Abstract
The mitochondrial inner membrane contains numerous multispanning integral proteins. The precursors of these hydrophobic proteins are synthesized in the cytosol and therefore have to cross the mitochondrial outer membrane and intermembrane space to reach the inner membrane. While the import pathways of noncleavable multispanning proteins, such as the metabolite carriers, have been characterized in detail by the generation of translocation intermediates, little is known about the mechanism by which cleavable preproteins of multispanning proteins, such as Oxa1, are transferred from the outer membrane to the inner membrane. We have identified a translocation intermediate of the Oxa1 preprotein in the translocase of the outer membrane (TOM) and found that there are differences from the import mechanisms of carrier proteins. The intermembrane space domain of the receptor Tom22 supports the stabilization of the Oxa1 intermediate. Transfer of the Oxa1 preprotein to the inner membrane is not affected by inactivation of the soluble TIM complexes. Both the inner membrane potential and matrix heat shock protein 70 are essential to release the preprotein from the TOM complex, suggesting a close functional cooperation of the TOM complex and the presequence translocase of the inner membrane. We conclude that mitochondria employ different mechanisms for translocation of multispanning proteins across the aqueous intermembrane space.
The mitochondrial inner membrane has to maintain the electrochemical proton gradient generated by the respiratory chain, while numerous metabolites and proteins need to be transported across or into this membrane in order to enable the organelle to fulfill its biological functions (18, 54, 76). As a consequence, the inner membrane of mitochondria is rich in specific proteins that mediate these transport steps (18, 53, 56). Among them are a large number of proteins with multiple hydrophobic transmembrane segments, so-called multispanning proteins. The mitochondrial genome encodes only a few multispanning proteins, belonging to the respiratory chain complexes (76, 78). Most multispanning proteins of the inner membrane are thus encoded by nuclear genes and synthesized on cytosolic polysomes. The proteins are recognized by receptors on the mitochondrial surface and are translocated by a general import pore (GIP) across the outer membrane. The translocase of the outer membrane (TOM) consists of a stable core complex (the GIP complex) and loosely associated receptor proteins (16, 26, 38, 49, 66). The mitochondrial inner membrane possesses two translocase complexes that mediate the import of precursor proteins. The presequence translocase of the inner membrane (TIM23 complex) mediates the import of all preproteins that carry cleavable amino-terminal targeting signals, termed presequences. The protein insertion complex (carrier translocase or TIM22 complex) is responsible for the insertion of multispanning membrane proteins that are synthesized without a presequence yet contain internal targeting signals (3, 30, 31, 38, 58, 65, 70, 72).
The metabolite carriers of the inner membrane, such as the abundant ADP/ATP carrier (AAC), as well as a few subunits of the TIM complexes, are representatives of the noncleavable multispanning proteins (3, 38, 63, 65). The precursors of carrier proteins are guided by a soluble hexameric complex of essential small Tim proteins, the Tim9-Tim10 complex, through the aqueous intermembrane space (1, 39, 41, 73). This soluble TIM complex binds to hydrophobic segments within the precursor proteins and is required to release the proteins from the TOM complex (10, 39, 73, 79). The carrier proteins are then transferred to the TIM22 complex, which assists in the translocation and insertion of the precursor proteins into the inner membrane in a strictly membrane potential-dependent manner (1, 17, 39, 41, 64, 73). A second soluble TIM complex of the intermembrane space has been found to assist in the transfer of the precursor of the multispanning protein Tim23 through the intermembrane space. This nonessential Tim8-Tim13 complex is homologous to the Tim9-Tim10 complex and promotes release of the Tim23 precursor from the outer membrane (11, 12, 40, 57).
Other multispanning inner membrane proteins, however, are synthesized with a cleavable presequence and, like soluble matrix proteins and inner membrane proteins with one or two transmembrane segments, employ the presequence translocase for their import. This translocase consists of two major parts: a complex with integral membrane proteins, including the pore-forming protein Tim23 (77), and an import motor complex at the matrix side (48, 55, 86). Two energy sources are needed to drive preproteins through the presequence translocase. The membrane potential activates the Tim23 channel and exerts an electrophoretic effect on the positively charged presequences (4, 24, 29, 47, 77). The essential matrix heat shock protein 70 (mtHsp70), also termed Ssc1 in yeast (34), forms the core of the import motor and functions in an ATP-dependent manner. Finally, the matrix-processing peptidase cleaves the translocated precursor proteins and selectively removes the presequences (3, 8, 20, 38). A representative of cleavable multispanning proteins is Oxa1 (oxidase assembly 1), which is synthesized with a 42-residue presequence (2, 7, 27). Oxa1 is the main component of a third translocase machinery of the mitochondrial inner membrane, the export machinery, which mediates the transport of mitochondrially encoded proteins and also some nuclear-encoded proteins from the matrix into the inner membrane (9, 25, 76). The mature 36-kDa Oxa1 contains five transmembrane segments. Herrmann et al. (27) reported that the precursor of Oxa1 is imported by the presequence translocase in a Δψ- and mtHsp70-dependent manner and inserts into the inner membrane from the matrix side.
Little is known about the transfer of the precursors of cleavable multispanning proteins from the outer membrane to the presequence translocase. The presence of several hydrophobic segments within these polypeptides should present similar demands upon their transport across the aqueous intermembrane space, as it does for the metabolite carriers. It has been reported that the import of Coq2 (polyprenyl diphosphate:4-HB transferase), a cleavable inner membrane protein with six putative transmembrane regions, involves the Tim9-Tim10 complex, suggesting that cleavable multispanning proteins are delivered to the presequence translocase via the soluble TIM complexes (46). A mechanistic characterization of the transport pathway, however, has not been possible due to the lack of suitable translocation intermediates.
Since an important advance in the analysis of carrier protein import into mitochondria has been the accumulation of a translocation intermediate in the outer membrane-intermembrane space, i.e., before insertion into the inner membrane (62, 67, 79), we attempted to obtain insight into the import mechanism of cleavable multispanning proteins by generation of a translocation intermediate. By reversible inactivation of the presequence translocase, we were able to accumulate a translocation intermediate of Oxa1 in the core of the TOM machinery, the GIP complex of the outer membrane. Surprisingly, transfer of the Oxa1 precursor through the intermembrane space is not impaired by inactivation of the soluble TIM complexes but depends on a close functional cooperation of the TOM complex and the TIM23-import motor machinery. Thus, mitochondria utilize different mechanisms to translocate hydrophobic proteins across the outer membrane and intermembrane space.
MATERIALS AND METHODS
Yeast strains, growth, and mitochondrial isolation conditions.
With the exception of AFY18, all yeast strains used were described previously: YPH499 (71), GB102 (MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim10::tim10-2) and PRY34 (MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim13::kanMX4 tim8::TRP1) (79), and PK82 (MATα his4-713 lys2 ura3-52 Δtrp1 leu2-3,112) and PK83 (MATα ade2-101 lys2 ura3-52 leu2-3,112 Δtrp1 ssc1-3[LEU2]) (21). Mitochondria were isolated from yeast strains grown at 30°C (AFY18, PRY34) or 24°C (GB102 and PK82) in YPG medium (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, and 3% [vol/vol] glycerol), resuspended in SEM (250 mM sucrose, 1 mM EDTA, and 10 mM MOPS [morpholinepropanesulfonic acid]-KOH [pH 7.2]) to a concentration of 10 mg/ml and stored at −80°C.
Generation of a yeast strain carrying tom22-2.
A PCR product containing the HIS3MX6 gene from pYM10 (36) with flanking sequences from Saccharomyces cerevisiae TOM22 that were complementary to the border sequences of the 3′ region truncated in tom22-2 (51) was generated by using primers AEF44 (5′-GACCACCACTGCTTTGTTACTCGGTGTGCCACTATCCTTATCTATACTTGCCGAACAATAGGGCGCGCCACTTCTA) and AEF45 (5′-CATGTATGGCTCCTTTTCTAAAACCCTCTCTTTTCTTTTACATCATTAAAATTAATGGCATCGATGAATTCGAGCTCG). The tom22-2 mutant was created by homologous recombination after transformation of YPH499 (71) with the PCR product and subsequent selection on minimal glucose medium containing all necessary growth supplements except histidine. The truncation of TOM22 (removal of the region encoding the C-terminal 31 amino acid residues) in strain AFY18 carrying the tom22-2 mutation was confirmed by PCR with genomic DNA as the template, DNA sequencing, and Western blotting of isolated mitochondrial proteins.
Cloning of OXA1 and COX18 for in vitro transcription-translation.
