Abstract
Genetic factors strongly influence interindividual variation in plasma high density lipoprotein cholesterol (HDL-C) levels, but the specific genetic polymorphisms that confer heritable variation in HDL-C levels have not been identified. In this study we examined the relationship between polymorphism in LIPC, the gene encoding hepatic lipase, and plasma HDL-C concentrations using a sequential approach comprising linkage analysis, DNA sequencing, and association studies. Linkage studies in 1465 American white subjects from 218 nuclear families indicated that allelic variation at, or closely linked to, the hepatic lipase gene accounts for a significant fraction (≈25%) of the variation in plasma HDL-C concentrations. The hepatic lipase gene was then sequenced in selected individuals, and four novel polymorphisms were identified in the 5′ flanking region of the gene. These polymorphisms were in complete linkage disequilibrium and thus identified a single novel allele. Association studies indicated that heterozygosity for the rare allele was associated with modestly increased concentrations of plasma HDL-C (41 ± 11 vs. 37 ± 10 mg/dl, P < 0.05) and apolipoprotein AI in men (131 ± 23 vs. 122 ± 21 mg/dl, P < 0.05) but not in women. Homozygosity for the rare allele was associated with markedly higher plasma HDL-C (63 ± 3 mg/dl) and apolipoprotein AI (153 ± 9 mg/dl) concentrations in men. The results of the association study were replicated in a second, independently ascertained sample. Taken together, the results of the linkage and association studies provide strong evidence that genetic variation in hepatic lipase activity is a major determinant of plasma HDL-C levels.
Epidemiologic studies have revealed that a low plasma concentration of high density lipoprotein cholesterol (HDL-C) is a major risk factor for coronary heart disease (1–3). Accordingly, the causes of low HDL levels have been investigated extensively. HDL metabolism is clearly modulated by environmental variables, and factors that confer coronary risk, such as cigarette smoking (4), obesity (5, 6), and lack of exercise (7), are frequently associated with low plasma HDL-C concentrations. HDL levels are also strongly influenced by genetic factors. Data from several family and twin studies (3, 8–12) indicate that genetic polymorphism accounts for between 40 and 60% of the interindividual variation in plasma HDL-C levels. Although heritability estimates based on family data are potentially confounded by the fact that relatives who live together share environment as well as genes, the similarity of plasma HDL levels in identical twins reared apart provides compelling evidence that genetic polymorphism is a major cause of interindividual variation in plasma HDL-C concentrations (9, 13).
Several studies have attempted to identify the specific genetic determinants of plasma HDL-C concentrations (14, 15). Plasma HDL-C concentrations do not segregate in Mendelian fashion in most families, so most studies have sought statistical associations between DNA sequence polymorphisms in candidate genes and plasma HDL-C concentrations in cohorts of unrelated patients (14, 15). To date, the fraction of variation in HDL-C levels that has been accounted for in these association studies is very small. More recent studies have used linkage methods to investigate the relationship between plasma HDL-C concentrations and candidate genes in extended families. Bu et al. (16) used sibling-pair analysis to test for linkage between plasma HDL-C concentrations and several candidate loci in 30 families solicited through patients with coronary atherosclerosis. In that study, markers linked to the cholesteryl ester transfer protein and apolipoprotein(a) loci were significantly associated with variation in plasma HDL-C concentrations. Mahaney et al. (17) used complex segregation and linkage analysis to investigate the genetic architecture of HDL-C in 526 Mexican-Americans from 25 randomly ascertained pedigrees. This analysis indicated that an unidentified major gene locus accounted for a small fraction of the variation in HDL-C. The locus was not linked to the genes encoding APOAI/CIII/AIV, hepatic lipase, lipoprotein lipase, or the low density lipoprotein receptor.
In a previous study of normotriglyceridemic whites, we used robust sibling-pair analysis to investigate the relationship between plasma HDL-C concentrations and polymorphism in the genes encoding hepatic lipase, apolipoprotein AI/CIII/AIV, and cholesteryl ester transfer protein (18). This analysis indicated that polymorphism at, or closely linked to, the genes encoding hepatic lipase and the apolipoprotein AI/CIII/AIV cluster accounted for a major fraction of the variation in plasma HDL-C concentrations. The effects of the two genes were not due to individuals with extreme trait levels, suggesting that common polymorphisms at these loci are systematically associated with variation in HDL-C concentrations. On the basis of these observations, we have undertaken further studies to (i) verify the findings in a larger sample, (ii) estimate variance components using likelihood methods for pedigrees, (iii) identify polymorphisms that are systematically associated with different HDL-C concentrations, and (iv) test the relationship between the polymorphisms identified and plasma HDL-C concentrations in association studies. In this report, we present strong evidence for linkage between LIPC (the gene encoding hepatic lipase) and HDL-C levels and identify a hepatic lipase allele that is systematically associated with high plasma HDL-C concentrations in men.
