Abstract
The muscle of Lawrence (MOL) is a bilaterally symmetrical muscle spanning the tergite of the fifth abdominal segment of adult male Drosophila melanogaster. It is not, however, a general feature of male-specific development within the subfamily Drosophilinae. Of 95 species surveyed within this subfamily, 67 exist with no MOL at all. By drawing comparisons with published cladograms of species relatedness, three conclusions regarding the evolutionary history of the MOL are made: (i) The MOL predates the major radiations of the genus Drosophila, given its presence in earlier-branching Chymomyza and Scaptodrosophila; the MOL has been subsequently excluded in at least one present species of each of these two primitive genera. (ii) Within the genus Drosophila the MOL is present sporadically in the radiation of the subgenus Sophophora, showing repetitive loss even in very close evolutionary lineages. (iii) The MOL may have been entirely excluded from the prolific radiation of the subgenus Drosophila. Thus the MOL shows a uniquely incongruous pattern of presence or absence relative to accepted drosophilid phylogeny.
Keywords: phylogeny
First described in Drosophila melanogaster, the muscle of Lawrence (MOL) is a large, bilaterally symmetrical muscle spanning the fifth tergite of the abdomen of adult males; it does not develop in females (1). Oddly, the MOL eluded decades of extensive genetic and biological investigation in D. melanogaster, including the definitive survey of adult musculature by Miller in 1950 (2). Only in 1984 was this muscle’s existence first revealed with the report of Lawrence (the muscle’s eponym) and Johnston (1), showing that formation of the MOL is sex-specific and is associated with developmental cues in the fifth abdominal segment (A5) of the male; homeotic mutations transforming either A4 or A6 to A5 induce the development of a MOL in the transformed segment (1, 3).
Development of the MOL depends not on the sex of its progenitor myoblasts, but rather the sex of the contacting motoneuron in A5. If the motoneuron is male, the MOL develops; if the motoneuron is female, the MOL fails to develop (3). Ablation of the ingrowing motoneuron in a male correspondingly blocks MOL development (4). Although use of the MOL to the adult male fly is unknown, other interesting observations have accumulated regarding its general biology, including genetic evidence that its development requires normal expression of fruitless (fru) (5, 6), a gene identified by its involvement in adult male courtship behavior (7) and as a sex determination gene that acts late in development (8, 9). In addition, formation of the MOL in A5 involves recruitment of the relatively rare 79B actin (10).
To gain insight into the evolutionary history of this muscle, we have analyzed its appearance within the subfamily Drosophilinae. Surprisingly, of 95 species surveyed, 67 have no MOL at all. By comparing our observations with generally accepted cladograms of species relatedness within the Drosophilinae (11–24), we provide evidence that the MOL was an existing feature of primitive forms within the early radiation of the subfamily. Also stemming from that comparison is our conclusion that several independent genetic exclusions of this muscle have occurred during the subsequent radiation of MOL-containing lines. This makes the MOL a rare example of an anatomical structure that has undergone independent, repeated loss among closely related evolutionary lines.
MATERIALS AND METHODS
All fly stocks were maintained on an instant cornmeal-based medium. Species other than D. melanogaster were obtained from the National Drosophila Species Resource Center, Bowling Green, OH.
The techniques for preparation of specimens and visualization of their musculature by birefringence under polarized light have been previously described (5). For each of the eight species of the melanogaster species subgroup, 20 males and 5 females were dissected and analyzed. For species outside the melanogaster species subgroup, 10 males and 5 females were dissected in each species, and then their musculature compared. For a species to be scored as MOL-absent, there was no discernible difference in the male and female A5 musculature. No MOL or MOL-like structure was ever observed in a female (n = 475 females dissected).
One set of experiments involved hybridization of males from a MOL-containing species to females of species whose males do not develop the MOL. Such crosses generally performed poorly and were optimized by crowding parents. For each hybrid cross, 10 males were assessed for their abdominal musculature. Their hybrid male genotypes were ascertained first by sterility with virgin females of both parental species (25) and then by verification of hybrid anatomy of the genital arch.