A PCR product carrying the coding sequence and an upstream promoter for SP6 polymerase was generated for S. cerevisiae OXA1 by using the primers PR71 (5′-GGATTTAGGTGACACTATAGAATACTGAAAAATTTAACCAGTGG) and PR72 (5′-ATAGAGCCTTTATTCATT) or for COX18 by using the primers PR73 (5′-GGATTTAGGTGACACTATAGAATACTGGCATTATGTTAAAGAGG) and PR74 (5′-CGTCAGGTTCACTCATCGTTGGT) and then cloned into the pCR-Blunt II-TOPO vector (Invitrogen). Radiolabeled precursor proteins were obtained by in vitro transcription from a plasmid (Oxa1) or by PCR (Cox18) by using SP6 polymerase and then translated in vitro by using rabbit reticulocyte lysate (Amersham) in the presence of [35S]methionine-cysteine essentially as described previously (68).
In vitro import of radiolabeled proteins.
Mitochondria were resuspended in import buffer (250 mM sucrose, 10 mM MOPS-KOH [pH 7.2], 80 mM KCl, 5 mM MgCl2, 3% [wt/vol] bovine serum albumin [BSA], and 5 mM methionine) that contained 2 mM ATP and 2 mM NADH. For import of Cox18, 350 mM KCl was used. Unless otherwise stated, import reaction mixtures also contained an ATP-regenerating system consisting of 5 mM creatine phosphate and 0.1 mg of creatine kinase per ml. The membrane potential was dissipated by addition of 1 μM valinomycin, 8 μM antimycin A, and 20 μM oligomycin. Import reactions were performed at 25°C (unless otherwise noted) by incubating mitochondria (25 to 75 μg of protein) with 5 to 15% (vol/vol) rabbit reticulocyte lysate containing 35S-labeled proteins. For import into ssc1-3 mitochondria and the corresponding wild type, a 15-min heat shock at 37°C was performed, where indicated, prior to import at 25°C. Import reactions were stopped by addition of 1 μM valinomycin. Where indicated, the mitochondria were protease treated by incubation with 50 μg of proteinase K per ml on ice for 15 min. The protease was inhibited by addition of 1 mM phenylmethylsulfonyl fluoride and incubated on ice for a further 10 min. Following a reisolation by centrifugation, the mitochondria were washed once with SEM, and the mitochondrial proteins were solubilized with the appropriate detergent-containing buffer and then resolved by sodium dodecyl sulfate (SDS)- or blue native (BN)-polyacrylamide gel electrophoresis (PAGE).
BN-PAGE and antibody shift BN-PAGE.
Separation of mitochondrial proteins by BN-PAGE was performed essentially as described previously (13, 79). Mitochondrial pellets (50 to 75 μg) were solubilized on ice for 15 min in 50 to 75 μl of prechilled buffer containing 20 mM Tris (pH 7.4), 0.1 mM EDTA, 50 mM NaCl, 10% (vol/vol) glycerol, 4 mM phenylmethylsulfonyl fluoride, and 1% (wt/vol) digitonin. Following a 15-min centrifugation at 12,000 × g and 4°C, 5 to 7.5 μl of sample buffer (5% Coomassie brilliant blue G-250, 500 mM ɛ-amino-n-caproic acid, and 100 mM bis-Tris [pH 7]) was added to the supernatants containing the solubilized mitochondrial proteins. These were then separated on a 6 to 16.5% polyacrylamide gradient gel at 4°C. For radiolabeled proteins, gels were destained, dried, and analyzed by digital autoradiography. For Western blotting, proteins were transferred to polyvinylidene difluoride membranes in transfer buffer (20 mM Tris, 150 mM glycine, 0.02% [wt/vol] SDS, and 20% [vol/vol] methanol), using a semidry blotting system.
For antibody shift BN-PAGE (79), immunoglobulin Gs (IgGs) were prepared by isolation from serum with protein A-Sepharose, lyophilization, and subsequent resuspension in import buffer prior to use (74). The Oxa1GIP intermediate was accumulated by import of radiolabeled Oxa1 for 20 min into mitochondria (50 μg protein) with a dissipated membrane potential generated by addition of 1 μM valinomycin, 8 μM antimycin A, and 20 μM oligomycin. Following reisolation and washing with SEM buffer, the mitochondria were resuspended in 100 μl of SEM buffer and incubated with the appropriate IgG on ice for 30 min with occasional mixing. After reisolation and washing, mitochondria were solubilized as described above and separated by BN-PAGE.
Chase of the Oxa1 intermediate.
Mitochondria were resuspended in low-BSA import buffer (250 mM sucrose, 10 mM MOPS-KOH [pH 7.2], 80 mM KCl, 5 mM MgCl2, 1% [wt/vol] BSA, and 5 mM methionine), and the Oxa1GIP was accumulated by import of radiolabeled Oxa1 under standard conditions for 20 min in the presence of 20 μM oligomycin and 60 μM carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone (FCCP). After transfer to ice, the mitochondria were reisolated, washed, and resuspended in SEM in the presence of 20 μM oligomycin and 60 μM FCCP or 1 μM valinomycin. The chase reaction was performed at 10°C by addition of standard import buffer supplemented with 2 mM ATP, 2 mM NADH, 5 mM creatine phosphate, 0.1 mg of creatine kinase per ml, and 20 μM oligomycin (diluting the FCCP to 6 μM). Reactions were stopped at the appropriate times with the addition of 1 μM valinomycin, and then the mixtures were treated as described above for separation by BN- or SDS-PAGE.
Miscellaneous.
SDS-PAGE was performed essentially as described previously (45). Radiolabeled proteins were detected by digital autoradiography (Molecular Dynamics). In some figures, nonrelevant gel lanes were excised by digital treatment. Western blotting was performed by standard techniques, and proteins were detected by enhanced chemiluminescence (Amersham).
RESULTS
The Oxa1 precursor accumulates in a 500-kDa complex in the absence of a membrane potential.
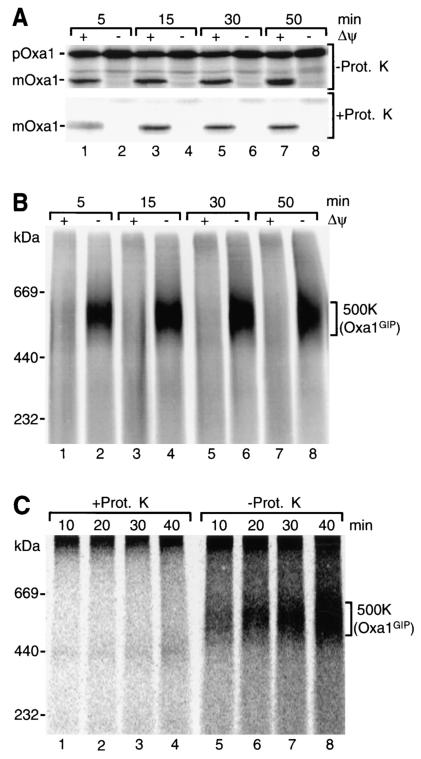
We synthesized the precursor of yeast Oxa1 in rabbit reticulocyte lysate in the presence of [35S]methionine-cysteine and imported it into isolated yeast mitochondria in the presence or absence of a membrane potential (Δψ) across the inner mitochondrial membrane. When the reisolated mitochondria were analyzed by SDS-PAGE, the expected result was obtained (27): In the presence of a Δψ, the Oxa1 precursor was processed to its mature form (Fig. 1A, upper panel, odd-numbered lanes). The processed form was largely protected against protease added to the isolated mitochondria (Fig. 1A, lower panel, odd-numbered lanes). When the Δψ was dissipated by the addition of the potassium ionophore valinomycin (in the presence of potassium in the medium), Oxa1 remained in its precursor form, which was degraded upon addition of protease (Fig. 1A, even-numbered lanes).
FIG. 1.
Accumulation of the precursor of Oxa1 in a 500-kDa complex of yeast mitochondria. 35S-labeled Oxa1 precursor was imported into wild-type mitochondria at 25°C in the presence or absence (addition of 1 μM valinomycin, 20 μM oligomycin, and 8 μM antimycin A) of a Δψ for the indicated times. (A) After import, mitochondria were either left untreated or treated with proteinase K (Prot. K), reisolated, and subjected to SDS-PAGE. The radiolabeled precursor (p) and mature (m) form of Oxa1 were visualized by digital autoradiography. (B) Following import of Oxa1 as for panel A, mitochondria were directly reisolated, solubilized in buffer containing 1% (wt/vol) digitonin, and analyzed by BN-PAGE. (C) Radiolabeled Oxa1 precursor was imported into wild-type mitochondria in the absence of a Δψ for the indicated times. Subsequently, samples were split and either left untreated or treated with proteinase K. After reisolation, mitochondria were solubilized and analyzed as for panel B.