METHODS
Subjects.
The study protocol was approved by the Institutional Review Board of the University of Texas Southwestern Medical Center at Dallas. Two groups of subjects were solicited independently. For linkage and variance components analysis and initial studies of association, a group of nuclear families was selected solely on the basis of family size (group I). For this group, families in which both parents and three or more children were available for sampling were recruited by advertisements in newspapers, churches, and health centers. Data from 80 of these families were the subject of a previous report (18). To perform confirmatory association studies in an independent sample (Group II), we recruited male relatives of probands with documented premature coronary atherosclerosis (defined as coronary artery bypass graft, coronary angioplasty, or angiographic evidence of greater than 75% stenosis of at least one major coronary artery occurring in men aged <60 years or in women aged <65 years).
Fasting blood samples were drawn into 10-ml vacuum tubes containing sodium EDTA. Plasma was separated by centrifugation and stored at 4°C until analysis. Genomic DNA was isolated from whole blood using commercial DNA isolation kits from Qiagen (Chatsworth, CA; catalog no. 10243). All participants completed a detailed questionnaire to furnish data on family history, past and current health status, diet and exercise habits, alcohol and tobacco use, and medications.
Assay of Plasma Lipids and Lipoproteins.
Plasma cholesterol and triglyceride concentrations were determined in duplicate by enzymatic assay using commercial reagents (Cholesterol/HP, Boehringer Mannheim, and Triglycerides GPO-TRINDER, Sigma). HDL-C was measured in the supernatant after precipitation of apolipoprotein B-containing lipoproteins with sodium phosphotungstate (0.555 mM)–MgCl2 (25 mM) (19). In 80 plasma samples, the mean HDL-C concentration determined using sodium phosphotungstate precipitation was 5 mg/dl lower than the mean value obtained using heparin manganese (46 mM) and was identical to the mean value obtained using heparin manganese (92 mM). The results from the three methods were highly correlated (r > 0.93). Intraassay variation was <3% for plasma cholesterol and triglyceride and <5% for plasma HDL-C. The plasma concentrations of apolipoprotein AI, the major protein constituent of HDL, were determined on a subset of the samples (n = 1200) by immunoturbidimetric assay (20) using antisera and standards purchased from Incstar (Stillwater, MN). The correlation coefficient between plasma HDL-C and apolipoprotein AI concentrations was 0.74, in excellent agreement with published reports (21–23).
Assay of Microsatellite Polymorphisms.
The four parental LIPC alleles of each nuclear family were distinguished using a single-strand conformation polymorphism and five microsatellite polymorphisms (LIPC, D15S98, D15S198, D15S117, and D15S643) located within 5 cM of the gene as described (18). In each family, at least two informative polymorphisms, including one within the gene, or one on each side of the gene was analyzed to preclude recombination between the polymorphism and the gene. Data from 23 individuals whose LIPC inheritance could not be unambiguously determined were excluded from the analysis.
DNA Sequencing.
The coding region, intron/exon boundaries, and 780 bp of 5′ flanking sequence of the hepatic lipase gene were sequenced using a standard protocol for cycle sequencing (24). DNA regions to be sequenced were PCR-amplified and purified using spin columns to remove excess PCR primers and buffer. Sequencing reactions were performed in 20-μl volumes containing 8 μl of terminator mix, 100–200 ng of DNA template, 3.2 pmol of primer, and 8 μl of water. Reactions were overlayed with 50 μl of mineral oil and subjected to 25 cycles of amplification. Extension products were purified using Centri-Sep spin columns (Princeton Separations, Adelphia, NJ), resuspended in loading buffer (containing deionized formamide, 25 mM EDTA, and 10 mg/liter blue dextran), heated to 90°C, and loaded onto an Applied Biosystems model 377 automated sequencer.
Assay of Hepatic Lipase Polymorphism.
A C-to-T substitution 514 bp upstream of the transcription initiation site created an NlaIII restriction site (5′-CATG-3′). To assay the polymorphism, a 299-bp fragment containing the restriction site was PCR-amplified using the primers 5′-AAGAAGTGTGTTTACTCTAGGATCA-3′ and 5′-GGTGGCTTCCACGTGGCTGCCTAAG-3′. The fragment was labeled by adding 0.3 pmol of [32P]dCTP to the PCR mixture. The PCR-amplified DNA fragment was digested by adding 10 units of NlaIII in 30 μl of New England Biolabs buffer 4 to the PCR and was electrophoresed on a 5% polyacrylamide gel.
Statistical Analysis. Heritability estimation.
