Abstract
Miz1 is a member of the POZ domain/zinc finger transcription factor family. In vivo, Miz1 forms a complex with the Myc oncoprotein and recruits Myc to core promoter elements. Myc represses transcription through Miz1 binding sites. We now show that the Miz1 gene is ubiquitously expressed during mouse embryogenesis. In order to elucidate the physiological function of Miz1, we have deleted the mouse Miz1 gene by homologous recombination. Miz1+/− mice are indistinguishable from wild-type animals; in contrast, Miz1−/− embryos are not viable. They are severely retarded in early embryonic development and do not undergo normal gastrulation. Expression of Goosecoid and Brachyury is detectable in Miz1−/− embryos, suggesting that Miz1 is not required for signal transduction by Nodal. Expression of p21Cip1, a target gene of Miz1 is unaltered; in contrast, expression of p57Kip2, another target gene of Miz1 is absent in Miz1−/− embryos. Miz1−/− embryos succumb to massive apoptosis of ectodermal cells around day 7.5 of embryonic development. Our results show that Miz1 is required for early embryonic development during gastrulation.
The MYC (or c-MYC) gene and two of its relatives, MYCL and MYCN, are causally involved in the genesis of a wide variety of human tumors (32). c-MYC encodes a transcription factor (Myc) that can both activate and repress transcription. Myc activates transcription as part of a heterodimeric complex with the Max protein (1, 23). The complex binds to specific sequences, termed E-boxes, and recruits both the Gcn5 and Tip60 histone acetylase complexes to E-box elements through interaction with the TRRAP protein (8, 16, 17, 25, 26). In addition, TRRAP-independent mechanisms of transcriptional activation have been demonstrated (30). These may involve interactions of Myc with the P-TEFb complex, which regulates transcriptional elongation (14). Both directed searches and a number of array analyses have identified a large number of genes that are activated by Myc in vivo (9, 27, 31, 45).
Similarly, a large number of genes have been identified that are repressed upon activation of Myc. However, the mechanisms of transcriptional repression by Myc have remained more elusive. For a number of genes, repression of Myc has been mapped to the core promoter, suggesting that Myc affects proteins that regulate transcription at or close to the start site of transcription (24). One suggestion has been that Myc directs the synthesis of a transcriptional repressor protein and thereby indirectly represses transcription, but such a repressor has not yet been identified. A second suggestion has been that Myc-Max complexes directly bind to the start site of one repressed gene, p27kip1 (49), but direct binding of Myc-Max complexes to start sites of other repressed genes has not been found. A third suggestion, therefore, has been that Myc is recruited to core promoters through protein-protein interactions with other transcription factors. A number of candidate interaction partners have been identified, including TFII-I (35), YY-1 (38), Smad2 (15), Sp1 (18), and Miz1 (34).
Recently, evidence has accumulated that three genes, p15Ink4b (37, 40), p21Cip1 (20, 36, 43, 48), and Mad4 (22), are repressed by Myc through interaction with Miz1. Miz1 is a transcription factor with 13 zinc fingers and a POZ/BTB domain at its amino terminus (4). Free Miz1 binds to the core promoter of all three genes and activates transcription. Upon binding to Myc, transcriptional activation by Miz1 is abolished, and the Myc-Miz1 complex acts as a transcriptional repressor; this is in part due to competition between p300 and Myc for binding to Miz1 (40). Array analyses and chromatin immunoprecipitation (ChIp) experiments suggest that several other genes that are repressed by Myc, including p57Kip2 (12) and C/EBPα (24), are directly repressed by Myc through interaction with Miz1 (V. Beuger and M. Eilers, unpublished observations).
These findings suggest that interaction with Miz1 plays a central role in transcriptional regulation and potentially transformation by Myc. In order to elucidate the physiological function of Miz1, we have knocked out the Miz1 gene by using conventional gene-targeting strategies. We now show that Miz1 is required for embryonic development around gastrulation.
MATERIALS AND METHODS
Gene targeting and generation of mutant mice.
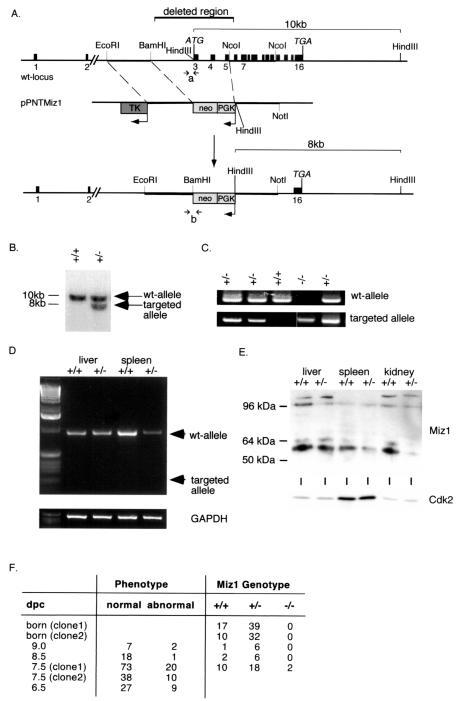
Genomic clones containing the murine Miz1 gene were isolated from an Sv129 genomic library and used to generate a targeting construct (see Fig. 2A) based on the vector pPNT (29). As 5′ homology region, a 2.5-kb EcoRI-BamHI fragment spanning a region 2 kb upstream of the first coding exon of the Miz1 locus was inserted between the herpes simplex thymidine kinase (TK) and the PGK-neomycin (PGKneo) cassette. A 2.3-kb NcoI fragment containing coding exons 4 to 10 of the murine Miz1 gene was cloned into pPNT by using its XhoI and the NotI sites flanking the PGKneo cassette at the 3′ end. The predicted targeting event with this vector replaces the first three coding exons and a 2-kb upstream sequence with PGKneo sequences. The targeting construct was linearized with NotI and electroporated into 129X1/SvJ ES cells. ES cells were selected with neomycin-ganciclovir. Positive ES clones were identified by the presence of an 8-kb fragment via Southern blotting with a probe spanning exon 16 of the murine Miz1 locus after HindIII cleavage of the DNA (Fig. 2B).