Upon import of Oxa1, the mitochondria were lysed with the nonionic detergent digitonin and separated by BN-PAGE in order to detect protein complexes. Processed, mature Oxa1 did not reveal a defined high-molecular-weight complex (Fig. 1B, odd-numbered lanes). When the import reactions were performed in the absence of Δψ, however, the Oxa1 precursor was found in a complex of about 500 kDa (500K complex) (Fig. 1B, even-numbered lanes). The 500K complex was degraded by proteinase K added to the mitochondria after the accumulation of Oxa1 (Fig. 1C, lanes 1 to 4), indicating that it was exposed at the mitochondrial surface.
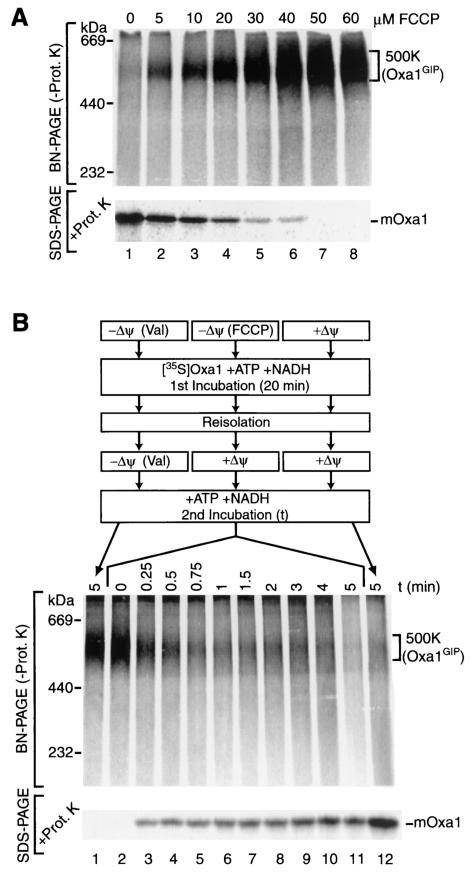
The mitochondrial membrane potential can be gradually reduced by addition of limiting amounts of a protonophore (24, 47, 64, 67). The Oxa1 precursor was incubated with mitochondria in the presence of increasing amounts of the protonophore FCCP; oligomycin was included to prevent generation of a Δψ by a reverse action of the F0F1-ATPase. The mitochondria were reisolated, split into two portions, and analyzed by two means (Fig. 2A): (i) BN-PAGE to assess the relative amounts of the Oxa1-containing 500K complex and (ii) protease treatment, followed by SDS-PAGE, to determine the level of imported, mature Oxa1. The gradual reduction of Δψ by increasing concentrations of FCCP caused a decrease in formation of mature Oxa1 and a concomitant increase in the formation of the 500K complex (Fig. 2A). Thus, the reduction in import due to lowering of the membrane potential correlates with the increased formation of the 500K intermediate, suggesting that the 500K complex may represent a translocation intermediate. In order to determine whether the 500K complex was a productive translocation intermediate, we investigated the competency of accumulated Oxa1 precursor to be chased to the inner membrane after reestablishing a membrane potential (Fig. 2B). The protonophore FCCP permits an efficient regeneration of Δψ upon reisolation of mitochondria and quenching of the remaining FCCP (67). Following arrest of the Oxa1 precursor as a 500K intermediate, under conditions of an FCCP-dissipated membrane potential, mitochondria were reisolated and regeneration of the Δψ was permitted for different times over a period of 5 min. Oxa1 was successfully chased from the 500K complex to a later import stage, as indicated by formation of mature Oxa1 in a time-dependent manner (Fig. 2B, lanes 2 to 11). As a control, a dissipated membrane potential was maintained throughout the course of the experiment, indicating the stability of the 500K complex (Fig. 2B, lane 1). As a second control, a full Δψ was present in both incubations, leading to an efficient import of Oxa1 (Fig. 2B, lane 12).
FIG. 2.
The precursor of Oxa1 forms a productive translocation intermediate. (A) Radiolabeled Oxa1 precursor was imported for 5 min at 25°C into mitochondria in the presence of FCCP, as indicated. The reaction mixtures were subsequently split in half and either subjected to proteinase K (Prot. K) treatment and SDS-PAGE or left untreated, solubilized in buffer containing 1% (wt/vol) digitonin, and subjected to BN-PAGE analysis. mOxa1, mature Oxa1. (B) Import of 35S-labeled Oxa1 precursor was performed either in the presence of a Δψ, in the presence of valinomycin (Val), or in the presence of 60 μM FCCP for 20 min at 25°C. After reisolation, mitochondria were resuspended in fresh import buffer and subjected to a second incubation at 10°C for the indicated times. During the second incubation, one sample remained in the presence of Δψ, one sample remained without a Δψ (Val), and samples that had previously received FCCP were now incubated in the presence of a reestablished Δψ. Samples were analyzed by BN-PAGE after solubilization or subjected to SDS-PAGE after proteinase K treatment.
We conclude that reestablishing a Δψ across the inner mitochondrial membrane allows for an efficient import of the arrested Oxa1 precursor to the inner membrane. These observations indicate that the 500K complex represents a productive translocation intermediate on the import pathway of Oxa1 into mitochondria.
The Oxa1 precursor is associated with the GIP complex of the outer membrane.
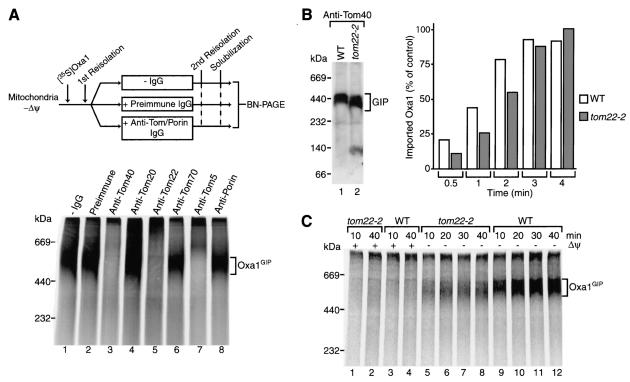
The exposure of the accumulated transport intermediate of Oxa1 on the mitochondrial surface, together with its approximate size of 500 kDa, suggested that it might be arrested in the TOM machinery (the GIP complex migrates at ∼440 kDa [79]). To directly determine whether Tom proteins associated with the arrested Oxa1 precursor, we performed antibody shift BN-PAGE (32, 79). Radiolabeled Oxa1 was accumulated in mitochondria in the absence of a Δψ. After reisolation, the mitochondria were incubated with antibodies directed against individual Tom proteins or control antibodies. The mitochondria were then lysed with digitonin and the complexes were separated by BN-PAGE (Fig. 3A). Association of the antibodies with their cognate antigen leads to a shift in complex size such that the antibody complexes migrate more slowly on BN-PAGE. Antibodies directed against subunits of the GIP complex, Tom40, Tom22, or Tom5 (14, 82), efficiently shifted the 500K complex to higher-molecular-weight species (Fig. 3A, lanes 3, 5, and 7). Multiple copies of Tom40 are present in the GIP complex, and thus the GIP-antibody complex is so large that it cannot be resolved by BN-PAGE (79). The 500K complex was not affected by antibodies directed against Tom70 and was affected only partially by antibodies directed against Tom20 (Fig. 3A, lanes 4 and 6), in good agreement with the loose association of these receptors with the GIP complex (14). Antibodies against the most abundant outer membrane protein, porin, as well as preimmune antibodies did not alter the mobility of the 500K complex. We conclude that the Oxa1 protein was arrested in the GIP complex of the outer membrane, which consists of Tom40, the central receptor Tom22, and small Tom proteins. The preprotein had obviously passed the initial stage of recognition on the mitochondrial surface, since it was not stably associated with the primary receptors Tom20 and Tom70. The quantitative shift of the 500K complex by antibodies against GIP subunits indicates that all arrested Oxa1 molecules are associated with the GIP complex. We thus refer to the 500K complex as the Oxa1GIP complex.
FIG. 3.