The heritability index of plasma HDL-C levels was estimated by regressing the average of the offspring plasma HDL-C values on the mid-parent values using weighted least-squares. Weights to adjust for unequal family sizes were calculated as suggested by Falconer (25), with an unweighted least-squares estimate used as the initial estimate of heritability in the weights.
Sibling-pair linkage analysis.
Initial testing for linkage was performed using the sibling-pair method described by Haseman and Elston (26). This procedure is based on simple genetic assumptions. Full siblings who share both parental alleles of a gene in common may be considered to have identical copies of the gene “by-descent.” Accordingly, if allelic variation in a candidate gene is associated with variation in a trait, then trait levels should tend to be more similar in siblings sharing both parental alleles at that locus than in siblings sharing neither parental allele in common. Conversely, if variation in a candidate gene does not influence a trait, then concordance for trait levels among siblings should be independent of the number of alleles of the candidate gene they share. In the model on which the procedure was originally based, genetic variance was due to a single biallelic gene, an assumption that is almost certainly violated for most traits. More recently, we proved that the procedure is also valid when genetic variance is due to multiple genes each having two or more alleles (Stoesz M., C.J.C., and G.R., unpublished work). The Haseman–Elston procedure regresses squared sibling-pair differences in trait levels (Δ2) on the proportion of alleles shared identical-by-descent (π) at the candidate locus. Regression parameters are estimated by ordinary least-squares, and a negative slope (decreasing Δ2 with increasing π) is considered evidence for linkage. Simulation studies have suggested that a one-sided t test of the slope parameter is unlikely to falsely reject the null hypothesis and is robust to nonindependent Δ2 values, which are inevitable in sibships of size 3 or greater (27, 28). Tests for linkage were performed using the observed data from all possible pairs.
To accommodate known limitations of the Haseman–Elston procedure, the data were reanalyzed after taking the following steps: (i) The method does not easily accommodate covariates, therefore the observed data were preadjusted for the effects of age and sex using Z scores as described (18). The timing of the pubertal decrease in HDL-C in males is difficult to establish, so all males between the ages of 12 and 16 years (inclusive) were omitted. (ii) To exclude possible confounding by environmental factors with a strong influence on HDL-C levels, individuals using hormones or lipid-lowering drugs or who had diabetes, monogenic dyslipidemia, or plasma triglyceride concentrations >200 mg/dl were excluded from the analysis. And (iii), the statistical test is based on a least-squares regression estimate that is sensitive to outliers, therefore values of Δ2 falling in the upper or lower 2.5 percentile at each π were excluded before regression analysis.
The squared sibling-pair difference (Δ2) on which the Haseman–Elston procedure is based does not satisfy the usual assumptions of the t test (29), so corroborative testing was performed using a nonparametric procedure as suggested by Kruglyak and Lander (30). The Jonckheere–Terpstra test (31) is appropriate in the present context because it can be used to evaluate whether the magnitude of Δ decreases with increasing π, as is expected under the alternative hypothesis of linkage.
Variance component analysis.
To test for linkage and estimate variance components using a more complete genetic model, we used a robust variance components procedure proposed by Hopper and Mathews (32) and developed by Amos (33). Like the Haseman–Elston method, the robust variance components method does not require specification of the genetic mechanism underlying the trait, requiring only identity-by-descent data at the candidate locus g. In addition, the Amos model more directly accommodates covariate effects and genetic covariances among siblings. This model attributes trait variance to covariate effects, genetic covariance due to a major gene locus (g), and residual polygenes (pg) and residual nongenetic effects (e). The variance components σg2 (major gene), σpg2 (polygenes), and σe2 (residual error) can be estimated using restricted maximum likelihood (34). To evaluate the null hypothesis of no linkage (σg2 = 0), a likelihood ratio test was used (35). The procedure was performed using all available data and was repeated after excluding individuals with secondary influences on HDL-C levels as described in (ii) above.
Association studies.
The association between the C-to-T polymorphism in the 5′ flanking region of the hepatic lipase gene and plasma HDL-C concentrations was tested in two independent samples. The first sample comprised all individuals from group I who did not use hormones or lipid-lowering drugs and who did not have diabetes, monogenic dyslipidemia, or plasma triglyceride concentrations >200 mg/dl. The polymorphism was assayed in the 50 men with the highest and in the 50 men with the lowest plasma HDL-C concentrations. A corresponding analysis was performed separately for women. The second sample included all men from group II who did not have premature coronary disease and who met the exclusion criteria used for group I (described above). The polymorphism was assayed in the 50 men with the highest and in the 50 men with the lowest plasma HDL-C concentrations. The association between genotype and HDL-C status (low or high) was tested using Fisher’s exact test.
RESULTS
Blood samples were drawn from 1465 individuals comprising 218 nuclear families selected solely on the basis of sibship size (group I). Ascertainment was essentially complete (>95%) for children over the age of 5 years. All participants identified themselves as white, not of Hispanic origin. Data from these individuals were used for heritability estimation, sibling pair linkage analysis, variance components analysis, and initial association studies.
