transcribed into cDNA. This cDNA was subjected to linear PCR amplification with specific primers for the indicated genes. (C) A5-1 cells were cultured in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of Dox in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 100 nM TSA for 24 h. Total protein was isolated, resolved by SDS-PAGE, and immunoblotted with antibodies as indicated. (D) Top panel: The cloned regulatory region of the rat RNRII promoter. Left panel: The A5-1 parental cell line was stably transfected with the rat RNRII promoter driving expression of the firefly luciferase gene. Cells were cultured in the presence or absence of Dox with or without 100 nM TSA as indicated, and relative luciferase activity was determined. Center panel: A5-1 cells were cultured in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of Dox and in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of TSA for 18 h. Cells were harvested, and RNRII RNA levels were determined by RT-PCR. Right panel: A5-1 cells were cultured in the presence (lanes 1 and 2) or absence (lanes 3 and 4) of Dox and the absence (lanes 1 and 3) or presence (lanes 2 and 4) of TSA for 24 h. Cells were harvested, and RNRII protein levels were determined by immunoblotting.
The expression of these targets was also monitored through investigation of endogenous RNA levels (Fig. 5B). Consistent with the reporter assays, we found that TSA ameliorated the attenuation of Cdc2, topoIIα, and TS RNA levels that is mediated by PSM-RB (Fig. 5B). Again at the RNA level, TSA failed to relieve the inhibition of cyclin A by PSM-RB. As expected, β-actin RNA levels were unaffected upon treatment with either Dox or TSA (Fig. 5B).
To determine whether the changes in RNA levels led to meaningful changes in protein levels, immunoblot analysis was performed. Analysis of cyclin A, Cdc2, topoIIα, and TS protein levels demonstrated PSM-RB-mediated attenuation (Fig. 5C, compare lanes 1 and 2). Consistent with the promoter and RNA analyses, treatment with TSA reversed the RB-mediated attenuation of Cdc2, topoIIα, and TS protein levels (Fig. 5C, compare lanes 3 and 4). However, TSA did not recover RB-mediated inhibition of cyclin A protein levels (Fig. 5C, compare lanes 3 and 4).
In addition to cyclin A, we analyzed the promoter activity of other genes associated with the RB/E2F-signaling axis and found that ribonucleotide reductase subunit II (RNRII) promoter activity was not recovered with TSA treatment (Fig. 5D). Specifically, an integrated RNRII reporter was repressed by PSM-RB, and this repression was not alleviated with TSA (Fig. 5D, left panel). Moreover, RNRII RNA (Fig. 5D, middle panel) and protein (Fig. 5D, right panel) levels were similarly reduced by RB action in the absence and presence of TSA. Thus, RNRII, like cyclin A, is also an HDAC-independent target of RB-mediated repression.
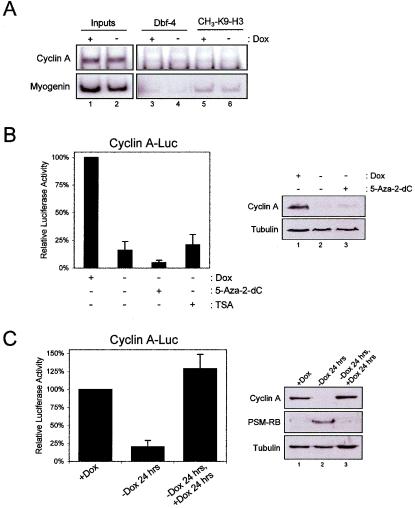
HDAC-independent mechanism of cyclin A repression.