Accumulation of the Oxa1 precursor in the GIP complex. (A) Radiolabeled Oxa1 precursor was accumulated in wild-type mitochondria in the absence of a Δψ. After reisolation, mitochondria were left untreated (lane 1), received preimmune IgGs (lane 2), or received IgGs directed against the indicated Tom proteins or porin. Subsequently, mitochondria were solubilized in digitonin buffer and analyzed by BN-PAGE and digital autoradiography. (B) Left panel, wild-type (WT) and tom22-2 mitochondria were solubilized in digitonin buffer, protein complexes were separated by BN-PAGE, and proteins were transferred to polyvinylidene difluoride membranes by Western blotting. The GIP complex was detected with anti-Tom40 antiserum. Right panel, the 35S-labeled Oxa1 precursor was imported into wild-type and tom22-2 mitochondria for the indicated times at 25°C in the presence of a Δψ. Mitochondria were treated with proteinase K and analyzed by SDS-PAGE, and quantification of the digital autoradiogram was performed with ImageQuant 1.2 (Molecular Dynamics). The amount of maximal import after 4 min was set to 100% (control). (C) Import of radiolabeled Oxa1 precursor into wild-type and tom22-2 mitochondria was performed for the indicated times at 25°C in the presence or absence of a Δψ. The mitochondria were then reisolated and solubilized in digitonin buffer. Samples were subsequently analyzed by BN-PAGE and digital autoradiography.
The GIP complex is used by all types of mitochondrial precursor proteins, regardless of whether they are synthesized with or without a presequence (15, 35, 44, 50, 82). Discrimination between preprotein types has, however, been found to occur on the trans side of the outer membrane. The central receptor Tom22 not only exposes a precursor binding domain to the cytosol but also exposes a domain of 4 kDa to the intermembrane space. The intermembrane space domain of Tom22 binds mitochondrial presequences and stimulates the import of presequence-containing preproteins (6, 33, 42, 51). Yeast mutants lacking the intermembrane space domain of Tom22, termed tom22-2 cells, are viable yet show a moderate reduction in the import of cleavable matrix proteins, in particular when combined with other impairments of mitochondrial function such as removal of cytosolic receptor domains (51). The tom22-2 mutation, however, affects neither the import of carrier proteins into energized mitochondria nor the accumulation of carrier intermediates (43, 51). The subunit composition of the GIP complex of tom22-2 compared to wild-type mitochondria is not altered, and its stability is only mildly affected when assayed by BN-PAGE, but the GIP complex migrates slightly faster due to the smaller size of Tom22 (82) (Fig. 3B, lanes 1 and 2). We generated a yeast strain carrying a stably integrated tom22-2, whereby the chromosomal TOM22 gene was truncated such that a shortened Tom22 lacking the intermembrane space domain was expressed. In the early, linear range of the import reaction, the import efficiency of Oxa1 into energized tom22-2 mitochondria was moderately reduced compared to that into wild-type mitochondria (Fig. 3B). Upon longer import times, the import yield reached that of wild-type mitochondria. When the Oxa1 precursor was accumulated in tom22-2 mitochondria in the absence of a Δψ, however, the amount of Oxa1GIP complex was strongly reduced, even after long incubation times (Fig. 3C, lanes 5 to 8).
This observation indicates that the intermembrane space domain of Tom22 helps to stably arrest the Oxa1 precursor in the GIP complex when transfer to the inner membrane is blocked. With fully energized mitochondria, the intermembrane space domain of Tom22 plays a stimulating role in Oxa1 import yet can be bypassed at longer incubation times. Similarly, Oxa1 precursor accumulated in tom22-2 mitochondria could be chased to the mature form upon reestablishment of a Δψ (data not shown). The intermembrane space domain of Tom22 is negatively charged, suggesting that an interaction with preproteins may be of an ionic, i.e., salt-sensitive, nature; however, Oxa1 is a hydrophobic protein. We addressed how the arrested precursor of Oxa1 was bound to the GIP by solubilizing mitochondria in the presence of increasing salt concentrations, yet we did not observe a significant difference between wild-type and tom22-2 mitochondria (data not shown), indicating that ionic forces do not play a major role in the interaction of Oxa1 with the intermembrane space domain of Tom22. A similar result was obtained with the precursor of Cox18 (see below).
Inactivation of the soluble TIM complexes of the intermembrane space does not inhibit the import of Oxa1.
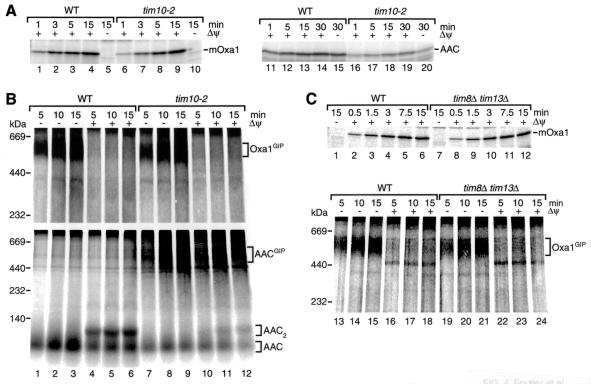
The Tim9-Tim10 complex is essential for carrier transport through the intermembrane space. The mutant allele tim10-2 impairs the function of this soluble TIM complex, causing a reduction of import of the AAC such that the AAC accumulates in a GIP intermediate complex of ∼500 kDa (79). We used tim10-2 mitochondria to investigate whether this soluble TIM complex plays a role in the import of Oxa1. tim10-2 mitochondria efficiently imported Oxa1 (Fig. 4A, lanes 6 to 9), while the import of AAC was significantly reduced (Fig. 4A, lanes 16 to 19). Moreover, the formation of the 500-kDa GIP intermediate of Oxa1 was indistinguishable in tim10-2 and wild-type mitochondria and occurred only in the absence of a Δψ (Fig. 4B, upper panel, lanes 1 to 3 and 7 to 9). In contrast, tim10-2 mitochondria accumulated a GIP intermediate of AAC independently of the presence or absence of a Δψ (Fig. 4B, lower panel, lanes 7 to 12) (79), whereas wild-type mitochondria accumulated a low-molecular-weight intermembrane space form of AAC in the absence of a Δψ (Fig. 4B, lower panel, lanes 1 to 3) and the assembled dimeric form of AAC in the presence of a Δψ (Fig. 4B, lower panel, lanes 4 to 6) (67). These results indicate that a functional impairment of the Tim9-Tim10 complex does not affect the import pathway of Oxa1.
FIG. 4.
The small TIM complexes of the intermembrane space are dispensable for Oxa1 import. (A) Radiolabeled Oxa1 precursor or AAC was imported at 15°C for the times indicated into wild-type (WT) or tim10-2 mitochondria in the presence of a Δψ, unless noted otherwise. After import, the mitochondria were subjected to proteinase K digestion and analyzed by SDS-PAGE. mOxa1, mature Oxa1. (B) Import of Oxa1 precursor (upper panel) and AAC (lower panel) at 25°C was performed as for panel A, except that proteinase K treatment was omitted and samples were analyzed by BN-PAGE after solubilization of mitochondria in digitonin buffer. (C) 35S-labeled Oxa1 precursor was imported in wild-type and tim8Δ tim13Δ mitochondria at 25°C. Mitochondria were either subjected to proteinase K digestion and SDS-PAGE analysis or directly reisolated, solubilized in digitonin buffer, and assayed by BN-PAGE for complex formation.
Similar results were obtained when the Oxa1 precursor was imported into mitochondria lacking the second soluble TIM complex, the Tim8-Tim13 complex, which is involved in the import of the Tim23 precursor (11, 12, 57). tim8Δ tim13Δ mitochondria imported Oxa1 like wild-type mitochondria (Fig. 4C, lanes 1 to 12), and the formation of the GIP intermediate of Oxa1 depended only on the absence of the membrane potential, independently of whether tim8Δ tim13Δ or wild-type mitochondria were used (Fig. 4C, lanes 13 to 15 and 19 to 21). We conclude that the Tim8-Tim13 complex is not required for the import of Oxa1.
mtHsp70 is required for release of the Oxa1 precursor from the GIP complex.