RESULTS AND DISCUSSION
The MOL Is Not a General Anatomical Feature of All Drosophila Males.
The MOL is shown in Fig. 1a as it normally appears in the dorsal abdomen of adult male D. melanogaster. Could this muscle be a general requirement for normal mating ability and fertility of all males within the genus? This question was first addressed only with great difficulty by generating two fru mutant D. melanogaster males who developed no MOL, yet were fertile (5). Here, the answer is a straightforward no. Almost three-fourths of the Drosophila species surveyed in this study showed no evidence of MOL development (61 of 84 species; Table 1, Fig. 2). Noting that there are more than 1,600 reported species of Drosophila (22), most of which are not easily available for analysis, our survey could not be exhaustive. Nonetheless, our results point first to an apparent division in MOL presence among the four largest species groups of the subgenus Sophophora. The MOL seems to appear exclusively within the obscura and melanogaster groups, whereas no species within the saltans and willistoni groups was found with a MOL (Fig. 2). The following points are particularly noteworthy.
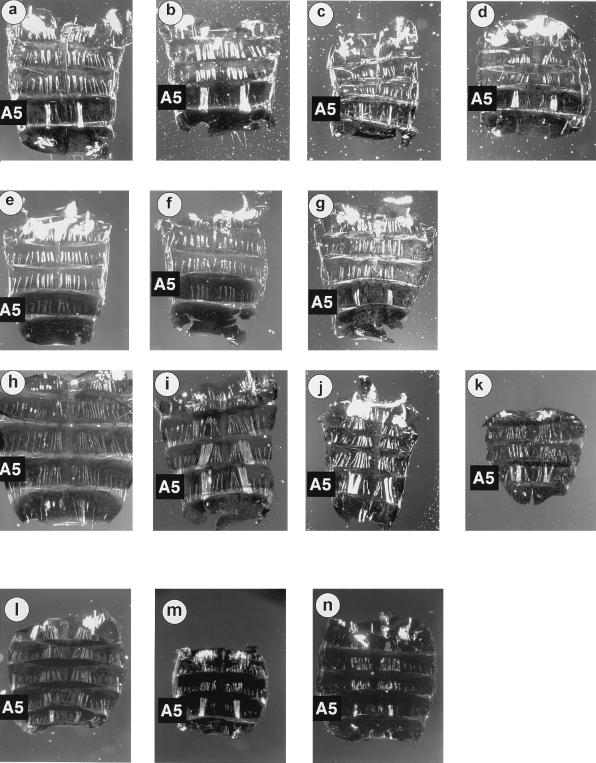
Figure 1.
Comparative dorsal abdominal musculature among adult drosophilid males and females. (a) D. melanogaster male. (b) D. mauritiana male. (c) D. simulans male. (d) D. sechellia male. (e) D. yakuba male. (f) D. erecta male. (g) Hybrid male from the cross D. mauritiana males × D. teissieri females (hybrid males from the cross D. mauritiana males × D. yakuba females also contained a MOL, not shown). (h) D. subobscura female. (i) D. subobscura male. (j) D. pseudoobscura male. (k) D. azteca male. (l) Chymomyza amoena female. (m) Chymomyza amoena male. (n) Scaptodrosophila lebanonensis male. When present, note especially the species-specific variation of the MOL in terms of fiber size and number. Consistent among all species with the MOL are its obvious length, size, position relative to the smaller, adjacent longitudinal fibers, and insertion in A6 (5), apart from the D. subobscura male, which shows a duplication of the MOL in A4 (i).
Table 1.