It is known that transcriptional repression elicited by RB can occur through the action of corepressors in addition to HDACs. Specifically, it has recently been demonstrated that the cyclin A promoter is irreversibly silenced during induced senescence (48). This pathway likely involves histone H3 lysine 9 methylation, heterochromatin protein 1 (HP1) recruitment, and DNA methylation of the promoter (34, 48, 52). Therefore, we initially analyzed the histone H3 lysine 9 methylation of the cyclin A promoter during RB-mediated arrest (Fig. 6A). Cells were cultured in the presence or absence of Dox to induce PSM-RB, and isolated chromatin was subjected to ChIP analysis using antibodies specific for histone H3 methylated on lysine 9. We observed histone H3 lysine 9 methylation on the myogenin promoter that is silenced in fibroblastic cells (Fig. 6A). In contrast, we failed to detect histone H3 lysine 9 methylation on the cyclin A promoter above background, suggesting that this silencing mechanism is not responsible for cyclin A repression. Consistent with this observation, culture in 5-aza-2-deoxcytidine, which blocks DNA methylation and reverses silencing (41, 45, 50, 80), failed to augment cyclin A promoter activity or protein levels in the presence of PSM-RB (Fig. 6B). Finally, we determined whether the RB-mediated repression of the cyclin A promoter was reversible. To do so, cells were cultured in the absence of Dox to induce PSM-RB, and cyclin A promoter activity was repressed (Fig. 6C, left panel). Readdition of Dox to the media resulted in the restoration of cyclin A promoter activity. Additionally, cyclin A protein levels were restored following the readdition of Dox (Fig. 6C, right panel). Together, these results indicate that stable epigenetic silencing mechanisms are not responsible for the observed RB-mediated repression of the cyclin A promoter.
FIG. 6.
The cyclin A promoter is not subjected to stable gene silencing. (A) A5-1 cells were cultured in the presence or absence of Dox as indicated. Chromatin was isolated and utilized in ChIP assays with antibodies specific for dimethylated K9 histone H3 (lanes 5 and 6). Input (lanes 1 and 2) and Dbf-4 (lanes 3 and 4) controls are shown. Chromatin was amplified with primers specific for the cyclin A and myogenin promoters, and products were detected by autoradiography. (B) A5-1 cells harboring the integrated cyclin A reporter were cultured in the presence of 5-aza-2-dC as described in Materials and Methods and then cultured in the absence of Dox for 24 h. Relative luciferase activity was determined by reporter assay (left panel), and endogenous protein levels were determined by immunoblotting (right panel). (C) A5-1 cells harboring the integrated cyclin A reporter were cultured in the presence or absence of Dox for 24 h. To attenuate PSM-RB, Dox was readministered to the indicated cultures. Relative luciferase activity was determined by reporter assay (left panel), and endogenous protein levels were determined by immunoblotting (right panel).
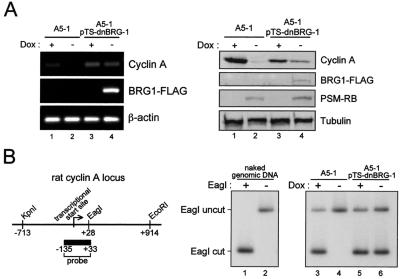
In addition to HDACs and silencing mechanisms, we and others have demonstrated that SWI/SNF activity plays a requisite role in the repression of the cyclin A promoter (70, 81). Consistent with these studies, we observed that, unlike 5-aza-2-deoxcytidine or TSA, ectopic expression of dominant negative BRG-1 (dnBRG-1) that inhibits SWI/SNF activity augments the expression of cyclin A RNA (Fig. 7A, left panel) and protein (Fig. 7A, right panel) levels in the presence of PSM-RB (Fig. 7A). These results indicate that chromatin remodeling represents a critical means through which cyclin A repression occurs. One possible explanation is that SWI/SNF functions in concert with RB to position nucleosomes near the transcription start site to inhibit transcription of the cyclin A promoter. Analysis of promoter structure was carried out in the presence or absence of PSM-RB by using the restriction enzyme accessibility assay in proximity to the transcription start site. We observed that chromatin from cells cultured in the absence of PSM-RB (+Dox) was readily digestible (Fig. 7B, lanes 3), whereas chromatin from cells expressing PSM-RB (−Dox) was resistant to enzyme cleavage (Fig. 7B, lanes 4). This change in chromatin structure was largely dependent upon SWI/SNF, as dnBRG-1 retarded the formation of the nuclease-resistant chromatin structure (Fig. 7B, lanes 6). These results indicate that chromatin remodeling occurs on the cyclin A promoter to mediate transcriptional repression.