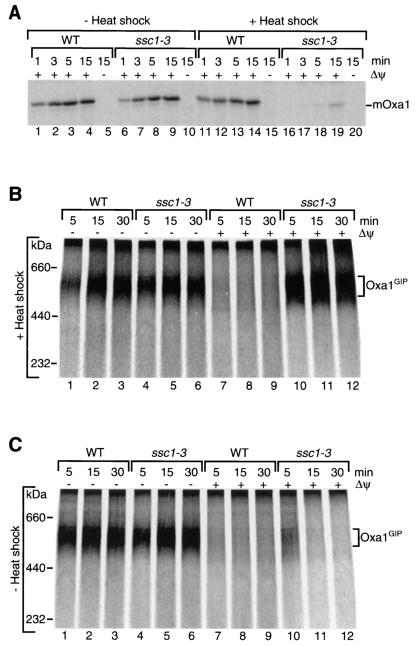
Transport of the Oxa1 precursor across the inner membrane requires mtHsp70 (Ssc1). ssc1-3 mutant mitochondria, isolated from a yeast strain with a point mutation in the SSC1 gene that leads to an amino acid substitution in the ATPase domain of the Hsp70 (21), are defective in the maturation of Oxa1 (27). The full mutant phenotype of the Ssc1-3 protein can be selectively induced by preincubation of the isolated mitochondria at 37°C (21, 85), leading to an almost complete block of Oxa1 import (Fig. 5A, lanes 16 to 19). In the absence of a heat shock, ssc1-3 mitochondria imported Oxa1 with an efficiency close to that of wild-type mitochondria (Fig. 5A, lanes 6 to 9). Since the mutant phenotype can be induced by a shift of the isolated mitochondria to the nonpermissive temperature and does not require a temperature shift of the growing cells, indirect effects of the ssc1-3 mutation on mitochondrial function are minimized.
FIG. 5.
Requirement for mtHsp70 (Ssc1) in Oxa1 import. (A) Import of the 35S-labeled Oxa1 precursor into wild-type (WT) and ssc1-3 mitochondria was performed after a 15-min temperature shift of the isolated mitochondria to 37°C (+ heat shock) or without a temperature shift (− heat shock) for the indicated times in the presence of a Δψ, unless indicated otherwise. Mitochondria were treated with proteinase K after import and analyzed by SDS-PAGE. mOxa1, mature Oxa1. (B) Imports of the Oxa1 precursor were performed after temperature shift of the isolated wild-type and ssc1-3 mitochondria to 37°C as described above. Proteinase K treatment was omitted; mitochondria were reisolated and solubilized, and complexes were assessed by BN-PAGE. (C) The experiment was performed as described for panel B except that wild-type and ssc1-3 mitochondria remained at 25°C for the entire import experiment.
We examined whether mtHsp70 was required only for the transport of Oxa1 across the inner membrane or whether it was also involved in the transfer of the preprotein across the outer membrane. When preincubated at the nonpermissive temperature, ssc1-3 mitochondria accumulated the Oxa1 precursor as a GIP intermediate not only in the absence of a Δψ but also in its presence (Fig. 5B, lanes 4 to 6 and 10 to 12), indeed suggesting that an inactivation of matrix Hsp70 leads to an arrest of Oxa1 in the outer membrane. The accumulation of Oxa1GIP in the presence of a membrane potential depended on the induction of the ssc1-3 mutant phenotype, since under permissive conditions efficient formation of the GIP intermediate was observed only in the absence of a Δψ (Fig. 5C, lanes 4 to 6 versus 10 to 12). It is important to note that ssc1-3 mitochondria are competent in generation of a Δψ. This has been shown with preproteins that depend only on a Δψ, and not on mtHsp70, for insertion into the inner membrane. The import of these preproteins, which contain a matrix-targeting signal as well as a hydrophobic stop transfer signal, is not inhibited by the ssc1-3 mutation under nonpermissive conditions (22, 23, 85). We conclude that a functional mtHsp70 is required for release of the Oxa1 precursor from the GIP complex of the outer membrane.
The multispanning inner membrane protein Cox18 forms a 500-kDa intermediate similar to that formed by Oxa1.
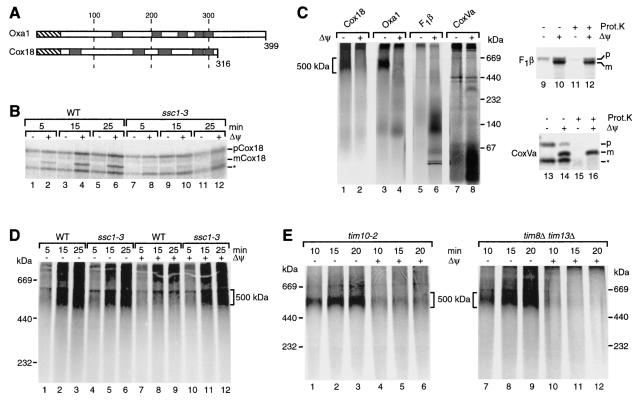
We considered that the accumulation of a stable GIP intermediate in the absence of a Δψ may be unique to the precursor of Oxa1. The mitochondrial inner membrane contains an integral membrane protein of 31 kDa, named Cox18 (for cytochrome c oxidase subunit 18), that is also involved in protein export but which functions independently of Oxa1 (28, 69, 75). Cox18 shows weak sequence similarity to Oxa1 and is also predicted to span the inner membrane five times, there are although considerable differences in the lengths of the connecting loops and the terminal segments (69, 75) (Fig. 6A). Import of the Cox18 precursor into mitochondria has not been reported so far. A sequence analysis suggested that Cox18 contains a presequence of about 43 amino acids (75). We synthesized and radiolabeled the precursor of yeast Cox18 in reticulocyte lysate and incubated it with isolated mitochondria. The precursor was proteolytically processed in a Δψ-dependent manner (Fig. 6B, lanes 2, 4, and 6). Its import was almost completely blocked in ssc1-3 mitochondria under nonpermissive conditions (Fig. 6B, lanes 7 to 12), indicating that the import of Cox18 required functional mtHsp70.
FIG. 6.
Formation of a translocation intermediate by the multispanning Cox18 protein. (A) Schematic comparison of the domain organizations of Oxa1 and Cox18 (primary structures). Hatched boxes, presequences; black boxes, transmembrane segments. (B) Radiolabeled Cox18 precursor (pCox18) was imported at 25°C for the indicated times in the presence or absence of a Δψ into wild-type (WT) and ssc1-3 mitochondria after a 15-min preshift of the mitochondria to 37°C. Mitochondria were subjected to proteinase K digestion and analyzed by SDS-PAGE. mCox18, mature Cox18. (C) 35S-labeled Cox18, Oxa1, F1β, and CoxVa precursor were imported into wild-type mitochondria in the presence or absence of a Δψ at 25°C for 20 min. After the import reaction, mitochondria were reisolated, solubilized in digitonin buffer, and analyzed by BN-PAGE, followed by digital autoradiography (samples 1 lanes 8). In addition, mitochondria that had been incubated with the precursors of F1β and CoxVa were analyzed by SDS-PAGE after treatment with or without proteinase K (Prot.K) (lanes 9 to 16). A fraction of the F1β precursor associates with mitochondria in the absence of a Δψ in an unspecific, nonproductive manner (59). Bands marked by an asterisk probably represent products of internal initiation of translation and are not specifically imported into mitochondria (22, 23). (D) Cox18 was imported into wild-type and ssc1-3 mitochondria for the indicated times in the presence or absence of a Δψ. Import was performed at 25°C for 15 min after a 15-min preincubation of the mitochondria at 37°C. Mitochondria were then reisolated and solubilized in digitonin buffer before separation by BN-PAGE. (E) Radiolabeled Cox18 precursor was imported in the presence or absence of a Δψ into tim10-2 mitochondria (left panel) and tim8Δ tim13Δ mitochondria (right panel) at 25°C for the indicated times. After import, the mitochondria were solubilized in digitonin buffer and analyzed by BN-PAGE.
To study a possible accumulation of the Cox18 precursor at the TOM machinery, we incubated the precursor with mitochondria in the presence or absence of Δψ, lysed the mitochondria with digitonin, and analyzed protein complexes by BN-PAGE. In the presence of a Δψ, a distinct high-molecular-weight complex was not observed for imported Cox18 (Fig. 6C, lane 2). In the absence of a Δψ, however, the Cox18 precursor efficiently accumulated in a 500-kDa complex like the precursor of Oxa1 (Fig. 6C, lanes 1 and 3). For comparison we used two additional cleavable preproteins: the matrix protein F1-ATPase subunit β (F1β), which does not contain a transmembrane segment, and the inner membrane protein subunit Va of cytochrome c oxidase (CoxVa), which contains a single transmembrane segment (22, 54). In the presence of a Δψ, both proteins were processed to their mature forms (Fig. 6C, lanes 10, 12, 14, and 16) and migrated mainly in the lower molecular weight range of the BN gel (Fig. 6C, lanes 6 and 8). In the absence of a Δψ, neither the precursor of F1β nor that of CoxVa was accumulated in a BN-PAGE complex comparable to that of Oxa1 or Cox18. Thus, for these preproteins with either no or one hydrophobic segment, no evidence was found for the formation of a stable complex with the TOM machinery when analyzed by BN-PAGE.