Heritability Estimation.
Regression of mean offspring on mid-parent HDL-C indicated that genetic factors accounted for 46 ± 9% of the variance in plasma HDL-C concentrations. Spousal HDL-C concentrations were not significantly correlated (r = 0.09, P = 0.36).
Sibling-Pair Linkage Analysis.
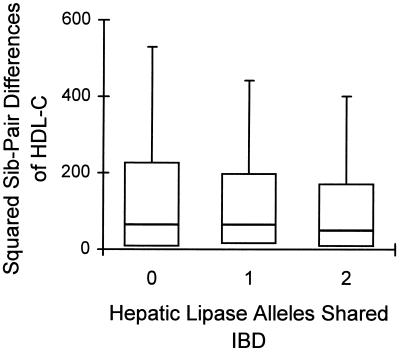
Analysis of the observed HDL-C data using all possible sibling-pairs revealed a clear decreasing trend in the squared sibling-paired differences of HDL-C levels with increasing proportion of hepatic lipase alleles shared in common (Fig. 1). Both parametric (t test) and nonparametric (Jonckheere–Terpstra test) tests for linkage between LIPC and plasma HDL-C concentrations were significant (Table 1). A second series of analyses was then performed to protect against spurious linkage due to covariate effects or outlying points. Exclusion of individuals with possible secondary causes of low or high HDL-C, adjustment for age and sex, exclusion of males aged 12 to 16 years, and exclusion of outlying points decreased the sample size to 1100 pairs, but the P values for linkage remained significant (Table 1).
Figure 1.
Distributions of squared sibling-pair differences of plasma HDL-C levels in siblings sharing none, one, or two hepatic lipase alleles identical-by-descent. Median values are indicated by bars within boxes; upper and lower quartiles are indicated by the tops and bottoms of the boxes, respectively; whiskers indicate the points closest to one-half of the interquartile range from the tops of the boxes.
Table 1.
Evidence for linkage between LIPC and plasma HDL-C concentrations
| Linkage analysis | Test | Data | n* | P |
|---|---|---|---|---|
| Sibling-pair | Haseman–Elston t test | observed data from all sibling-pairs | 1995 | 0.01 |
| Sibling-pair | Jonckheere–Terpstra | observed data from all sibling-pairs | 1995 | 0.01 |
| Sibling-pair | Haseman–Elston t test | age- and sex-adjusted†, restricted data‡ | 1100 | 0.03 |
| Sibling-pair | Jonckheere–Terpstra | age- and sex-adjusted†, restricted data‡ | 1100 | 0.01 |
| Sibling-pair | Haseman–Elston t test | observed data from CC homozygotes | 1097 | 0.03 |
| Variance component | likelihood ratio | all individuals | 1413 | 0.02 |
| Variance component | likelihood ratio | restricted data‡ | 987 | 0.01 |
| Variance component | likelihood ratio | CC homozygotes | 750 | 0.02 |
For sibling-pair analyses, n = number of sibling pairs. For variance components analysis, n = number of individuals.
Observed data were preadjusted for the effects of age and sex using Z scores.
Males between the ages of 12 and 16 years (inclusive) and individuals using hormones or lipid-lowering drugs or who had diabetes, monogenic dyslipidemia, or plasma triglyceride concentrations >200 mg/dl were excluded from the analysis.
Variance Component Analysis.
The likelihood ratio test comparing models with and without the hepatic lipase gene indicated that the variance component due to LIPC was significantly greater than 0 (Table 1). Variance component analysis indicated that polymorphism at the hepatic lipase locus accounted for 25% of the variation in plasma HDL-C concentrations.
Identification of DNA Sequence Polymorphism.
Four polymorphisms (−250 G-to-A, −514 C-to-T, −710 T-to-C, and −763 A-to-G) were identified in the 780-bp region 5′ of the transcription initiation sequence. The sequence changes were confirmed by restriction digestion with the enzymes DraI, NlaIII, AvaII, and SphI, respectively. Analysis of 13 individuals who were homozygous for −514 T and 12 individuals homozygous for −514 C indicated that the four polymorphisms were in complete linkage disequilibrium; therefore, subsequent analyses were performed using only the −514 polymorphism. The frequency of LIPC alleles with T at position −514 was found to be 0.15 in 272 unrelated individuals. The observed genotype frequencies (198 CC, 67 CT, and 7 TT) were consistent with Hardy–Weinberg equilibrium (197 CC, 69 CT, and 6 TO). Sequencing of both strands of the coding region and intron/exon boundaries in two homozygotes for the −514 T allele revealed no further polymorphisms.
Association Studies.