FIG. 7.
RB-mediated repression of the cyclin A promoter involves chromatin remodeling. (A) Parental A5-1 cells (lanes 1 and 2) or A5-1 cells engineered to inducibly coexpress dnBRG-1 and PSM-RB (lanes 3 and 4) were cultured in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of Dox for 24 h. Cells were harvested, and protein and RNA levels were determined by RT-PCR (left panel) or immunoblotting (right panel) as indicated. (B) Left panel: genomic structure of the rat cyclin A locus. Right panel: Southern blot analysis of genomic DNA isolated from A5-1 cells (lanes 1 and 2). Parental A5-1 cells (lanes 3 and 4) or A5-1 cells engineered to inducibly express dnBRG-1 (lanes 5 and 6) were cultured in the presence or absence of Dox as indicated. Permeabilized nuclei were subjected to digestion with EagI, and then isolated genomic DNA was subjected to cleavage with KpnI and EcoRI. Restriction fragments were detected by radioactive Southern blotting.
DISCUSSION
RB represses the expression of multiple genes involved in cell cycle transitions. This transcriptional control has been attributed to multiple corepressors recruited by RB. Here we specifically focused on elucidating the role of HDACs in RB-mediated transcriptional repression of four critical targets (cyclinA, Cdc2, topoIIα, and TS). We demonstrate that active RB leads to histone deacetylation on the promoters of these four genes, and the observed deacetylation is dependent on HDAC activity. This action of HDACs was required for the transcriptional repression of Cdc2, topoIIα, and TS genes, as TSA reversed RB-mediated repression. However, this action was promoter specific, as TSA failed to recover cyclin A or RNRII levels. Analysis of the HDAC-independent repression of cyclin A indicated that it was not due to epigenetic silencing mechanisms but was reversible and involved chromatin remodeling. Importantly, we demonstrate that the cell cycle-inhibitory action of RB is independent of HDAC enzymatic activity. Together, these results demonstrate the intricate interplay between RB and HDACs in transcriptional regulation and cell cycle control.
Role of HDACs in RB-mediated transcriptional repression.
HDACs represent corepressors that are identified as interacting with RB (6, 39, 40). However, relatively few studies have analyzed their role in RB-mediated transcriptional repression and cell cycle control. Originally, it was demonstrated that HDAC activity is associated with RB, but only in the context of a synthetic promoter was histone deacetylation observed (39). Subsequent studies have shown that specific RB/E2F target genes in fact undergo changes in promoter histone acetylation during the cell cycle (47, 58). Our results shown here demonstrate that RB-mediated repression leads to histone deacetylation at all promoters analyzed. In principle, such an effect could be due to either recruitment of HDAC or the inhibition of HAT activity. Inhibition of HAT as a mechanism for promoter deacetylation is not without merit, as E2F proteins likely employ HAT-dependent mechanisms for gene activation (74). By using the HDAC inhibitor TSA, we could specifically demonstrate that the deacetylation of these promoters is dependent on the enzymatic activity associated with class I HDAC molecules (43). The effect of RB on histone acetylation is not due to a bulk-deacetylation phenomenon, as total cellular levels of acetylated histone are not changed by the expression of PSM-RB. Interestingly, while TSA does lead to bulk histone hyperacetylation, it does not lead intrinsically to the hyperacetylation of histones at the E2F/RB-regulated promoters studied here. This could be because the histones at these promoters are already hyperacetylated or because HDAC is not associated with the promoter in asynchronously proliferating cells. Such a hypothesis is supported by the finding that we observed HDAC1 specifically associated with the Cdc2 promoter when PSM-RB was expressed. Clearly, in the context of RB-mediated repression, the mechanism of histone deacetylation is HDAC dependent and can be reversed by TSA on all promoters analyzed. The inhibition of histone deacetylation by TSA had functional consequences in the context of Cdc2, topoIIα, and TS gene expression. Interestingly, even in cases in which TSA only partially reversed RB-mediated histone deacetylation, TSA was capable of fully restoring promoter activity (i.e., the cdc2 promoter). Such a finding, for which there is precedent in the literature (60, 83), suggests that only a moderate level of histone acetylation is required for transcriptional activity on specific promoters. In fact, TSA fully restored the promoter activity, endogenous RNA, and protein levels of Cdc2, topoIIα, and TS in the presence of PSM-RB. Such a finding is critical, as it demonstrates that HDAC activity represents the sole means through which RB mediates repression of these targets. In contrast, we failed to observe recovery of cyclin A expression when HDAC activity was inhibited in presence of RB, even though promoter histones were acetylated following TSA treatment.