In order to determine if the Cox18 precursor was arrested in a manner similar to that of the Oxa1 precursor, we studied the formation of the 500K complex of Cox18 with ssc1-3 mitochondria. Indeed, the Cox18 precursor accumulated as a 500K complex in ssc1-3 mitochondria not only in the absence of a Δψ but also in the presence of a Δψ (Fig. 6D, lanes 5, 6, 11, and 12). An efficient arrest of the Cox18 precursor in the 500K complex required the induction of nonpermissive conditions (Fig. 6D) and was not observed with ssc1-3 mitochondria under permissive conditions (data not shown). We conclude that dissipation of the membrane potential as well as the inactivation of mtHsp70 causes an accumulation of the Cox18 precursor in a 500K complex, thus resembling the import pathway observed for Oxa1.
We also addressed whether the soluble TIM complexes of the intermembrane space were involved in the transport of Cox18. In the presence of a Δψ, the transport of Cox18 into the inner mitochondrial membrane was affected neither in tim10-2 mitochondria nor in tim8Δ tim13Δ mitochondria (data not shown). In contrast to the GIP intermediate of carrier proteins such as AAC, but similar to what we observed for Oxa1 (Fig. 4), the 500K intermediate of Cox18 was formed in tim10-2 mitochondria or tim8Δ tim13Δ mitochondria only in the absence of a Δψ (Fig. 6E). Accordingly, none of the soluble TIM complexes of the intermembrane space contributed to the formation of the Cox18 transport intermediate.
DISCUSSION
The transport of nuclear-encoded multispanning inner mitochondrial membrane proteins is especially demanding to cells. During transport of these proteins through the aqueous surroundings of the cytosol and the mitochondrial intermembrane space, the multiple hydrophobic regions must be shielded in order to prevent protein aggregation. The precursor proteins are bound and protected by molecular chaperones in the cytosol and delivered to the TOM import machinery of the mitochondrial outer membrane (5, 84, 88). The identification of soluble TIM complexes with putative chaperone-like functions provided important insight into how noncleavable multispanning membrane proteins with internal targeting signals, such as the metabolite carriers, are transported through the intermembrane space (10-12, 39, 41, 57, 73, 79, 83).
Here we addressed the transport pathway taken by cleavable inner membrane proteins containing multiple transmembrane regions across the outer mitochondrial membrane and the intermembrane space by analyzing the transport requirements of the precursors of Oxa1 and Cox18. Upon depletion of the membrane potential Δψ across the inner membrane, the preproteins form a stable intermediate of 500 kDa at the core of the TOM machinery, the GIP complex. Remarkably, the preprotein-GIP interaction is so stable that it is not dissociated by an electrophoretic run of several hours. In contrast to these hydrophobic proteins, soluble mitochondrial matrix proteins and cleavable proteins with single transmembrane spans were not found to form a stable GIP intermediate. They are probably released from the TOM machinery when subsequent transport into the presequence translocase is blocked (19, 37, 80, 81). Thus, at the level of the GIP complex, the transport characteristics of presequence-containing multispanning proteins differ from those of matrix proteins and single-membrane-spanning proteins.
Previous studies indicated that the C terminus of the central receptor Tom22, which protrudes into the intermembrane space, serves as a trans binding site for presequences once they emerge from the Tom40 pore (6, 33, 42, 51). A lack of this intermembrane space domain strongly reduced the formation of a stable Oxa1-GIP intermediate in the absence of a membrane potential. With fully energized mitochondria, the import was only moderately delayed, but it reached wild-type levels after longer import times. Thus, when transport across the inner membrane is blocked by depletion of Δψ, the intermembrane space domain of Tom22 is required to stably hold the Oxa1 preprotein in the GIP complex. In energized mitochondria, the preprotein is efficiently pulled by the presequence translocase of the inner membrane, and thus the requirement for the intermembrane space domain of Tom22 can be bypassed.
Following the transport route of Oxa1 and Cox18 further, from the TOM machinery to the presequence translocase, we analyzed the dependence of their transport on the soluble TIM complexes of the intermembrane space. While the metabolite carriers strictly require the Tim9-Tim10 complex for transport across the outer mitochondrial membrane and through the intermembrane space (79), a functional impairment of this complex in tim10-2 mutant mitochondria did not influence Oxa1 or Cox18 transport, including formation of the GIP intermediate. While it cannot be fully excluded that the essential Tim9-Tim10 complex may participate in import of cleavable multispanning inner membrane proteins, the strikingly different effect of the tim10-2 mutation on the import of Oxa1/Cox18 and carrier proteins indicates that different mechanisms exist for the transport of these hydrophobic proteins through the GIP complex and intermembrane space. Moreover, the Tim8-Tim13 complex is not required for the import of Oxa1 and Cox18.
The Δψ is necessary for all mitochondrial proteins to cross or be transported into the inner membrane. Interestingly, multispanning inner membrane proteins are arrested at different locations along their import pathway when the Δψ is lowered. The noncleavable precursors of metabolite carriers accumulate at the Tim9-Tim10 complex in the intermembrane space yet do not form a stable intermediate with the GIP complex (67, 79), while the cleavable preproteins Oxa1 and Cox18 are arrested as a GIP translocation intermediate. A GIP intermediate of Oxa1 and Cox18 was also generated when the mitochondrial import motor was inactivated by using a temperature-conditional allele of mtHsp70 (Ssc1). The release of Oxa1 or Cox18 from the GIP complex thus requires a fully active presequence translocase with two energy sources, the membrane potential and the ATP-dependent mtHsp70 import motor. These two driving forces are probably needed to actively pull the preproteins out of the TOM machinery. In contrast, the carrier proteins are released from the TOM machinery by the action of the Tim9-Tim10 complex without a need for an energized inner membrane; a stable GIP intermediate of carrier proteins is thus observed upon inactivation of the Tim9-Tim10 complex (79). The membrane potential is subsequently needed at the level of the protein insertion complex of the inner membrane for integration of the carriers into the membrane (39, 41, 60, 61, 64, 72, 73). We conclude that the carrier proteins are released from the TOM machinery and transferred through the intermembrane space in several steps, while the release of cleavable multispanning proteins like Oxa1 and Cox18 involves a tight cooperation of the presequence translocase with the TOM machinery. Since even the inactivation of the matrix import motor causes a stable arrest of the preproteins in the GIP complex, it is very unlikely that the import of these proteins includes a membrane-free intermembrane space intermediate. A putative import component that could potentially stabilize intermediates of cleavable preproteins in the intermembrane space would be Tim50, the recently discovered subunit of the presequence translocase that possesses a large intermembrane space domain (23, 52, 87). However, since Tim50 already interacts with preproteins in the absence of a Δψ and before the action of mtHsp70, it is apparently not sufficient for the release of Oxa1 or Cox18 from the TOM complex, an action that strictly depends on both Δψ and functional mtHsp70.
We conclude that mitochondria possess more than one pathway for translocation of highly hydrophobic proteins through the intermembrane space: a pathway via soluble TIM complexes for noncleavable precursor proteins and a pathway involving a close functional cooperation of the TOM and presequence translocase for the cleavable preproteins of Oxa1 and Cox18. The cleavable precursor of Coq2 may even switch between both pathways (46); however, the identification of a translocation intermediate for this preprotein will be required in order to address the roles of individual transport components in its import pathway at a mechanistic level.
Acknowledgments
We thank C. Meisinger and S. D. Emr for discussion and support and E. Schiebel for PCR templates.
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 388, the Max Planck Research Award/Alexander von Humboldt Foundation, and the Fonds der Chemischen Industrie/BMBF.