FIG. 2.
Disruption of the Miz1 gene. (A) Targeting strategy. The panel shows the structure of the murine genomic locus of Miz1, the replacement vector (pPNT-Miz1), and the structure of the targeted allele. (B) Southern blot documenting successful targeting of the Miz1 locus in ES cells. Shown is a hybridization using the probe indicated in panel A to a HindIII digest of genomic DNA. The expected fragments are shown in panel A. (C) PCR-based genotyping of E7.5 embryos using primer pairs specific for either the wild-type (wt) or the targeted Miz1 locus. The primers used are indicated in panel A as “a” for the wild-type and “b” for the knockout allele. Shown are results derived from intercrosses of Miz1+/− animals. (D) Absence of a transcript from the targeted allele. Shown are results from reverse transcriptase PCR assays using primers located in exons 2 and 7 of Miz1 from RNA isolated from liver and spleen of heterozygous animals. An arrow indicates the predicted size of the PCR product from the knockout allele. (E) Absence of a truncated protein in heterozygous embryos. The panel shows a Western blot of lysates of the indicated organs from either wild-type or Miz1+/− animals with an anti-Miz1 monoclonal antibody (10E2). (F) Phenotypes and genotype of neonates and embryos from the heterozygous intercrosses. Many of the abnormal embryos could not be genotyped due to their size and/or beginning resorption. Therefore, the ratios of the genotypes are skewed. dpc, day postcoitus.
Cells from two different ES cell clones were injected into C57BL/6 blastocysts and implanted in pseudopregnant females. Chimeric mice were backcrossed to C57BL/6 mice. Progeny from these crosses were genotyped by multiplex PCR with the wild-type-specific primer mMiz1 (5′-GTCTCATGGGCTGGCTGGCTACCTG-3′), the common primer mMiz2 (5′-CTGAGGAAGGACGGGAGGCTGGAT-3′), and the neo-specific primer PB3L (5′-GTGCTACTTCCATTTGTCACGTCCTG-3′). Reverse transcriptase PCRs were performed with primers located in exons 2 (5′-AGGCGGGGAGCCGAGCTG-3′) and 7 (5′-GTGCCGGCAGACTCTTCATTCTCC-3′), respectively.
Western blot analysis was performed with lysates derived from different organs of Miz1+/+ or Miz1+/− heterozygous mice using 10E2, a monoclonal antibody raised against a fragment encompassing amino acids 269 to 803 of the Miz1 protein.
Embryo dissection and histological analysis.
Timed matings were conducted with Miz1+/− mice. Females with copulation plugs were considered to be at embryonic development day 0.5 (E0.5) of gestation. Pregnant females were sacrificed at different time points of gestation, and the embryos were dissected from maternal tissue, examined, photographed, and genotyped by nested PCR for both the wild-type and targeted alleles (primer, wild-type external upper, 5′-CCCCATGCCTACCCCTTTCTACCT-3′; wild-type external lower, 5′-GCAGCGGCCTTCTCGTCTTTG-3′; wild-type internal upper, 5′-GAGCCTGTAACTGCCCTTTCA-3′; wild-type internal lower, 5′-CCTGCAGCCTCCACATCACAA-3′; neo external upper, 5′-GCGAAGGGGCCACCAAAGAACG-3′; neo external lower, 5′-AGGGGATGGGGACTGGCAATGAA-3′; neo internal upper, 5′-TTGGCGCCTACCGGTGGATGTTGG-3′; neo internal lower, 5′-ACGAGGGTGCGGAGGGAGACGA-3′).
For histological preparations, embryos in deciduae were fixed in 4% paraformaldehyde overnight at 4°C. Tissues were processed as described previously (3). Sections were cut from paraffin blocks and stained with hematoxylin and eosin.
Immunohistochemistry.
Paraffin sections were deparaffinized in xylene and rehydrated. After being blocked in phosphate-buffered saline (PBS) with 3% bovine serum albumin for 60 min, sections were incubated with a 1:100 dilution of anti-p57Kip2 antibody (Santa Cruz Biotechnology) overnight at 4°C. Sections were then washed three times for 10 min in PBS, incubated with 1:500 biotinylated rabbit anti-goat antibody (DAKO) for 45 min, washed again, incubated with horseradish peroxidase (HRP)-conjugated streptavidin (DAKO) for 15 min, and washed again. Sections were then incubated with aminoethylcarbazole color substrate for 15 min and mounted with Mowiol. For p15 staining, an anti-p15ink4b serum (provided by David Parry) was used as a primary antibody, and a biotinylated goat anti-rabbit antibody was used as a secondary antibody. Immunostaining against p21Cip1 was performed as described previously (43) with the Santa Cruz anti-p21Cip1 antibody (M-19).
In situ hybridization.
Whole-mount in situ hybridizations were carried out with digoxigenin-labeled RNA probes as described in reference 5. cDNA probes for Brachyury and Goosecoid were a kind gift from Martin Blum, Stuttgart, Germany. For Miz1, a murine RNA probe spanning a 383-bp HincII-SmaI fragment was used. In situ hybridizations on sections were processed by using the identical Miz1 probe following a protocol described at http://stratuslifesci.ucla.edu/hhmi/derobertis/. In situ hybridization for c-myc was performed with sense and antisense RNA probes spanning the first 500 bases of c-myc cDNA.
BrdU labeling of embryos.
Pregnant females at E7.5 were injected intraperitoneally with 50 mg of bromodeoxyuridine (BrdU) and were sacrificed 2 h later. Deciduae were removed, fixed in 4% paraformaldehyde overnight at 4°C, and processed as described previously (3). Five-micrometer sections were processed for anti-BrdU labeling. After deparaffinization and rehydration sections were incubated in 50% formamide-1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% Tween at 70°C for 30 min, microwaved in 10 mM citric acid (pH 6.0) for 10 min, and incubated with anti-BrdU antibody (Amersham) overnight at 4°C. Biotinylated antimouse antibodies were used as secondary antibodies; streptavidin-HRP was used as described above. Sections were counterstained with Hoechst 33258 and mounted with Mowiol.
TUNEL analysis of embryos.