HDAC-independent mechanisms of transcriptional repression.
The findings that we observe with cyclin A gene regulation indicate that RB utilizes mechanisms in addition to histone deacetylation to mediate repression. Such a conclusion is not without precedent (44, 81). However, to definitively make such a conclusion, it is critical to determine that inhibition of HDAC activity actually reversed the histone deacetylation on the promoter. We clearly observed that TSA was sufficient to efficiently reverse RB-mediated histone deacetylation on the cyclin A promoter. Thus, the failure of TSA to reverse RB-mediated repression of the cyclin A promoter is due to a mechanism that is clearly distinct from HDAC activity. Additionally, while we did not explicitly evaluate the RNRII promoter, it behaved in a manner similar to that of cyclin A in that its repression was independent of HDAC activity. Therefore, it seems likely that repression of cyclin A and RNRII by RB is dependent on other chromatin-modifying factors.
RB has been shown to associate with other chromatin-modifying enzymes to mediate transcriptional repression. Specifically, recent studies indicate that RB target genes (including cyclin A) are silenced through a mechanism involving histone methylation and HP1 chromatin association (48, 52). Such silencing, which is observed in senescent cells, is irreversible (48, 52). Here we find that the repression of cyclin A by PSM-RB does not involve irreversible silencing mechanisms, as we failed to detect any influence of histone or DNA methylation on RB-mediated repression of the cyclin A promoter. Additionally, the repression of the cyclin A promoter was readily reversible. Such a result is consistent with the ability of quiescent cells with the cyclin A promoter repressed to reenter the cell cycle (48) and with the failure to detect histone methylation or HP1 chromatin association with the cyclin A promoter in quiescent cells (48).
In addition to silencing mechanisms and HDACs, RB is known to associate with components of the SWI/SNF chromatin remodeling complex, and the activity of SWI/SNF is required for the repression of cyclin A. For example, PSM-RB or expression of p16ink4a to activate endogenous RB in BRG-1- or BRM-deficient cell lines does not attenuate cyclin A and RNRII levels or lead to cell cycle arrest (references 70 and 81 and our unpublished data). One means through which SWI/SNF acts is by facilitating histone acetylation and deacetylation. This action of SWI/SNF is not sufficient for cyclin A repression, as the promoter remained repressed even with histones acetylated. Rather, our results suggest that SWI/SNF is functioning to modify chromatin structure to inhibit transcription. Consistent with this model, we find that the nuclease accessibility in the region of the cyclin A transcription start site is inhibited during RB-mediated repression. Thus, chromatin topology, not histone modification, is likely sufficient for RB-mediated repression events on the cyclin A promoter.
HDAC activity is not required for RB-mediated cell cycle inhibition.