REFERENCES
- 1.Adam, A., M. Endres, C. Sirrenberg, F. Lottspeich, W. Neupert, and M. Brunner. 1999. Tim9, a new component of the TIM22.54 translocase in mitochondria. EMBO J. 18:313-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, M., M. Behrens, K. Esser, G. Michaelis, and E. Pratje. 1994. PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol. Gen. Genet. 245:272-278. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, M. F., S. Hofmann, W. Neupert, and M. Brunner. 2000. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 10:25-31. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, M. F., C. Sirrenberg, W. Neupert, and M. Brunner. 1996. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 87:33-41. [DOI] [PubMed] [Google Scholar]
- 5.Beddoe, T., and T. Lithgow. 2002. Delivery of nascent polypeptides to the mitochondrial surface. Biochim. Biophys. Acta 1592:35-39. [DOI] [PubMed] [Google Scholar]
- 6.Bolliger, L., T. Junne, G. Schatz, and T. Lithgow. 1995. Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 14:6318-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnefoy, N., F. Chalvet, P. Hamel, P. P. Slonimski, and G. Dujardin. 1994. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol. 239:201-212. [DOI] [PubMed] [Google Scholar]
- 8.Chacinska, A., N. Pfanner, and C. Meisinger. 2002. How mitochondria import hydrophilic and hydrophobic proteins. Trends Cell Biol. 12:299-303. [DOI] [PubMed] [Google Scholar]
- 9.Chen, M., K. Xie, F. Jiang, L. Yi, and R. E. Dalbey. 2002. YidC, a newly defined evolutionarily conserved protein, mediates membrane protein assembly in bacteria. Biol. Chem. 383:1565-1572. [DOI] [PubMed] [Google Scholar]
- 10.Curran, S. P., D. Leuenberger, W. Oppliger, and C. M. Koehler. 2002. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 21:942-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran, S. P., D. Leuenberger, E. Schmidt, and C. M. Koehler. 2002. The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 158:1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, A. J., N. B. Sepuri, J. Holder, A. E. Johnson, and R. E. Jensen. 2000. Two intermembrane space TIM complexes interact with different domains of Tim23p during its import into mitochondria. J. Cell Biol. 150:1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekker, P. J., F. Martin, A. C. Maarse, U. Bömer, H. Müller, B. Guiard, M. Meijer, J. Rassow, and N. Pfanner. 1997. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16:5408-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker, P. J., M. T. Ryan, J. Brix, H. Müller, A. Hönlinger, and N. Pfanner. 1998. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 18:6515-6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diekert, K., A. I. de Kroon, U. Ahting, B. Niggemeyer, W. Neupert, B. de Kruijff, and R. Lill. 2001. Apocytochrome c requires the TOM complex for translocation across the mitochondrial outer membrane. EMBO J. 20:5626-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endo, T., and D. Kohda. 2002. Functions of outer membrane receptors in mitochondrial protein import. Biochim. Biophys. Acta 1592:3-14. [DOI] [PubMed] [Google Scholar]
- 17.Endres, M., W. Neupert, and M. Brunner. 1999. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J. 18:3214-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ernster, L., and G. Schatz. 1981. Mitochondria: a historical review. J. Cell Biol. 91:227s-255s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esaki, M., T. Kanamori, S. Nishikawa, and T. Endo. 1999. Two distinct mechanisms drive protein translocation across the mitochondrial outer membrane in the late step of the cytochrome b2 import pathway. Proc. Natl. Acad. Sci. USA 96:11770-11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gakh, O., P. Cavadini, and G. Isaya. 2002. Mitochondrial processing peptidases. Biochim. Biophys. Acta 1592:63-77. [DOI] [PubMed] [Google Scholar]
- 21.Gambill, B. D., W. Voos, P. J. Kang, B. Miao, T. Langer, E. A. Craig, and N. Pfanner. 1993. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol. 123:109-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gärtner, F., W. Voos, A. Querol, B. R. Miller, E. A. Craig, M. G. Cumsky, and N. Pfanner. 1995. Mitochondrial import of subunit Va of cytochrome c oxidase characterized with yeast mutants. J. Biol. Chem. 270:3788-3795. [DOI] [PubMed] [Google Scholar]
- 23.Geissler, A., A. Chacinska, K. N. Truscott, N. Wiedemann, K. Brandner, A. Sickmann, H. E. Meyer, C. Meisinger, N. Pfanner, and P. Rehling. 2002. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111:507-518. [DOI] [PubMed] [Google Scholar]
- 24.Geissler, A., T. Krimmer, U. Bömer, B. Guiard, J. Rassow, and N. Pfanner. 2000. Membrane potential-driven protein import into mitochondria: the sorting sequence of cytochrome b2 modulates the ΔΨ-dependence of translocation of the matrix-targeting sequence. Mol. Biol. Cell 11:3977-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hell, K., W. Neupert, and R. A. Stuart. 2001. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 20:1281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrmann, J. M., and W. Neupert. 2000. Protein transport into mitochondria. Curr. Opin. Microbiol. 3:210-214. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann, J. M., W. Neupert, and R. A. Stuart. 1997. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 16:2217-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hikkel, I., Y. Gbelska, Q. J. van der Aart, G. Lubecu, and J. Subik. 1997. Cloning and characterization of KlCOX18, a gene required for activity of cytochrome oxidase in Kluyveromyces lactis. Curr. Genet. 32:267-272. [DOI] [PubMed] [Google Scholar]
- 29.Huang, S., K. S. Ratliff, and A. Matouschek. 2002. Protein unfolding by the mitochondrial membrane potential. Nat. Struct. Biol. 9:301-307. [DOI] [PubMed] [Google Scholar]
- 30.Jensen, R. E., and C. D. Dunn. 2002. Protein import into and across the mitochondrial inner membrane: role of the TIM23 and TIM22 translocons. Biochim. Biophys. Acta 1592:25-34. [DOI] [PubMed] [Google Scholar]
- 31.Jensen, R. E., and A. E. Johnson. 2001. Opening the door to mitochondrial protein import. Nat. Struct. Biol. 8:1008-1010. [DOI] [PubMed] [Google Scholar]
- 32.Johnston, A. J., J. Hoogenraad, D. A. Dougan, K. N. Truscott, M. Yano, M. Mori, N. J. Hoogenraad, and M. T. Ryan. 2002. Insertion and assembly of human tom7 into the preprotein translocase complex of the outer mitochondrial membrane. J. Biol. Chem. 277:42197-42204. [DOI] [PubMed] [Google Scholar]
- 33.Kanamori, T., S. Nishikawa, M. Nakai, I. Shin, P. G. Schultz, and T. Endo. 1999. Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc. Natl. Acad. Sci. USA 96:3634-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang, P. J., J. Ostermann, J. Shilling, W. Neupert, E. A. Craig, and N. Pfanner. 1990. Requirement for Hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348:137-143. [DOI] [PubMed] [Google Scholar]
- 35.Kiebler, M., R. Pfaller, T. Söllner, G. Griffiths, H. Horstmann, N. Pfanner, and W. Neupert. 1990. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature 348:610-616. [DOI] [PubMed] [Google Scholar]
- 36.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 37.Knox, C., E. Sass, W. Neupert, and O. Pines. 1998. Import into mitochondria, folding and retrograde movement of fumarase in yeast. J. Biol. Chem. 273:25587-25593. [DOI] [PubMed] [Google Scholar]
- 38.Koehler, C. M. 2000. Protein translocation pathways of the mitochondrion. FEBS Lett. 476:27-31. [DOI] [PubMed] [Google Scholar]
- 39.Koehler, C. M., E. Jarosch, K. Tokatlidis, K. Schmid, R. J. Schweyen, and G. Schatz. 1998. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279:369-373. [DOI] [PubMed] [Google Scholar]
- 40.Koehler, C. M., D. Leuenberger, S. Merchant, A. Renold, T. Junne, and G. Schatz. 1999. Human deafness dystonia syndrome is a mitochondrial disease. Proc. Natl. Acad. Sci. USA 96:2141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koehler, C. M., S. Merchant, W. Oppliger, K. Schmid, E. Jarosch, L. Dolfini, T. Junne, G. Schatz, and K. Tokatlidis. 1998. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 17:6477-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komiya, T., S. Rospert, C. Koehler, R. Looser, G. Schatz, and K. Mihara. 1998. Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the ‘acid chain' hypothesis. EMBO J. 17:3886-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kübrich, M., J. Rassow, W. Voos, N. Pfanner, and A. Hönlinger. 1998. The import route of ADP/ATP carrier into mitochondria separates from the general import pathway of cleavable preproteins at the trans side of the outer membrane. J. Biol. Chem. 273:16374-16381. [DOI] [PubMed] [Google Scholar]