Listing of species analyzed for MOL phenotype
| Genus Subgenus Species group: Species |
|---|
| Chymomyza (MOL+): amoena |
| Chymomyza (MOL−): procnemis |
| Drosophila |
| Dorsilopha (MOL−): busckii |
| Drosophila (MOL−) |
| annulimana: aracatacas; cardini: cardini; funebris: funebris; immigrans: immigrans, nasuta; melanica: melanica; mesophragmatica: gaucha; modified mouth parts: mimica; nannoptera: nannoptera; quinaria: palustris; repleta: arizonensis, mercatorum mercatorum, mulleri; robusta: robusta; tripunctata: tripunctata; virilis: virilis |
| Sophophora |
| melanogaster (MOL+): elegans, eugracilis, lucipennis, lutescens, mauritiana, melanogaster, mimetica, paralutea, prostipennis, pseudotakahashii, pulchrella, rajasekari, sechellia, simulans, takahashii |
| melanogaster (MOL−): ananassae, auraria, baimaii, bipectinata, ercepeae, erecta, ficusphila, jambulina, kikkawai, lacteicornis, lini, malerkotliana, mayri, orena, pallidosa, parabipectinata, pennae, phaeopleura, pseudoananassae, punjabiensis, quadraria, rufa, seguyi, teissieri, varians, yakuba |
| obscura (MOL+): ambigua, azteca, bifasciata, miranda, persimilis, pseudoobscura, subobscura, tolteca |
| obscura (MOL−): affinis |
| saltans (MOL−): austrosaltans, emarginata, lusaltans, milleri, neocordata, prosaltans, saltans, sturtevanti, subsaltans |
| willistoni (MOL−): capricorni, equinoxialis, fumipennis, nebulosa, paulistorum, succinea, tropicalis, willistoni |
| Hawaiian “Drosophila” (Idiomyia) (MOL−): crucigera, gymnobasis |
| Hirtodrosophila (MOL−): pictiventris |
| Scaptodrosophila (MOL+): dimorpha, lebanonensis lebanonensis, pattersoni, stonei |
| Scaptodrosophila (MOL−): latifasciaeformis |
| Zaprionus (MOL−): tuberculatus |
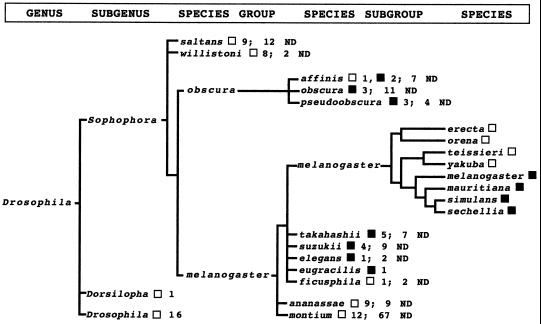
Figure 2.
Distribution of the MOL in Drosophila species analyzed in this study. Note that a complete cladogram is shown for the melanogaster species subgroup only. This is a consensus cladogram based on allozyme, chromosomal and behavioral comparisons (12); a very similar phylogeny based on DNA hybridization criteria also has been reported (13). See references below for specific details regarding other groupings. □, MOL absent; ▪, MOL present; ND, not dissected. To interpret particular results, the saltans species group is explained as example: Of 21 total species within the group, 9 of 9 dissected contained no MOL; 12 species were not dissected. For species outside the melanogaster species subgroup n = 10 males and 5 females dissected per species; within the melanogaster species subgroup n = 20 males and 5 females per species. Groupings are based on the following general literature: melanogaster subgroup (11–13); melanogaster group (14); saltans group (15); willistoni group (16); obscura group (17–19); obscura and melanogaster group cluster (20, 21); general reviews on evolution within the family Drosophilidae (20–24).
Not every species of the obscura and melanogaster groups that was examined had a MOL. Whereas 8 of 9 species dissected from the obscura species group contained a MOL, only 15 of 41 from the melanogaster species group did (Fig. 2).
Conspicuously within the melanogaster group, dissection of half the species of the ananassae species subgroup, and about one-sixth of the many species of the montium species subgroup, did not reveal a species with the MOL (Fig. 2). Likewise, analysis of nearly half the species of the saltans and nearly all the species of the willistoni species groups revealed no MOL (Fig. 2). Consequently, these may represent sophophoran lines in which the MOL is completely missing.