Another finding from our studies is that inhibition of HDAC activity by TSA does not rescue cells from arrest imposed by RB. This result is quite surprising, as TSA exerts pronounced effects on gene expression but has no detectable effect on RB-mediated cell cycle inhibition. Most likely, failure of TSA to block RB-mediated repression of cyclin A and similarly regulated genes (e.g., RNRII) is responsible for this phenomenon. Cyclin A is required for traversing the cell cycle (19, 55), and thus lack of cyclin A expression alone might well explain the failure of cells to proliferate when HDAC activity is inhibited. An important extension of our work is based on the current use of HDAC inhibitors in clinical trials for treatment of several forms of cancer (33). In this context, HDAC inhibitors are believed to reactivate genes that have been inappropriately silenced during tumor progression and thus inhibit tumor growth. Based on our findings that TSA does not reverse RB-mediated cell cycle arrest, we conclude that the treatment of tumors with HDAC inhibitors will not have the undesired effect of inactivating the RB pathway of cell cycle inhibition.
In summary, transcriptional repression of cell cycle genes by RB is a complex process involving multiple chromatin-modifying factors. Here we find that one class of factors, HDACs, play a critical role in the transcriptional repression programs elicited by RB. This action of HDACs is promoter specific and as such is not required for RB-mediated cell cycle inhibition.
Acknowledgments
We are grateful to Karen Knudsen, Christopher Mayhew, Steven Angus, Craig Burd, Michael Markey, and Christin Petre for critical reading of the manuscript. We thank Shelly Barton, Anthony Imbalzano, Bernard Weissman, and members of the Knudsens' laboratories for helpful discussions. Sandy Schwemberger provided technical assistance with flow cytometry analysis.
This work was supported by grants to E.S.K. from the National Cancer Institute (no. CA82525).
H.S. and D.A.S. contributed equally to this work.
REFERENCES
- 1.Angus, S. P., A. F. Fribourg, M. P. Markey, S. L. Williams, H. F. Horn, J. DeGregori, T. F. Kowalik, K. Fukasawa, and E. S. Knudsen. 2002. Active RB elicits late G1/S inhibition. Exp. Cell Res. 276:201-213. [DOI] [PubMed] [Google Scholar]
- 2.Bartek, J., J. Bartkova, and J. Lukas. 1997. The retinoblastoma protein pathway in cell cycle control and cancer. Exp. Cell Res. 237:1-6. [DOI] [PubMed] [Google Scholar]
- 3.Bartkova, J., J. Lukas, and J. Bartek. 1997. Aberrations of the G1- and G1/S-regulating genes in human cancer. Prog. Cell Cycle Res. 3:211-220. [DOI] [PubMed] [Google Scholar]
- 4.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 5.Botz, J., K. Zerfass-Thome, D. Spitkovsky, H. Delius, B. Vogt, M. Eilers, A. Hatzigeorgiou, and P. Jansen-Durr. 1996. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol. Cell. Biol. 16:3401-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 7.Bremner, R., B. L. Cohen, M. Sopta, P. A. Hamel, C. J. Ingles, B. L. Gallie, and R. A. Phillips. 1995. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol. Cell. Biol. 15:3256-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chellappan, S. P., S. Hiebert, M. Mudryj, J. M. Horowitz, and J. R. Nevins. 1991. The E2F transcription factor is a cellular target for the RB protein. Cell 65:1053-1061. [DOI] [PubMed] [Google Scholar]
- 9.Chen, T. T., and J. Y. Wang. 2000. Establishment of irreversible growth arrest in myogenic differentiation requires the RB LXCXE-binding function. Mol. Cell. Biol. 20:5571-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Classon, M., and E. Harlow. 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2:910-917. [DOI] [PubMed] [Google Scholar]