Analysis of the plasma lipid and lipoprotein levels of 272 unrelated individuals (from group I) revealed a small effect of the CT genotype on plasma HDL-C and apolipoprotein AI levels among men (Table 2) but not in women. Only two men were homozygotes for the TT genotype, and both had very high plasma HDL-C and apolipoprotein AI levels. No effect of LIPC genotype on total cholesterol or triglyceride concentrations was observed in either sex (Table 2). To test the association between the −514 T allele of hepatic lipase and elevated or decreased plasma HDL-C concentrations among individuals in group I, the frequencies of the three possible genotypes (CC, CT, and TT) were compared in the 50 men with the lowest HDL-C concentrations and in the 50 men with the highest HDL-C concentrations. The association between LIPC genotype and HDL-C status (high or low) was significant at the 0.01 level (Table 3). No homozygotes for the −514 T allele were found in the low HDL-C group whereas eight were found in the high HDL-C group. The mean plasma HDL-C and apolipoprotein AI concentrations of these eight men were 63 ± 3 mg/dl and 153 ± 9 mg/dl, respectively. The frequency of CT heterozygotes was similar in the two groups. A similar analysis performed among women in group I revealed no association between the −514 C-to-T polymorphism and HDL-C concentrations (Table 3).
Table 2.
Lipoprotein levels associated with LIPC genotypes
| Men
|
Women
|
|||||
|---|---|---|---|---|---|---|
| CC | CT | TT | CC | CT | TT | |
| n | 101 | 30 | 2 | 97 | 37 | 5 |
| Cholesterol | 197 ± 39 | 209 ± 32 | 201 ± 45 | 195 ± 40 | 190 ± 39 | 199 ± 40 |
| Triglyceride | 159 ± 110 | 146 ± 74 | 78 ± 9 | 106 ± 48 | 116 ± 67 | 108 ± 50 |
| HDL-C | 37 ± 10 | 41 ± 11* | 65 ± 2 | 50 ± 14 | 48 ± 13 | 56 ± 20 |
| Apo AI | 122 ± 21 | 131 ± 23* | 160 ± 3 | 140 ± 30 | 140 ± 25 | 160 ± 40 |
Values are means ± SD. Units are mg/dl. Apo, apolipoprotein.
Mean HDL-C and apolipoprotein AI levels are higher in men with CT genotype than in men with CC genotype (unpaired t test, P < 0.05). The P values remained significant after exclusion of individuals who had plasma triglyceride concentrations >200 mg/dl. None of the other lipid or lipoprotein levels differed with genotype in men or in women.
Table 3.
Association between LIPC genotype and plasma HDL-C concentrations
| Group I men*
|
Group I women
|
Group II men*
|
||||
|---|---|---|---|---|---|---|
| Low HDL-C | High HDL-C | Low HDL-C | High HDL-C | Low HDL-C | High HDL-C | |
| n | 50 | 50 | 50 | 50 | 50 | 50 |
| HDL-C | 26 ± 3 | 65 ± 9 | 30 ± 3 | 77 ± 8 | 29 ± 3 | 53 ± 8 |
| CC, n | 36 | 27 | 30 | 28 | 36 | 28 |
| CT, n | 14 | 15 | 17 | 18 | 14 | 15 |
| TT, n | 0 | 8 | 3 | 4 | 0 | 7 |
Association between LIPC genotype and HDL-C status (high or low) was significant at the 0.01 level.
Given the different findings in men and women from group I, a second association study was undertaken in an independent group (group II) comprising 441 male relatives of individuals with premature coronary atherosclerosis. The frequencies of the CC, CT, and TT genotypes were compared in the 50 men with the lowest HDL-C concentrations and in the 50 men with the highest HDL-C concentrations. The association between LIPC genotype and HDL-C status (high or low) was significant at the 0.01 level (Table 3). None of the 50 men with the lowest HDL-C were homozygous for the hepatic lipase −514 T allele whereas 7 of the 50 men in the high HDL-C group were homozygous for this allele. The mean plasma HDL-C and apolipoprotein AI concentrations of these seven men were 56 ± 11 mg/dl and 147 ± 14 mg/dl, respectively.
Linkage and Variance Components Analysis in CC Homozygotes.
To determine whether the polymorphisms identified in this study accounted for all of the effect of hepatic lipase genotype on plasma HDL-C concentrations, sibling-pair linkage analysis and variance components analysis were performed in 115 families in which both parents were homozygous for the −514 C allele. Both tests revealed significant evidence for linkage between LIPC and HDL-C levels (Table 1).