Also conspicuous was our finding that this unusual pattern of MOL presence and absence carried through even to the melanogaster species subgroup, in which 4 of the 8 species had the MOL, but 4 did not (Fig. 1 a–d vs. e and f; Fig. 2). Remarkably, however, hybrid males can be generated in certain crosses between one of these MOL containing species and a species with no MOL (Fig. 1g; also see below).
When present, the melanogaster species group MOL was homogeneous in appearance and within the narrow range of forms as displayed by the melanogaster species subgroup (Fig. 1 a–d). By comparison, the obscura species group MOL varied widely in appearance. The most striking case was D. subobscura in which the MOL is duplicated into A4 (Fig. 1 h vs. i). The MOL of D. pseudoobscura is more like that of D. melanogaster (Fig. 1 j vs. a), whereas the MOL of D. azteca is more weakly formed (Fig. 1k).
MOL Presence, Then Absence: A Case of Repeated Evolutionary Loss.
Phylogeny and the MOL within the subfamily Drosophilinae. Whether assessing criteria as diverse as morphological characteristics or DNA sequence, only a few drosophilid groups raise controversy regarding their phylogenetic placement (cf. 20, 22–24). Fig. 3 shows a current drosophilid phylogeny, based on extensive morphological comparisons (23), and includes the corresponding MOL analysis.
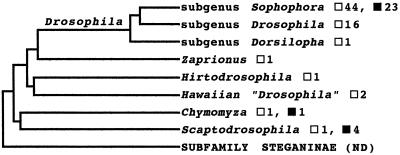
Figure 3.
Phylogenetic relationship of the major genera within the subfamily Drosophilinae and MOL analysis (23). Hirtodrosophila and Zaprionus have been alternatively placed within the Drosophila genus radiation (24). □, MOL absent, ▪, MOL present in a particular group. A single lineage with both symbols contains species with and without the MOL.
The Chymomyza and Scaptodrosophila genera represent two of the earliest radiations within the subfamily Drosophilinae (23); present-day species of both show the MOL (Fig. 3, Table 1). The early split of these two genera has been substantiated by molecular comparison of introns and coding sequences of the superoxide dismutase (Sod) gene (24). Thus we conclude that the MOL, as an anatomical structure, predated all the major Drosophila radiations. By their phylogeny, both Chymomyza and Scaptodrosophila conspicuously show evidence of independent loss of the MOL (Fig. 3, Table 1). There is also a distinct phenotypic variability of the MOL within these two genera. The Chymomyza MOL appears more like that of the melanogaster species subgroup (compare first the Chymomyza female vs. male, Fig. 1 l and m; then Fig. 1 m vs. a–d). By comparison, the MOL fibers of Scaptodrosophila species are consistently shorter, approximately the length of adjoining longitudinal fibers (Fig. 1n is representative of the four Scaptodrosophila species that were found with a MOL; Table 1).
Subsequent to the Chymomyza-Scaptodrosophila radiations, but still before those of the genus Drosophila, are branches leading to the genera Hawaiian “Drosophila” (or Idiomyia), Hirtodrosophila and Zaprionus, as well as the subgenus Dorsilopha (23). Our anatomical analysis of these four taxa revealed no MOL (Fig. 3, Table 1). Such a pattern again necessitates independent MOL losses, at the least, at three more evolutionary branch points (Fig. 3).
Hirtodrosophila and Zaprionus have been alternatively placed, based on their Sod gene structure, within the radiation leading to the subgenus Drosophila (24). This might reveal the constancy of a single evolutionary lineage in which the MOL has been entirely excluded, since again, no species of any of these three lines was found with a MOL (Figs. 2 and 3). This includes 16 species of the subgenus Drosophila, representing 13 species groups (21). But whether these entire taxa are MOL-less is speculative, because our conclusion is based on these few species (Table 1, Fig. 3). For example, the subgenus Drosophila radiation has been by far the most prolific, evolving more than 800 species (22).