- 11.Dahiya, A., M. R. Gavin, R. X. Luo, and D. C. Dean. 2000. Role of the LXCXE binding site in Rb function. Mol. Cell. Biol. 20:6799-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalton, S. 1992. Cell cycle regulation of the human cdc2 gene. EMBO J. 11:1797-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Serna, I. L., K. A. Carlson, D. A. Hill, C. J. Guidi, R. O. Stephenson, S. Sif, R. E. Kingston, and A. N. Imbalzano. 2000. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 20:2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Serna, I. L., K. A. Carlson, and A. N. Imbalzano. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27:187-190. [DOI] [PubMed] [Google Scholar]
- 15.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 16.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 17.Flemington, E. K., S. H. Speck, and W. G. Kaelin, Jr. 1993. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc. Natl. Acad. Sci. USA 90:6914-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furukawa, Y., Y. Terui, K. Sakoe, M. Ohta, and M. Saito. 1994. The role of cellular transcription factor E2F in the regulation of cdc2 mRNA expression and cell cycle control of human hematopoietic cells. J. Biol. Chem. 269:26249-26258. [PubMed] [Google Scholar]
- 19.Furuno, N., N. den Elzen, and J. Pines. 1999. Human cyclin A is required for mitosis until mid prophase. J. Cell Biol. 147:295-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng, Y., E. N. Eaton, M. Picon, J. M. Roberts, A. S. Lundberg, A. Gifford, C. Sardet, and R. A. Weinberg. 1996. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene 12:1173-1180. [PubMed] [Google Scholar]
- 21.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 22.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 23.Harbour, J. W., R. X. Luo, S. A. Dei, A. A. Postigo, and D. C. Dean. 1999. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98:859-869. [DOI] [PubMed] [Google Scholar]
- 24.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 25.Hassig, C. A., and S. L. Schreiber. 1997. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr. Opin. Chem. Biol. 1:300-308. [DOI] [PubMed] [Google Scholar]
- 26.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 27.Helin, K., C. L. Wu, A. R. Fattaey, J. A. Lees, B. D. Dynlacht, C. Ngwu, and E. Harlow. 1993. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 7:1850-1861. [DOI] [PubMed] [Google Scholar]
- 28.Herrera, R. E., T. P. Makela, and R. A. Weinberg. 1996. TGF-β-induced growth inhibition in primary fibroblasts requires the retinoblastoma protein. Mol. Biol. Cell 7:1335-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huet, X., J. Rech, A. Plet, A. Vie, and J. M. Blanchard. 1996. Cyclin A expression is under negative transcriptional control during the cell cycle. Mol. Cell. Biol. 16:3789-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaelin, W. G., Jr. 1997. Recent insights into the functions of the retinoblastoma susceptibility gene product. Cancer Investig. 15:243-254. [DOI] [PubMed] [Google Scholar]
- 31.Karlseder, J., H. Rotheneder, and E. Wintersberger. 1996. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol. 16:1659-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knudsen, E. S., C. Buckmaster, T. T. Chen, J. R. Feramisco, and J. Y. Wang. 1998. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 12:2278-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer, O. H., M. Gottlicher, and T. Heinzel. 2001. Histone deacetylase as a therapeutic target. Trends Endocrinol. Metab. 12:294-300. [DOI] [PubMed] [Google Scholar]
- 34.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 35.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 36.Lai, A., J. M. Lee, W. M. Yang, J. A. DeCaprio, W. G. Kaelin, Jr., E. Seto, and P. E. Branton. 1999. RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol. Cell. Biol. 19:6632-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai, A., R. C. Marcellus, H. B. Corbeil, and P. E. Branton. 1999. RBP1 induces growth arrest by repression of E2F-dependent transcription. Oncogene 18:2091-2100. [DOI] [PubMed] [Google Scholar]