E7.5 embryos in deciduae were fixed in 4% paraformaldehyde and processed as described above. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end-labeling (TUNEL) assays were performed on sections with the TUNEL AP kit purchased from Roche Diagnostics. Sections were counterstained with Hoechst 33258 and mounted with Mowiol.
ChIp.
ChIp was performed as described previously (8) with the antibodies (all from Santa Cruz) Myc (N262), Max (C17), and Gadd45 (H-165), as well as with Miz1 polyclonal antiserum (kind gift of Bob Tjian and Joe Ziegelbauer). Primer sequences are available upon request.
RESULTS
Miz1 is expressed ubiquitously during embryonic development.
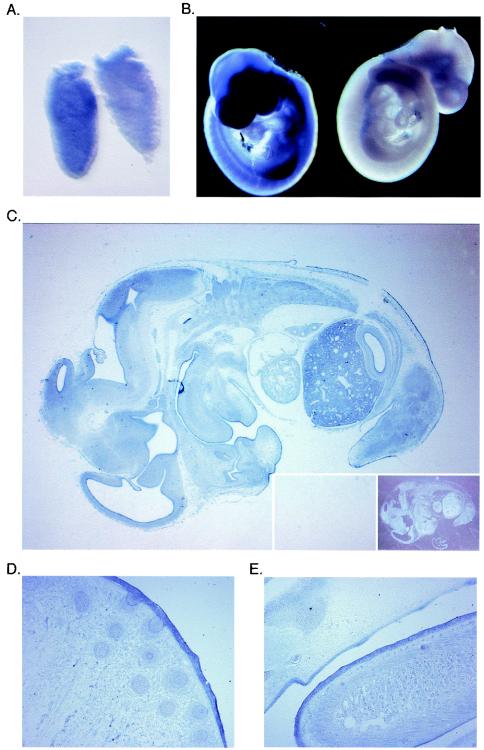
In order to define the expression pattern of Miz1 during embryonic development, in situ hybridization was performed with digoxigenin-labeled antisense RNA probes; a sense probe was used as a control. Expression of Miz1 was detectable as early as 6.5 days postcoitum (E6.5) in whole-mount hybridizations (Fig. 1A). Miz1 is expressed ubiquitously during early embryogenesis from E6.5 to E8.5 (data not shown). At E9.5, enhanced expression is detectable in the limb buds; in contrast, expression of Miz1 is lower in the heart (Fig. 1B).
FIG. 1.
Expression of Miz1 during embryogenesis. (A and B) Whole-mount in situ hybridization documenting expression of Miz1 mRNA in embryos at E6.5 (A) and E9.5 (B). In each panel, the embryo on the left was hybridized with an antisense probe, and the embryo on the right was hybridized with a sense control probe. (C) In situ hybridization documenting expression of Miz1 mRNA in a section of an E13.5 embryo. The inserts show hybridization with a sense probe as the negative control, on the left side illuminated as the antisense section and on the right side as a dark-field picture. (D and E) In situ hybridization documenting expression of Miz1 mRNA in epithelia of an E15.5 embryo. The pictures show a sagittal section of the nose with primordia of the vibrissae (D) and a sagittal section of the tongue (E).
In order to determine whether expression of Miz1 was restricted to specific organs later in development, in situ hybridization was performed on sections of embryos at E13.5 (Fig. 1C). At E13.5, specific hybridization signals were detected in all organs analyzed, including skin, brain and ganglia, liver, muscles, and all epithelia. A similar expression patterns was observed at E15.5 (data not shown). The highest signal intensities were observed in the liver, brain, and in several epithelia, including skin, the olfactory epithelium, and epithelia of the gastrointestinal tract (Fig. 1D and E). Expression of Miz1 was weaker in the heart and in the lung. Taken together, the data show that Miz1 is expressed ubiquitously from early embryogenesis to organogenesis, with a preference of expression in neural and epidermal tissues.
Targeted disruption of the Miz1 gene.
Genomic clones containing the murine Miz1 gene were isolated from an Sv129 genomic library (see Materials and Methods) and used to generate a targeting vector (pPNT Miz1) that replaces the first three coding exons of the Miz1 gene comprising the POZ domain and a transcriptional activation domain with a neomycin resistance cassette (Fig. 2A). The vector was transfected into ES cells, and clones were selected by neomycin-ganciclovir selection. Positive ES clones were confirmed by Southern blotting and injected into blastocysts. Two independent founder lines were derived from different ES cell clones. The knockout of the Miz1 gene was confirmed both by Southern blotting (Fig. 2B) and by genotyping with PCR with primers specific for either the wild-type or the knockout allele (Fig. 2C). The phenotypes described occurred in mice derived from either ES cell clone (Fig. 2F). Repeated attempts to target the second allele of Miz1 by using either elevated concentrations of neomycin or a hygromycin-containing targeting allele were not successful (K. Peukert, unpublished observations).
Comparison of published cDNA and expressed sequence tag sequences with the recently published mouse genomic sequence (44) revealed the presence of two noncoding exons 14 and 17 kb upstream of the first ATG, raising the possibility that a truncated Miz1 mRNA might be expressed from the targeted allele. In order to examine this possibility, reverse transcriptase PCRs were performed with primers located in exons 2 and 7, respectively (Fig. 2D). In both spleen and liver, the expected product for the wild-type allele was detected in Miz1+/+ mice and, in smaller amounts, in Miz1+/− mice. No transcript from the targeted allele was detectable; similar results were obtained with RNA from different organs under various PCR conditions, including some that favor the synthesis of shorter products (data not shown). A putative N-terminally-truncated protein translated from the targeted allele starting at the first in-frame ATG in exon 4 would be expected to have a molecular mass of 64.9 kDa; Western blotting with a monoclonal antibody that recognizes an epitope that is carboxy terminal to the first in-frame methionine present in the targeted allele revealed no evidence for the presence of a truncated protein in lysates from several organs of Miz1+/− mice (Fig. 1E). Taken together, the findings strongly suggest that the presence of the neomycin cassette inhibits transcription from the targeted allele. We concluded that the targeting strategy generated a null allele of Miz1.