DISCUSSION
There is considerable evidence that hepatic lipase activity is an important determinant of plasma HDL-C concentrations. Clinical studies consistently have found an inverse relationship between hepatic lipase activity measured in postheparin plasma and plasma HDL-C concentrations (24, 36–39), and drugs, such as anabolic steroids, that increase hepatic lipase activity cause a proportional reduction in plasma levels of HDL-C (40). A study of monozygotic and fraternal twins indicated that hepatic lipase activity is strongly influenced by genetic factors (41), but the effect of polymorphism in the hepatic lipase gene on plasma HDL-C concentrations has received little attention. In this study, we used a sequential approach comprising linkage analysis, DNA sequencing, and association studies to investigate the role of genetic variation in LIPC, the gene encoding hepatic lipase. Two primary observations were established. First, linkage and variance components analysis indicated that allelic variation at, or closely linked to, the hepatic lipase gene accounts for a significant fraction (≈25%) of the variation in plasma HDL-C concentrations. Second, a hepatic lipase allele with systematic effects on plasma HDL-C concentrations was identified. Taken together, these two observations provide strong evidence that genetic variation in hepatic lipase activity is a major determinant of plasma HDL-C levels.
The relationship between hepatic lipase activity and plasma HDL-C concentrations has been reported widely, but direct evidence for linkage between LIPC and HDL-C has not been published previously. Our preliminary analysis of 80 nuclear families (comprising a subset of the current data) indicated that allelic variation in hepatic lipase accounted for 25% of the variation in plasma HDL-C concentrations, but the sample size did not provide sufficient power for statistically significant linkage (18). Therefore, the first goal of the present study was to determine whether polymorphism in, or closely linked to, the hepatic lipase gene is a significant cause of variation in plasma HDL-C concentrations. Sibling-pair linkage analysis under the least restrictive conditions (i.e., using the raw data) resulted in significant P values using both parametric and nonparametric tests. Significant evidence for linkage also was obtained when the sibling-pair procedure was applied under the most conservative conditions (exclusion of individuals with secondary factors known to influence HDL-C, adjustment of the data for the effects of age and sex, and exclusion of outlying points). Essentially identical results were obtained when the data were analyzed using a more complete genetic model evaluated by maximum likelihood methods. Thus, it is highly unlikely that the linkage observed between LIPC and plasma HDL-C concentrations is an artifact of the analytical methods used or of covariates or outliers. Accordingly, the results of the linkage analysis indicate that polymorphism in the hepatic lipase gene, or in a closely linked gene, causes interindividual variation in plasma HDL-C levels.
To identify DNA polymorphisms that may confer heritable variation in plasma HDL-C concentrations, we sequenced 780 base pairs of the 5′ flanking region and the exons and intron/exon boundaries of the gene in selected individuals. The first polymorphism identified was a C-to-T substitution 514 base pairs upstream of the transcription initiation site. Substitution of T for C at this position in the sequence introduced an NlaIII restriction site that was used to confirm the results of the sequencing and provide a simple assay for association studies. The −514 T allele was far less common than the −514 C allele; therefore, we examined the effects of the polymorphism by comparing the prevalence of TT homozygotes among individuals with high or low HDL-C concentrations. To control for spurious associations between the TT genotype and HDL-C concentrations, data from individuals with secondary conditions such as elevated plasma triglyceride concentrations, diabetes, or smoking (which are known to influence HDL-C concentrations) were excluded from the analysis, and two separate studies were performed in individuals ascertained by different criteria. These studies provided direct evidence for an association between the TT genotype and high plasma HDL-C concentrations in men. Heterozygosity for the −514 T allele was associated with a modest increase in plasma HDL-C concentrations. The effect of the −514 T allele on HDL-C concentrations was thus consistent with a recessive pattern of inheritance, common to several enzyme deficiencies. No effect of the TT genotype was evident in women. The different effects of the polymorphism on HDL-C levels of men and women may reflect gender-specific expression of the polymorphic alleles. Alternatively, the rate-limiting factors determining plasma HDL-C concentrations may differ among men and women.
We did not measure hepatic lipase activity in the present study, so we only speculate about the effects of the −514 polymorphism on the activity of this enzyme. Indeed, we cannot exclude the possibility that the −514 T is in linkage disequilibrium with an HDL-raising allele of another, as yet unidentified gene that is closely linked to LIPC. Given the well established inverse relationship between hepatic lipase activity and HDL-C concentrations, however, it seems most likely that the −514 T is associated with low hepatic lipase activity either by directly affecting hepatic lipase expression or through linkage disequilibrium with another polymorphism that directly decreases hepatic lipase activity. To determine whether LIPC alleles containing −514 T encode a dysfunctional hepatic lipase protein, the coding region and the intron/exon junctions of the hepatic lipase gene were sequenced in two homozygotes for the T allele. Sequencing of both the coding and noncoding strands did not reveal any polymorphisms in the coding region or in the intron/exon boundaries. Therefore, it is most unlikely that the effects of the T allele were caused by production of an abnormal hepatic lipase protein. Three additional polymorphisms were identified in the 5′ flanking region. Each of these polymorphisms appeared to be in complete linkage disequilibrium with the −514 T allele, constituting a single haplotype. The association between the −514 T allele and plasma HDL-C concentrations may be due to any of these polymorphisms acting individually or in combination or to linkage disequilibrium between these polymorphisms and another, as yet identified sequence change(s).