- 38.Lee, Y., and L. F. Johnson. 2000. Transcriptional control elements of the rat thymidylate synthase promoter: evolutionary conservation of regulatory features. Exp. Cell Res. 258:53-64. [DOI] [PubMed] [Google Scholar]
- 39.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 40.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 41.Mal, A., and M. L. Harter. 2003. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc. Natl. Acad. Sci. USA 100:1735-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markey, M. P., S. P. Angus, M. W. Strobeck, S. L. Williams, R. W. Gunawardena, B. J. Aronow, and E. S. Knudsen. 2002. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 62:6587-6597. [PubMed] [Google Scholar]
- 43.Marks, P., R. A. Rifkind, V. M. Richon, R. Breslow, T. Miller, and W. K. Kelly. 2001. Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer 1:194-202. [DOI] [PubMed] [Google Scholar]
- 44.Meloni, A. R., E. J. Smith, and J. R. Nevins. 1999. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl. Acad. Sci. USA 96:9574-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merlo, A., J. G. Herman, L. Mao, D. J. Lee, E. Gabrielson, P. C. Burger, S. B. Baylin, and D. Sidransky. 1995. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat. Med. 1:686-692. [DOI] [PubMed] [Google Scholar]
- 46.Morris, E. J., and N. J. Dyson. 2001. Retinoblastoma protein partners. Adv. Cancer Res. 82:1-54. [DOI] [PubMed] [Google Scholar]
- 47.Morrison, A. J., C. Sardet, and R. E. Herrera. 2002. Retinoblastoma protein transcriptional repression through histone deacetylation of a single nucleosome. Mol. Cell. Biol. 22:856-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narita, M., S. Nunez, E. Heard, M. Narita, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 49.Nevins, J. R. 1998. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 9:585-593. [PubMed] [Google Scholar]
- 50.Nguyen, C. T., D. J. Weisenberger, M. Velicescu, F. A. Gonzales, J. C. Lin, G. Liang, and P. A. Jones. 2002. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 62:6456-6461. [PubMed] [Google Scholar]
- 51.Nicolas, E., V. Morales, L. Magnaghi-Jaulin, A. Harel-Bellan, H. Richard-Foy, and D. Trouche. 2000. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J. Biol. Chem. 275:9797-9804. [DOI] [PubMed] [Google Scholar]
- 52.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 53.Ogris, E., I. Mudrak, and E. Wintersberger. 1993. Distinct amounts of polyomavirus large T antigen are required for different functions of the protein. Oncogene 8:1277-1283. [PubMed] [Google Scholar]
- 54.Ohtani, K., J. DeGregori, and J. R. Nevins. 1995. Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl. Acad. Sci. USA 92:12146-12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pagano, M., R. Pepperkok, F. Verde, W. Ansorge, and G. Draetta. 1992. Cyclin A is required at two points in the human cell cycle. EMBO J. 11:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pattenden, S. G., R. Klose, E. Karaskov, and R. Bremner. 2002. Interferon-γ-induced chromatin remodeling at the CIITA locus is BRG1 dependent. EMBO J. 21:1978-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearson, B. E., H. P. Nasheuer, and T. S. Wang. 1991. Human DNA polymerase α gene: sequences controlling expression in cycling and serum-stimulated cells. Mol. Cell. Biol. 11:2081-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rayman, J. B., Y. Takahashi, V. B. Indjeian, J. H. Dannenberg, S. Catchpole, R. J. Watson, H. te Riele, and B. D. Dynlacht. 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 16:933-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reed, S. I. 1997. Control of the G1/S transition. Cancer Surv. 29:7-23. [PubMed] [Google Scholar]