Among the offspring of intercrosses between Miz1+/− animals, both Miz1+/+ and Miz1+/− animals were detected at the expected ratio and were phenotypically indistinguishable (Fig. 2E) (data not shown). No Miz1−/− animals were born from either of the founder lines. In order to determine the precise time point at which Miz1−/− embryos died, we set up timed matings and analyzed the embryos by PCR-based protocols. No Miz1−/− embryos were found at E12, E9, or E8.5. Morphologically, we detected a few severely retarded embryos at E9 and E8.5; in addition, we found remnants of resorbed embryos. At days E7.5 and E6.5, between 20 and 25% of all embryos were morphologically abnormal. We succeeded in genotyping two of the abnormal embryos and found that they were indeed Miz1−/− (Fig. 2C). We concluded that Miz1 is required for embryonic development beyond E7.5.
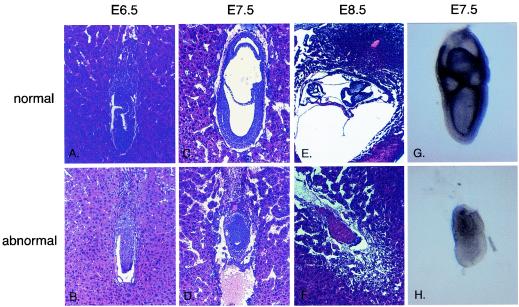
In the abnormal embryos at E6.5, both the extraembryonic and embryonic regions were smaller in size and reduced in cell number. The proamniotic cavity, which is already established in normal embryos, was missing in mutant embryos (Fig. 3A and B). At E7.5, normal embryos had completed gastrulation and formed three embryonic cavities. Abnormal embryos were generally much smaller than control embryos, and 93% of them failed to gastrulate (Fig. 3C and D). Both extraembryonic and embryonic parts gave rise to compact structures consisting of ectodermal and endodermal tissue with reduced if apparent embryonic cavities. Traces of a third germ layer were detected in some embryos with abnormal morphology. In some cases, first signs of resorption became apparent. Both PCR genotyping of abnormal embryos (Fig. 2C) and in situ hybridization (Fig. 3G and H) confirmed that phenotypically abnormal embryos were indeed Miz1 deficient.
FIG. 3.
Phenotype of Miz1−/− embryos. Histological sections of embryos generated from crosses between Miz1 heterozygous mice. (A and B) Sagittal section of E6.5 normal (A) and abnormal (B) embryos. (C and D) Sagittal section of E7.5 normal (C) and abnormal (D) embryos. (E and F) Transverse section of E8.5 embryo, which had completed the process of turning (E), and sagittal section of E8.5 mutant embryo in the process of resorption (F). The abnormal embryo (F) is depicted in a twofold-higher magnification than its normal littermate. (G and H) Whole-mount RNA in situ hybridization of normal and mutant embryos with a Miz1 probe.
At E8.5, normal embryos had developed a heart, neural ectoderm, and somites. At this stage, sections of some deciduae showed only remnants of implanted embryos, which were never larger than abnormal embryos at E7.5, suggesting that the death occurred around this time point (Fig. 3E and F).
Proliferation and apoptosis in Miz1−/− embryos.
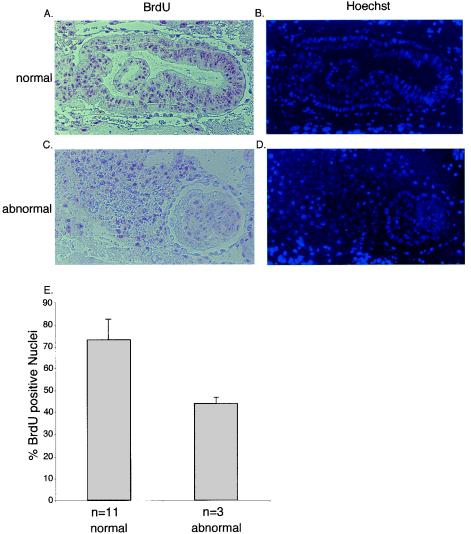
At E7.5, abnormal embryos consisted of a significantly smaller number of cells than normal embryos (data not shown). In order to establish the cause of lethality of Miz1−/− embryos, we determined both the rate of proliferation by using BrdU incorporation and the extent of apoptosis by using TUNEL assays in E7.5 embryos (Fig. 4 and 5).
FIG. 4.
BrdU incorporation in E7.0 embryos. Panels A and C document staining of sections of normal (A) and abnormal (C) E7.5 embryos with antibodies directed against BrdU; animals were injected with BrdU 2 h before fixation. The right panels (B and D) show counterstaining with Hoechst 33285 to visualize nuclei. Panel E shows a quantitation of the results obtained by calculating the percentage of BrdU-incorporating cells of 11 normal and 3 abnormal embryos. The error bars represent the standard deviation.
FIG. 5.
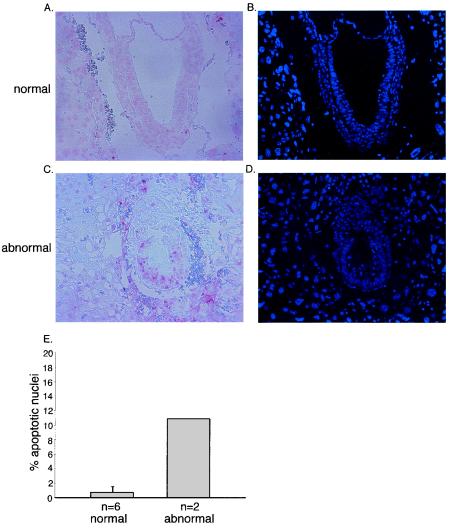
Apoptosis in E7.5 embryos. Panels A and C document TUNEL staining of morphologically normal (A) and abnormal (C) embryos at E7.5. Panels B and D document staining of the nuclei with Hoechst 33285. Panel E shows a quantitation of the results. The percentages of apoptotic nuclei were calculated for six normal and two abnormal embryos.