To determine whether linkage between LIPC and plasma HDL-C concentrations was due entirely to the allele defined by −514 T, linkage studies were performed in families in which the T allele was not present (both parents were CC homozygotes). These analyses indicated significant linkage between LIPC and HDL-C concentrations. Thus, although homozygosity for the T genotype has a marked effect on the HDL-C concentration of an individual, the allele defined by this polymorphism does not fully account for the effects of hepatic lipase polymorphism on plasma HDL-C concentrations in the population. Accordingly, other hepatic lipase alleles with systematic effects on plasma HDL-C levels are likely to be present in the population. We anticipate that further sequencing of the hepatic lipase gene will reveal additional polymorphisms that confer heritable variation in plasma HDL-C concentrations.
Acknowledgments
We gratefully acknowledge Betty Pham and Sijing Niu for excellent technical assistance and Dick Verstraete and Ned Warner for recruiting the families. This work was supported by National Institutes of Health Grants HL-53917 and HL-29252.
ABBREVIATION
- HDL-C
high density lipoprotein cholesterol
References
- 1.Miller N E, Miller G J. Lancet. 1975;i:1033. doi: 10.1016/s0140-6736(75)91977-7. [DOI] [PubMed] [Google Scholar]
- 2.National Cholesterol Education Program. NIH Publication No. 93–3095. Bethesda, MD: National Institutes of Health; 1993. [Google Scholar]
- 3.Bucher K D, Friedlander Y, Kaplan E B, Namboodiri K K, Kark J D, Eisenberg S, Stein Y, Rifkind B M. Genet Epidemiol. 1988;5:17–33. doi: 10.1002/gepi.1370050103. [DOI] [PubMed] [Google Scholar]
- 4.Craig W Y, Palomaki G E, Haddow J E. Br Med J. 1996;298:784–788. doi: 10.1136/bmj.298.6676.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutherland W H, Temple W A, Nye E R, Herbison G P. Am J Clin Nutr. 1980;33:2581–2587. doi: 10.1093/ajcn/33.12.2581. [DOI] [PubMed] [Google Scholar]
- 6.Wannamethee G, Shaper A G. J Epidemiol Commun Health. 1992;46:197–202. doi: 10.1136/jech.46.3.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schieken R M. Ann NY Acad Sci. 1991;623:269–274. doi: 10.1111/j.1749-6632.1991.tb43736.x. [DOI] [PubMed] [Google Scholar]
- 8.Perusse L, Despres J, Tremblay A, Leblanc C, Talbot J, Allard C, Bouchard C. Arteriosclerosis. 1989;9:308–318. doi: 10.1161/01.atv.9.3.308. [DOI] [PubMed] [Google Scholar]
- 9.Heller D A, De Faire D, Pedersen N L, Dahlen G, McClearn G E. N Engl J Med. 1993;328:1150–1156. doi: 10.1056/NEJM199304223281603. [DOI] [PubMed] [Google Scholar]
- 10.Rice T, Vogler G P, Perry T S, Laskarzewski P M, Rao D C. Hum Hered. 1991;41:107–121. doi: 10.1159/000153987. [DOI] [PubMed] [Google Scholar]
- 11.Austin M A, King M C, Bawol R D, Hulley S B, Friedman G D. Am J Epidemiol. 1987;125:308–318. doi: 10.1093/oxfordjournals.aje.a114531. [DOI] [PubMed] [Google Scholar]
- 12.Rao D C, Laskarzewski P M, Morrison J A, Khoury P, Kelly K, Wette R, Russell J, Glueck C J. Am J Hum Genet. 1982;34:888–903. [PMC free article] [PubMed] [Google Scholar]
- 13.Heller D A, Pedersen N L, de Faire U, McClearn G E. Am J Hum Genet. 1994;55:1255–1267. [PMC free article] [PubMed] [Google Scholar]
- 14.Lusis A J. J Lipid Res. 1988;29:397–429. [PubMed] [Google Scholar]
- 15.Humphries S E. Atherosclerosis. 1988;72:89–108. doi: 10.1016/0021-9150(88)90069-x. [DOI] [PubMed] [Google Scholar]