- 60.Rietveld, L. E., E. Caldenhoven, and H. G. Stunnenberg. 2002. In vivo repression of an erythroid-specific gene by distinct corepressor complexes. EMBO J. 21:1389-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schulze, A., K. Zerfass, D. Spitkovsky, S. Middendorp, J. Berges, K. Helin, P. Jansen-Durr, and B. Henglein. 1995. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc. Natl. Acad. Sci. USA 92:11264-11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sellers, W. R., and W. G. Kaelin, Jr. 1997. Role of the retinoblastoma protein in the pathogenesis of human cancer. J. Clin. Oncol. 15:3301-3312. [DOI] [PubMed] [Google Scholar]
- 63.Sellers, W. R., J. W. Rodgers, and W. G. Kaelin, Jr. 1995. A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc. Natl. Acad. Sci. USA 92:11544-11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shan, B., T. Durfee, and W. H. Lee. 1996. Disruption of RB/E2F-1 interaction by single point mutations in E2F-1 enhances S-phase entry and apoptosis. Proc. Natl. Acad. Sci. USA 93:679-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 66.Shimizu, M., E. Ichikawa, U. Inoue, T. Nakamura, T. Nakajima, H. Nojima, H. Okayama, and K. Oda. 1995. The G1/S boundary-specific enhancer of the rat cdc2 promoter. Mol. Cell. Biol. 15:2882-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shimizu, M., Y. Nomura, H. Suzuki, E. Ichikawa, A. Takeuchi, M. Suzuki, T. Nakamura, T. Nakajima, and K. Oda. 1998. Activation of the rat cyclin A promoter by ATF2 and Jun family members and its suppression by ATF4. Exp. Cell Res. 239:93-103. [DOI] [PubMed] [Google Scholar]
- 68.Singh, P., J. Coe, and W. Hong. 1995. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature 374:562-565. [DOI] [PubMed] [Google Scholar]
- 69.Slansky, J. E., and P. J. Farnham. 1996. Transcriptional regulation of the dihydrofolate reductase gene. Bioessays 18:55-62. [DOI] [PubMed] [Google Scholar]
- 70.Strobeck, M. W., K. E. Knudsen, A. F. Fribourg, M. F. DeCristofaro, B. E. Weissman, A. N. Imbalzano, and E. S. Knudsen. 2000. BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl. Acad. Sci. USA 97:7748-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strobeck, M. W., D. N. Reisman, R. W. Gunawardena, B. L. Betz, S. P. Angus, K. E. Knudsen, T. F. Kowalik, B. E. Weissman, and E. S. Knudsen. 2002. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J. Biol. Chem. 277:4782-4789. [DOI] [PubMed] [Google Scholar]
- 72.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 73.Tommasi, S., and G. P. Pfeifer. 1995. In vivo structure of the human cdc2 promoter: release of a p130-E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol. Cell. Biol. 15:6901-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trouche, D., A. Cook, and T. Kouzarides. 1996. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 24:4139-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, J. Y., E. S. Knudsen, and P. J. Welch. 1994. The retinoblastoma tumor suppressor protein. Adv. Cancer Res. 64:25-85. [DOI] [PubMed] [Google Scholar]
- 76.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 77.Weintraub, S. J., K. N. Chow, R. X. Luo, S. H. Zhang, S. He, and D. C. Dean. 1995. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature 375:812-815. [DOI] [PubMed] [Google Scholar]
- 78.Wells, J., K. E. Boyd, C. J. Fry, S. M. Bartley, and P. J. Farnham. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20:5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoon, J. H., J. K. Kim, G. B. Rha, M. Oh, S. H. Park, R. H. Seong, S. H. Hong, and S. D. Park. 1999. Sp1 mediates cell proliferation-dependent regulation of rat DNA topoisomerase IIα gene promoter. Biochem. J. 344: 367-374. [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang, C. L., T. A. McKinsey, and E. N. Olson. 2002. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol. Cell. Biol. 22:7302-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]
- 82.Zhang, H. S., A. A. Postigo, and D. C. Dean. 1999. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFβ, and contact inhibition. Cell 97:53-61. [DOI] [PubMed] [Google Scholar]
- 83.Zhang, Y., and M. L. Dufau. 2002. Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J. Biol. Chem. 277:33431-33438. [DOI] [PubMed] [Google Scholar]