Abnormal embryos showed clearly reduced incorporation of BrdU relative to normal embryos at E7.0 (Fig. 4A to E). At E6.5 and E7.0, no significant number of apoptotic cells was found in either morphologically normal or abnormal embryos, although abnormal embryos were clearly detectable and had a lower cell number (data not shown). We concluded that the lower cell number of abnormal embryos at this early stage of development is due to impaired cell proliferation. In contrast, Miz1−/− embryos showed a high percentage of apoptotic cells at E7.5 and E8.0; consistent with previous studies, very few apoptotic cells were found in morphologically normal embryos (Fig. 5A to E). Previous studies have shown that apoptosis in normal embryos is limited to ectodermal cells that are not in contact with the basal membrane (11); in contrast, multiple ectodermal cells that contact the basal membrane underwent apoptosis in Miz1−/− embryos (Fig. 5C). We concluded that Miz1 is required for proliferation and for survival of ectodermal cells during gastrulation.
Induction of mesoderm occurs in Miz1−/− embryos.
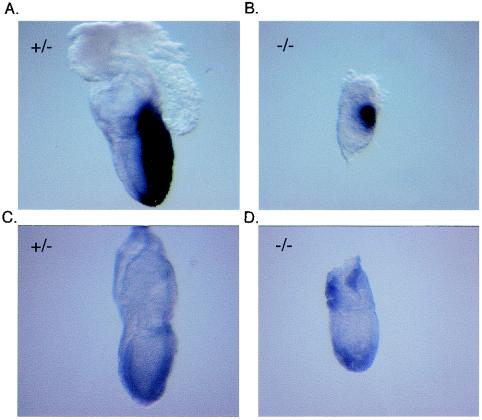
One potential reason for a failure to complete gastrulation is a defect in mesoderm formation. Induction of mesoderm formation depends, among other factors, on the transforming growth factor β (TGF-β) family member Nodal (10) and both Smad2 and Smad4 (39, 46). Since Miz1 has been implicated in signaling through TGF-β (37), we wondered whether there was a requirement for Miz1 in signaling through Nodal during mesoderm formation. Morphologically, a primitive streak was detectable in some but not all aberrant embryos (data not shown). In order to extend this observation, we used in situ hybridization with antisense probes for Brachyury (47) (Fig. 6A and B) and Goosecoid (7) (Fig. 6C and D), two genes specifically expressed in mesoderm, to determine whether mesoderm induction had taken place. Expression of both genes was detectable in both morphologically normal embryos and some of the abnormal embryos at E7.5 (Fig. 6). Genotyping with specific primers confirmed that the morphologically abnormal embryos shown indeed had Miz1 deleted. We concluded that Miz1 is not absolutely required for mesoderm induction.
FIG. 6.
Expression of Brachyury and Goosecoid in E7.5 embryos. The panels document whole-mount in situ hybridization with either a Brachyury (A and B) or a Goosecoid (C and D) antisense probe of E7.5 embryos genotyped as Miz1−/− (B and D) and of two of their Miz1+/− littermates (A and C).
Expression of target genes of Miz1.
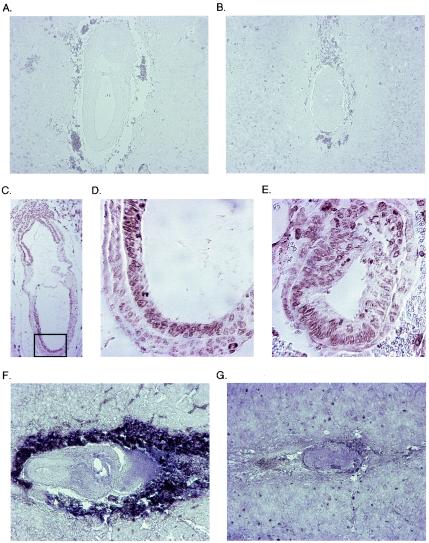
Experiments using small interfering RNA have shown that Miz1 is required for p21Cip1 induction in response to DNA damage (20); furthermore, Miz1 has been implicated in the expression of p15Ink4b by TGF-β in keratinocytes (37). Also, we speculated that Miz1 might be required for expression of c-myc, since Myc is known to suppress its own transcription (33). We therefore tested whether expression of any of these three genes is affected in Miz1−/− embryos. Consistent with previous reports (50), expression of p15Ink4b was not detectable in either normal or abnormal E7.5 embryos (Fig. 7A and B). p21Cip1 was expressed ubiquitously at low levels in normal E7.5 embryos, with a more intense staining in anterior ectodermal nuclei. A similar pattern was observed in Miz1−/− embryos (Fig. 7C, D, and E). c-myc is expressed ubiquitously at E6.5 and later becomes more restricted to the ectoplacental cone and the extraembryonic part of ectoderm and mesoderm (13). This pattern was clearly visible in normal embryos, whereas the expression in abnormal embryos at E7.5 resembled the pattern described for E.65 normal embryos, most likely due to the retardation in development (Fig. 7F and G).
FIG. 7.
Expression of p15Ink4b, p21Cip1, and c-myc in normal and abnormal mouse embryos. The upper two panels document staining with an anti-p15Ink4b antibody of either a normal (A) or an abnormal (B) antibody. Note the absence of staining in the embryo, but the presence of staining in the surrounding decidua. The middle panels show staining of morphologically normal (C and D) or abnormal (E) E7.5 embryos with an anti-p21Cip1 antibody. The embryo in panel C is shown in a lower magnification; the frame marks the area shown in panel D at the same magnification as the abnormal embryo in section E. The lower two panels show in situ hybridizations with a c-myc antisense probe of either a normal (F) or an abnormal (G) E7.5 embryo. Note that the intense staining surrounding the normal embryo derives from the peroxidase present in erythrocytes and does not reflect c-myc expression.