- 16.Bu X, Warden C H, Xia Y R, De Meester C, Puppione D L, Teruya S, Lokensgard B, Daneshmand S, Brown J, Gray R J, Rotter J I, Lusis A J. Hum Genet. 1994;93:639–648. doi: 10.1007/BF00201563. [DOI] [PubMed] [Google Scholar]
- 17.Mahaney M C, Blangero J, Rainwater D L, Comuzzie A G, VandeBerg J L, Stern M P, MacCluer J W, Hixson J E. Arterioscler Thromb Vasc Biol. 1995;15:1730–1739. doi: 10.1161/01.atv.15.10.1730. [DOI] [PubMed] [Google Scholar]
- 18.Cohen J C, Wang Z, Grundy S M, Stoesz M R, Guerra R. J Clin Invest. 1994;94:2377–2384. doi: 10.1172/JCI117603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assmann G, Schriewer H, Schmitz G, Hagele E O. Clin Chem. 1983;29:2026–2030. [PubMed] [Google Scholar]
- 20.Sandkamp M, Tambyrajah B, Schriewer H, Assmann G. J Clin Chem Clin Biochem. 1988;26:685–688. doi: 10.1515/cclm.1988.26.11.685. [DOI] [PubMed] [Google Scholar]
- 21.Patsch W, Sharrett A R, Sorlie P D, Davis C E, Brown S A. Am J Epidemiol. 1992;136:546–557. doi: 10.1093/oxfordjournals.aje.a116532. [DOI] [PubMed] [Google Scholar]
- 22.Phillips N R, Havel R J, Kane J P. Am J Epidemiol. 1982;116:302–313. doi: 10.1093/oxfordjournals.aje.a113414. [DOI] [PubMed] [Google Scholar]
- 23.Contois J, McNamara J R, Lammi-Keefe C, Wilson P W, Massov T, Schaefer E J. Clin Chem. 1996;42:507–514. [PubMed] [Google Scholar]
- 24.Applebaum-Bowden D, Haffner S M, Wahl P W, Hoover J J, Warnick G R, Albers J J, Hazzard W R. Arteriosclerosis. 1985;5:273–282. doi: 10.1161/01.atv.5.3.273. [DOI] [PubMed] [Google Scholar]
- 25.Falconer D S. Quantitative Genetics. 3rd Ed. Harrow: Longman; 1989. pp. 148–186. [Google Scholar]
- 26.Haseman J K, Elston R C. Behav Genet. 1972;2:3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- 27.Blackwelder W C, Elston R C. Commun Stat. 1982;11:449–484. [Google Scholar]
- 28.Amos C I, Elston R C, Wilson A F, Bailey-Wilson J E. Genet Epidemiol. 1989;6:435–449. doi: 10.1002/gepi.1370060306. [DOI] [PubMed] [Google Scholar]
- 29.Kruglyak L, Lander E S. Am J Hum Genet. 1995;57:439–454. [PMC free article] [PubMed] [Google Scholar]
- 30.Kruglyak L, Lander E S. Genetics. 1995;139:1421–1428. doi: 10.1093/genetics/139.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehmann E L. Nonparametrics: Statistical Methods Based on Ranks. San Francisco: Holden–Day; 1975. pp. 232–238. [Google Scholar]
- 32.Hopper J L, Mathews J D. Ann Hum Genet. 1982;46:373–383. doi: 10.1111/j.1469-1809.1982.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 33.Amos C I. Am J Hum Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- 34.Harville D A. J Am Stat Assoc. 1977;72:320–340. [Google Scholar]
- 35.Self S G, Liang K L. J Am Stat Assoc. 1987;82:605–610. [Google Scholar]
- 36.Kuusi T, Saarinen P, Nikkila E A. Atherosclerosis. 1980;36:589–593. doi: 10.1016/0021-9150(80)90251-8. [DOI] [PubMed] [Google Scholar]
- 37.Jackson R L, Yates M T, McNerney C A, Kashyap M L. Horm Metab Res. 1990;22:289–294. doi: 10.1055/s-2007-1004904. [DOI] [PubMed] [Google Scholar]
- 38.Kuusi T, Ehnholm C, Viikari J, Harkonen R, Vartiainen E, Puska P, Taskinen M R. J Lipid Res. 1989;30:1117–1126. [PubMed] [Google Scholar]
- 39.Blades B, Vega G L, Grundy S M. Arterioscler Thromb. 1993;13:1227–1235. doi: 10.1161/01.atv.13.8.1227. [DOI] [PubMed] [Google Scholar]
- 40.Kantor M A, Bianchini A, Bernier D, Sady S P, Thompson P D. Med Sci Sports Exercise. 1985;17:462–465. doi: 10.1249/00005768-198508000-00010. [DOI] [PubMed] [Google Scholar]
- 41.Kuusi T, Kesaniemi Y A, Vuoristo M, Miettinen T A, Koskenvuo M. Arteriosclerosis. 1987;7:421–425. doi: 10.1161/01.atv.7.4.421. [DOI] [PubMed] [Google Scholar]