- 44.Kurz, M., H. Martin, J. Rassow, N. Pfanner, and M. T. Ryan. 1999. Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol. Biol. Cell 10:2461-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 46.Leuenberger, D., N. A. Bally, G. Schatz, and C. M. Koehler. 1999. Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 18:4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin, J., K. Mahlke, and N. Pfanner. 1991. Role of an energized inner membrane in mitochondrial protein import. ΔΨ drives the movement of presequences. J. Biol. Chem. 266:18051-18057. [PubMed] [Google Scholar]
- 48.Matouschek, A., N. Pfanner, and W. Voos. 2000. Protein unfolding by mitochondria. The Hsp70 import motor. EMBO Rep. 1:404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meisinger, C., J. Brix, K. Model, N. Pfanner, and M. T. Ryan. 1999. The preprotein translocase of the outer mitochondrial membrane: receptors and a general import pore. Cell. Mol. Life Sci. 56:817-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meisinger, C., M. T. Ryan, K. Hill, K. Model, J. H. Lim, A. Sickmann, H. Müller, H. E. Meyer, R. Wagner, and N. Pfanner. 2001. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small Tom proteins, and import receptors. Mol. Cell. Biol. 21:2337-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moczko, M., U. Bömer, M. Kübrich, N. Zufall, A. Hönlinger, and N. Pfanner. 1997. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol. 17:6574-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mokranjac, D., S. A. Paschen, C. Kozany, H. Prokisch, S. C. Hoppins, F. E. Nargang, W. Neupert, and K. Hell. 2003. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 22:816-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nelson, D. R., C. M. Felix, and J. M. Swanson. 1998. Highly conserved charge-pair networks in the mitochondrial carrier family. J. Mol. Biol. 277:285-308. [DOI] [PubMed] [Google Scholar]
- 54.Neupert, W. 1997. Protein import into mitochondria. Annu. Rev. Biochem. 66:863-917. [DOI] [PubMed] [Google Scholar]
- 55.Neupert, W., and M. Brunner. 2002. The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 3:555-565. [DOI] [PubMed] [Google Scholar]
- 56.Palmieri, L., F. M. Lasorsa, A. Vozza, G. Agrimi, G. Fiermonte, M. J. Runswick, J. E. Walker, and F. Palmieri. 2000. Identification and functions of new transporters in yeast mitochondria. Biochim. Biophys. Acta 1459:363-369. [DOI] [PubMed] [Google Scholar]
- 57.Paschen, S. A., U. Rothbauer, K. Kaldi, M. F. Bauer, W. Neupert, and M. Brunner. 2000. The role of the TIM8-13 complex in the import of Tim23 into mitochondria. EMBO J. 19:6392-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfanner, N., and A. Geissler. 2001. Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2:339-349. [DOI] [PubMed] [Google Scholar]
- 59.Pfanner, N., H. K. Müller, M. A. Harmey, and W. Neupert. 1987. Mitochondrial protein import: involvement of the mature part of a cleavable precursor protein in the binding to receptor sites. EMBO J. 6:3449-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfanner, N., and W. Neupert. 1987. Distinct steps in the import of ADP/ATP carrier into mitochondria. J. Biol. Chem. 262:7528-7536. [PubMed] [Google Scholar]
- 61.Pfanner, N., and W. Neupert. 1985. Transport of proteins into mitochondria: a potassium diffusion potential is able to drive the import of ADP/ATP carrier. EMBO J. 4:2819-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfanner, N., M. Tropschug, and W. Neupert. 1987. Mitochondrial protein import: nucleoside triphosphates are involved in conferring import-competence to precursors. Cell 49:815-823. [DOI] [PubMed] [Google Scholar]
- 63.Rassow, J., P. J. T. Dekker, S. van Wilpe, M. Meijer, and J. Soll. 1999. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J. Mol. Biol. 286:105-120. [DOI] [PubMed] [Google Scholar]
- 64.Rehling, P., K. Model, K. Brandner, P. Kovermann, A. Sickmann, H. E. Meyer, K. W., R. Wagner, K. N. Truscott, and N. Pfanner. 2003. Protein insertion into the mitochondria inner membrane by a twin-pore translocase. Science 299:1747-1751. [DOI] [PubMed] [Google Scholar]
- 65.Rehling, P., N. Pfanner, and C. Meisinger. 2003. Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane—a guided tour. J. Mol. Biol. 326:639-657. [DOI] [PubMed] [Google Scholar]
- 66.Ryan, K. R., and R. E. Jensen. 1995. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell 83:517-519. [DOI] [PubMed] [Google Scholar]
- 67.Ryan, M. T., H. Müller, and N. Pfanner. 1999. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 274:20619-20627. [DOI] [PubMed] [Google Scholar]
- 68.Ryan, M. T., W. Voos, and N. Pfanner. 2001. Assaying protein import into mitochondria. Methods Cell Biol. 65:189-215. [DOI] [PubMed] [Google Scholar]
- 69.Saracco, S. A., and T. D. Fox. 2002. Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol. Biol. Cell 13:1122-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schnell, D. J., and D. N. Hebert. 2003. Protein translocons. Multifunctional mediators of protein translocation across membranes. Cell 112:491-505. [DOI] [PubMed] [Google Scholar]
- 71.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sirrenberg, C., M. F. Bauer, B. Guiard, W. Neupert, and M. Brunner. 1996. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature 384:582-585. [DOI] [PubMed] [Google Scholar]
- 73.Sirrenberg, C., M. Endres, H. Fölsch, R. A. Stuart, W. Neupert, and M. Brunner. 1998. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature 391:912-915. [DOI] [PubMed] [Google Scholar]
- 74.Söllner, T., J. Rassow, and N. Pfanner. 1991. Analysis of mitochondrial protein import using translocation intermediates and specific antibodies. Methods Cell Biol. 34:345-358. [DOI] [PubMed] [Google Scholar]
- 75.Souza, R. L., N. S. Green-Willms, T. D. Fox, A. Tzagoloff, and F. G. Nobrega. 2000. Cloning and characterization of COX18, a Saccharomyces cerevisiae PET gene required for the assembly of cytochrome oxidase. J. Biol. Chem. 275:14898-14902. [DOI] [PubMed] [Google Scholar]
- 76.Stuart, R. 2002. Insertion of proteins into the inner membrane of mitochondria: the role of the Oxa1 complex. Biochim. Biophys. Acta 1592:79-87. [DOI] [PubMed] [Google Scholar]
- 77.Truscott, K. N., P. Kovermann, A. Geissler, A. Merlin, M. Meijer, A. J. Driessen, J. Rassow, N. Pfanner, and R. Wagner. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8:1074-1082. [DOI] [PubMed] [Google Scholar]
- 78.Truscott, K. N., N. Pfanner, and W. Voos. 2001. Transport of proteins into mitochondria. Rev. Physiol. Biochem. Pharmacol. 143:81-136. [DOI] [PubMed] [Google Scholar]
- 79.Truscott, K. N., N. Wiedemann, P. Rehling, H. Müller, C. Meisinger, N. Pfanner, and B. Guiard. 2002. Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol. Cell. Biol. 22:7780-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ungermann, C., B. Guiard, W. Neupert, and D. M. Cyr. 1996. The Δψ- and Hsp70/MIM44-dependent reaction cycle driving early steps of protein import into mitochondria. EMBO J. 15:735-744. [PMC free article] [PubMed] [Google Scholar]
- 81.Ungermann, C., W. Neupert, and D. M. Cyr. 1994. The role of Hsp70 in conferring unidirectionality on protein translocation into mitochondria. Science 266:1250-1253. [DOI] [PubMed] [Google Scholar]
- 82.van Wilpe, S., M. T. Ryan, K. Hill, A. C. Maarse, C. Meisinger, J. Brix, P. J. Dekker, M. Moczko, R. Wagner, M. Meijer, B. Guiard, A. Hönlinger, and N. Pfanner. 1999. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401:485-489. [DOI] [PubMed] [Google Scholar]
- 83.Vial, S., H. Lu, S. Allen, P. Savory, D. Thornton, J. Sheehan, and K. Tokatlidis. 2002. Assembly of Tim9 and Tim10 into a functional chaperone. J. Biol. Chem. 277:36100-36108. [DOI] [PubMed] [Google Scholar]
- 84.Voos, W. 2003. A new connection. Chaperones meet a mitochondrial receptor. Mol. Cell 11:1-3. [DOI] [PubMed] [Google Scholar]
- 85.Voos, W., B. D. Gambill, B. Guiard, N. Pfanner, and E. A. Craig. 1993. Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J. Cell Biol. 123:119-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Voos, W., and K. Röttgers. 2002. Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim. Biophys. Acta 1592:51-62. [DOI] [PubMed] [Google Scholar]
- 87.Yamamoto, H., M. Esaki, T. Kanamori, Y. Tamura, S. Nishikawa, and T. Endo. 2002. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell 111:519-528. [DOI] [PubMed] [Google Scholar]
- 88.Young, J. C., N. J. Hoogenraad, and F. U. Hartl. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112:41-50. [DOI] [PubMed] [Google Scholar]