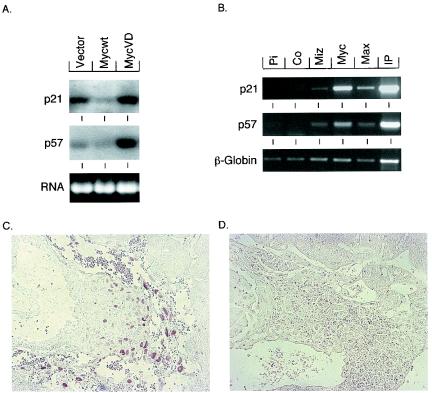
Recent microarray analyses have shown that p57Kip2 is a target for regulation by the Myc-Miz1 complex in fibroblasts (V. Beuger and M. Eilers, unpublished observations). Northern blotting showed that expression of p57Kip2 was repressed by Myc, but not by MycV394D, an allele of Myc that is unable to bind to Miz1 (20) (Fig. 8A). Furthermore, ChIP showed that Miz1 is bound to the start site of the p57Kip2 gene in vivo (Fig. 8B). Expression of p57Kip2 is restricted to the ectoplacental cone at this stage of development (42). Specific staining with anti-p57Kip2 antibodies was detected in normal embryos, but not in abnormal embryos (Fig. 8C and D). We concluded that Miz1 is required for expression of p57Kip2, but not for expression of p21Cip1 and c-myc during early embryonic development.
FIG. 8.
p57Kip2 is a target gene of Miz1 and is absent in Miz−/− embryos. (A) Northern blots demonstrating differential expression of the indicated genes in primary mouse embryo fibroblasts infected with viruses of the indicated genotypes. MycV394D is an allele of Myc that does not bind to Miz1 (20). (B) ChIp experiments documenting in vivo binding of Myc and Miz1 to DNA surrounding the start site of transcription of the P57KIP2and P21CIP1 genes. For P57KIP2, a fragment between nucleotides −139 and +65 relative to the major start site of transcription was amplified, and for P21CIP1, a fragment between nucleotides −61 and +353 was amplified. The experiment was performed with HeLa cells as described by Herold et al. (20). Panels C and D document staining of the ectoplacental cone of morphologically normal (C) or abnormal (D) E7.5 embryos with anti-p57Kip2 antibodies.
DISCUSSION
Biochemical evidence implicates Miz1 as an interaction partner of the Myc oncoprotein and shows that Myc can repress specific genes through the interaction with Miz1. Specifically, Miz1 has been implicated in two pathways that negatively regulate cell proliferation: the responses to TGF-β (37, 40) and to DNA damage (20, 36). Recent data also support a role for Miz1 in the differentiation of epithelial cells of the colon and of hematopoietic cells (22, 43, 48).
The physiological role of Miz1 has not been identified up to now; as a first step, we now report the phenotype of Miz1−/− embryos. We show that Miz1 is required for proper embryonic development and that Miz1−/− embryos fail to undergo proper gastrulation. Since Miz1 is a negative regulator of cell proliferation in fibroblasts, one might expect that Miz1−/− embryos die as a result of deregulated proliferation. However, we did not observe this phenotype. In contrast, the percentage of proliferating cells is higher in wild-type than in Miz1−/− embryos, reflecting the rapid growth of normal embryos at this stage of development. Therefore, we did not obtain evidence that Miz1 negatively regulates cell proliferation during gastrulation.
Of the pathways in which Miz1 has been implicated, signaling by a member of the TGF-β family, Nodal, is active during gastrulation and is required for the formation of the primitive streak. In contrast to epithelial cells, however, Nodal signaling does not regulate cell proliferation and does not regulate expression of p15Ink4b in early embryos. Our data show that induction of mesoderm is intact, both Goosecoid and Brachyury are expressed, and a primitive streak is visible in some in Miz1−/− embryos. This is in contrast to knockout phenotypes of the TGF-β family member Nodal (10), of several Smad genes (19, 41), and of TGF-β receptor family genes (6) that disrupt mesoderm induction during gastrulation. Taken together, the data strongly suggest that signal transduction through Nodal and Smad function is unimpaired in Miz1−/− embryos. Our data are consistent with the “dual-input” model initially proposed by Seoane et al. (37). The model suggests that Miz1 is not a direct part of the TGF-β signaling cascade, but specifically regulates a subset of TGF-β target genes through interaction with Myc. Activation of Miz1-dependent transactivation by TGF-β occurs indirectly as a response to the reduced expression of Myc. Since there is no evidence that Myc is involved in signaling through Nodal signaling, the model predicts that Miz1 is not involved in signal transduction through Nodal.
Miz1 has been implicated in the regulation of several target genes, most notably the cell cycle inhibitors p15Ink4b, p21Cip1, and p57Kip2. p15Ink4b is not expressed at this early stage in development (50). Loss of Miz1 has a differential effect on the expression of p21Cip1 and p57Kip2. We find that Miz1 is not required for expression of p21Cip1 in the developing mouse embryo; in contrast, expression of p57Kip2 in the ectoplacental cone depends on Miz1. The situation for p21Cip1 contrasts to the induction of p21Cip1 in response to DNA damage: experiments using either transient (20) or stable ablation (M. Wanzel and M.E., unpublished) of Miz1 with RNA interference show that Miz1 is required for induction of p21Cip1 in this setting. One possible explanation would be that homologous proteins exist in mouse. However, the recently published sequence of the mouse genome reveals no gene with a homology to Miz1 that extends beyond the similarity to other POZ domain/zinc finger proteins. Therefore, Miz1 is not generally required for transcription of p21Cip1. Similarly, the finding that Miz1−/− embryos die after multiple rounds of division suggests that Miz1 is not generally required for initiator-dependent transcription, but rather is a specific transcription factor that transmits specific signals (e.g., in response to DNA damage) to the promoters it binds to.
Finally, we show that Miz1−/− embryos succumb to massive apoptosis of ectodermal cells around E8. This may reflect a nonspecific response, since a number of knockout mice show this effect (28). Alternatively, Miz1 might be involved in a survival pathway that relays adhesion-dependent survival signals to ectodermal cells. If so, this might explain the strongly reduced adhesion of both fibroblasts and stem cells (2) expressing deregulated Myc and the correlation between binding to Miz1, lack of adhesion and apoptosis that is observed in fibroblast models expressing deregulated Myc (21). Further work with conditional knockout models of Miz1 will be necessary to decide between these alternatives.
Acknowledgments
The work reported in this article was supported by the Deutsche Forschungsgemeinschaft (DFG) and the European Community.
We would like to thank Antje Grzeschiczek and Waltraud Ackermann for expert technical assistance, Andreas Trumpp and Dave Parry for the gift of antibodies, Stefan Gaubatz for critical comments on the manuscript, and Martin Blum for a lot of help, advice, and plasmids.
REFERENCES
- 1.Amati, B., M. W. Brooks, N. Levy, T. D. Littlewood, G. I. Evan, and H. Land. 1993. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 72:233-245. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, I., and F. M. Watt. 2001. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr. Biol. 11:558-568. [DOI] [PubMed] [Google Scholar]
- 3.Bancroft, J. D., and A. Stevens. 1990. Theory and practice of histological techniques, 3rd ed. Churchill Livingstone, Edinburgh, United Kingdom.
- 4.Bardwell, V. J., and R. Treisman. 1994. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 8:1664-1677. [DOI] [PubMed] [Google Scholar]
- 5.Belo, J. A., T. Bouwmeester, L. Leyns, N. Kertesz, M. Gallo, M. Follettie, and E. M. De Robertis. 1997. Cerberus-like is a secreted factor with neutralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech. Dev. 68:45-57. [DOI] [PubMed] [Google Scholar]
- 6.Beppu, H., M. Kawabata, T. Hamamoto, A. Chytil, O. Minowa, T. Noda, and K. Miyazono. 2000. BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev. Biol. 221:249-258. [DOI] [PubMed] [Google Scholar]
- 7.Blum, M., S. J. Gaunt, K. W. Cho, H. Steinbeisser, B. Blumberg, D. Bittner, and E. M. De Robertis. 1992. Gastrulation in the mouse: the role of the homeobox gene goosecoid. Cell 69:1097-1106. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard, C., O. Dittrich, A. Kiermaier, K. Dohmann, A. Menkel, M. Eilers, and B. Luscher. 2001. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 15:2042-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coller, H. A., C. Grandori, P. Tamayo, T. Colbert, E. S. Lander, R. N. Eisenman, and T. R. Golub. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conlon, F. L., K. M. Lyons, N. Takaesu, K. S. Barth, A. Kispert, B. Herrmann, and E. J. Robertson. 1994. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development 120:1919-1928. [DOI] [PubMed] [Google Scholar]
- 11.Coucouvanis, E., and G. R. Martin. 1995. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell 83:279-287. [DOI] [PubMed] [Google Scholar]
- 12.Dauphinot, L., C. De Oliveira, T. Melot, N. Sevenet, V. Thomas, B. E. Weissman, and O. Delattre. 2001. Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2and c-Myc expression. Oncogene 20:3258-3265. [DOI] [PubMed] [Google Scholar]
- 13.Downs, K. M., G. R. Martin, and J. M. Bishop. 1989. Contrasting patterns of c-myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 3:860-869. [DOI] [PubMed] [Google Scholar]
- 14.Eberhardy, S. R., and P. J. Farnham. 2002. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 277:40156-40162. [DOI] [PubMed] [Google Scholar]
- 15.Feng, X. H., Y. Y. Liang, M. Liang, W. Zhai, and X. Lin. 2002. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B). Mol. Cell 9:133-143. [DOI] [PubMed] [Google Scholar]
- 16.Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gartel, A. L., X. Ye, E. Goufman, P. Shianov, N. Hay, F. Najmabadi, and A. L. Tyner. 2001. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. USA 98:4510-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamamoto, T., H. Beppu, H. Okada, M. Kawabata, T. Kitamura, K. Miyazono, and M. Kato. 2002. Compound disruption of smad2 accelerates malignant progression of intestinal tumors in apc knockout mice. Cancer Res. 62:5955-5961. [PubMed] [Google Scholar]
- 20.Herold, S., M. Wanzel, V. Beuger, C. Frohme, D. Beul, T. Hillukkala, J. Syvaoja, H. P. Saluz, F. Hänel, and M. Eilers. 2002. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell 10:509-521. [DOI] [PubMed] [Google Scholar]
- 21.James, L., and R. Eisenman. 2002. Myc and Mad bHLHZ domains possess identical DNA-binding specificities but only partially overlapping functions in vivo. Proc. Natl. Acad. Sci. USA 99:10429-10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kime, L., and S. C. Wright. 2003. Mad4 is regulated by a transcriptional repressor complex that contains Miz-1 and c-Myc. Biochem. J. 370:291-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kretzner, L., E. M. Blackwood, and R. N. Eisenmann. 1992. Myc and Max proteins possess distinct transcriptional activities. Nature 359:426-429. [DOI] [PubMed] [Google Scholar]
- 24.Li, L., C. Nerlov, G. Prendergast, D. MacGregor, and E. B. Ziff. 1994. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 13:4070-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 26.McMahon, S. B., M. A. Wood, and M. D. Cole. 2000. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 20:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menssen, A., and H. Hermeking. 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. USA 99:6274-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller, A. G., M. Moser, R. Kluge, S. Leder, M. Blum, R. Buttner, H.-G. Joost, and A. Schürmann. 2002. Embryonic lethality caused by apoptosis during gastrulation in mice lacking the gene of the ADP-ribosylation factor-related protein 1. Mol. Cell. Biol. 22:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Napirei, M., H. Karsunky, B. Zevnik, H. Stephan, H. G. Mannherz, and T. Moroy. 2000. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat. Genet. 25:177-181. [DOI] [PubMed] [Google Scholar]
- 30.Nikiforov, M. A., S. Chandriani, J. Park, I. Kotenko, D. Matheos, A. Johnsson, S. B. McMahon, and M. D. Cole. 2002. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol. Cell. Biol. 22:5054-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Hagan, R. C., N. Schreiber-Agus, K. Chen, G. David, J. A. Engelman, R. Schwab, L. Alland, C. Thomson, D. R. Ronning, J. C. Sacchettini, P. Meltzer, and R. A. DePinho. 2000. Gene-target recognition among members of the myc superfamily and implications for oncogenesis. Nat. Genet. 24:113-119. [DOI] [PubMed] [Google Scholar]
- 32.Oster, S. K., C. S. Ho, E. L. Soucie, and L. Z. Penn. 2002. The myc oncogene: MarvelouslY complex. Adv. Cancer Res. 84:81-154. [DOI] [PubMed] [Google Scholar]
- 33.Penn, L. J. Z., M. W. Brooks, E. M. Laufer, and H. Land. 1990. Negative autoregulation of c-myc transcription. EMBO J. 9:113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peukert, K., P. Staller, A. Schneider, G. Carmichael, F. Hanel, and M. Eilers. 1997. An alternative pathway for gene regulation by Myc. EMBO J. 16:5672-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy, A. L., C. Carruthers, T. Gutjahr, and R. G. Roeder. 1993. Direct role for Myc in transcription initiation mediated by interactions with TFII-I. Nature 365:359-361. [DOI] [PubMed] [Google Scholar]
- 36.Seoane, J., H. V. Le, and J. Massague. 2002. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419:729-734. [DOI] [PubMed] [Google Scholar]
- 37.Seoane, J., C. Pouponnot, P. Staller, M. Schader, M. Eilers, and J. Massague. 2001. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat. Cell Biol. 3:400-408. [DOI] [PubMed] [Google Scholar]
- 38.Shrivastava, A., S. Saleque, G. V. Kalpana, S. Artandi, S. P. Goff, and K. Calame. 1993. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science 262:1889-1891. [DOI] [PubMed] [Google Scholar]
- 39.Sirard, C., J. L. de la Pompa, A. Elia, A. Itie, C. Mirtsos, A. Cheung, S. Hahn, A. Wakeham, L. Schwartz, S. E. Kern, J. Rossant, and T. W. Mak. 1998. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 12:107-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staller, P., K. Peukert, A. Kiermaier, J. Seoane, J. Lukas, H. Karsunky, T. Moroy, J. Bartek, J. Massague, F. Hanel, and M. Eilers. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 3:392-399. [DOI] [PubMed] [Google Scholar]
- 41.Takaku, K., M. Oshima, H. Miyoshi, M. Matsui, M. F. Seldin, and M. M. Taketo. 1998. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell 92:645-656. [DOI] [PubMed] [Google Scholar]
- 42.Tamai, Y., T. Ishikawa, M. R. Bosl, M. Mori, M. Nozaki, H. Baribault, R. G. Oshima, and M. M. Taketo. 2000. Cytokeratins 8 and 19 in the mouse placental development. J. Cell Biol. 151:563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Wetering, M., E. Sancho, C. Verweij, W. de Lau, I. Oving, A. Hurlstone, K. van der Horn, E. Batlle, D. Coudreuse, A. P. Haramis, M. Tjon-Pon-Fong, P. Moerer, M. van den Born, G. Soete, S. Pals, M. Eilers, R. Medema, and H. Clevers. 2002. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111:241-250. [DOI] [PubMed] [Google Scholar]
- 44.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, S. E. Antonarakis, J. Attwood, R. Baertsch, J. Bailey, K. Barlow, S. Beck, E. Berry, B. Birren, T. Bloom, P. Bork, M. Botcherby, N. Bray, M. R. Brent, D. G. Brown, S. D. Brown, C. Bult, J. Burton, J. Butler, R. D. Campbell, P. Carninci, S. Cawley, F. Chiaromonte, A. T. Chinwalla, D. M. Church, M. Clamp, C. Clee, F. S. Collins, L. L. Cook, R. R. Copley, A. Coulson, O. Couronne, J. Cuff, V. Curwen, T. Cutts, M. Daly, R. David, J. Davies, K. D. Delehaunty, J. Deri, E. T. Dermitzakis, C. Dewey, N. J. Dickens, M. Diekhans, S. Dodge, I. Dubchak, D. M. Dunn, S. R. Eddy, L. Elnitski, R. D. Emes, P. Eswara, E. Eyras, A. Felsenfeld, G. A. Fewell, P. Flicek, K. Foley, W. N. Frankel, L. A. Fulton, R. S. Fulton, T. S. Furey, D. Gage, R. A. Gibbs, G. Glusman, S. Gnerre, N. Goldman, L. Goodstadt, D. Grafham, T. A. Graves, E. D. Green, S. Gregory, R. Guigo, M. Guyer, R. C. Hardison, D. Haussler, Y. Hayashizaki, L. W. Hillier, A. Hinrichs, W. Hlavina, T. Holzer, F. Hsu, A. Hua, T. Hubbard, A. Hunt, I. Jackson, D. B. Jaffe, L. S. Johnson, M. Jones, T. A. Jones, A. Joy, M. Kamal, E. K. Karlsson, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 45.Watson, J. D., S. K. Oster, M. Shago, F. Khosravi, and L. Z. Penn. 2002. Identifying genes regulated in a Myc-dependent manner. J. Biol. Chem. 277:36921-36930. [DOI] [PubMed] [Google Scholar]
- 46.Weinstein, M., X. Yang, C. Li, X. Xu, J. Gotay, and C. X. Deng. 1998. Failure of egg cylinder elongation and mesoderm induction in mouse embryos lacking the tumor suppressor smad2. Proc. Natl. Acad. Sci. USA 95:9378-9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkinson, D. G., S. Bhatt, and B. G. Herrmann. 1990. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature 343:657-659. [DOI] [PubMed] [Google Scholar]
- 48.Wu, S., C. Cetinkaya, M. J. Munoz-Alonso, N. von der Lehr, F. Bahram, V. Beuger, M. Eilers, J. Leon, and L. G. Larsson. 2003. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene 22:351-360. [DOI] [PubMed] [Google Scholar]
- 49.Yang, W., J. Shen, M. Wu, M. Arsura, M. FitzGerald, Z. Suldan, D. W. Kim, C. S. Hofmann, S. Pianetti, R. Romieu-Mourez, L. P. Freedman, and G. E. Sonenshein. 2001. Repression of transcription of the p27(Kip1) cyclin-dependent kinase inhibitor gene by c-Myc. Oncogene 20:1688-1702. [DOI] [PubMed] [Google Scholar]
- 50.Zindy, F., D. E. Quelle, M. F. Roussel, and C. J. Sherr. 1997. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 15:203-211. [DOI] [PubMed] [Google Scholar]