Could the MOL be a general feature of dipteran development? It might be possible ultimately to reveal the MOL’s evolutionary origin by analyzing ever-earlier radiations. This should include species of the drosophilid subfamily Steganinae (20, 23), before moving outside Drosophilidae into other families of the order Diptera. We have broached this question with dissection of one tephritid (Rhagoletis completa) and one muscid (Musca domestica); both had no MOL.
Phylogeny and the MOL within the subgenus Sophophora. The simplest evolutionary model would predict that with the appearance of an anatomical structure such as the MOL it might be transmitted in a linear fashion to all species radiating from that point forward in time. Thus, if the MOL were present as early as the Chymomyza and Scaptodrosophila radiations, one ought to uncover a direct lineage in which all radiating species have the structure.
This is certainly the case with male-specific sex combs. This is an anatomical feature found in all species of the obscura and melanogaster species groups, but in no other Drosophila (21). Regarding the MOL, this evolutionary scenario does not hold, from the primitive genera of the subfamily, to the melanogaster species subgroup (Figs. 2 and 3). Consequently we conclude that the sporadic and frequent absence of the MOL as we have recorded in the radiation of the entire subfamily Drosophilinae is due to evolutionary loss of the muscle not once, but repeatedly. This becomes especially evident within the subgenus Sophophora.
Perhaps the most striking loss is seen within the close lineage of the eight species of the melanogaster species subgroup. The cladogram displayed in Fig. 2 for this subgroup is a composite based on allozyme analysis at many enzyme loci, with complementing chromosomal and behavioral comparisons (11, 12), and if correct, necessitates two independent evolutionary losses of the MOL (at the branch point of the erecta cluster and at the branch point of the yakuba cluster; Fig. 2). Note that a very similar cladogram of the subgroup has been derived from DNA hybridization studies (13); this grouping also requires two independent exclusions of the MOL.
The only constancy of lineage appears within four species subgroups considered close to the melanogaster species subgroup: takahashii, suzukii, elegans and eugracilis (12). Of the 29 total species in this cluster, the 11 we have surveyed all contain a similar-appearing MOL. But if the grouping here of the ficusphila species subgroup is correct (12), then another incidence of evolutionary loss must be invoked (Fig. 2).
A similar case arises within the obscura species group (Fig. 2). With about one-fourth of the reported species in this group analyzed (17–19), only one was found to be lacking the MOL; this must have involved yet another independent case of MOL loss. As noted, the appearance of the MOL in this group is much more variable than in the melanogaster group (Fig. 1). This could correlate with the early branching of these two groups during the sophophoran radiation (24). For whatever reason, the form of the obscura MOL is not fixed and conserved, and this variability should provide important insight as additional features of the MOL, its physiology, and evolutionary history are investigated.
Our analysis of the melanogaster species-group branch-point, leading to the ananassae and montium species subgroups, indicates at least one more point of MOL loss (Fig. 2). Taken together, these instances of independent MOL loss within the Sophophora radiation correlate with the MOL losses we have documented in Chymomyza and Scaptodrosophila.
Alternative hypotheses regarding the evolution of this muscle are less tenable: that the MOL has arisen independently more than once, or that published cladograms—based on extensive morphological, chromosomal, behavioral, allozyme, and DNA sequence analysis (11–24)—are fraught with errors.
When present in a given species, the MOL phenotype was fully penetrant and generally did not vary among males. One exception was D. miranda, a member of the obscura species group (Table 1). Nine of 10 males had a similar MOL in A5, whereas one male had only a unilateral MOL. Another exception was Scaptodrosophila lebanonensis. In this species nine of 10 males showed clustering and/or thickening of fibers in A5 (as in the male pictured in Fig. 1n), whereas one male showed a complete absence of the MOL. Given their general rarity, these could be indications of species segregating more than one MOL phenotype in males.
Hybrid Analysis Within the melanogaster Species Subgroup.
Given that four species of the melanogaster species subgroup have the MOL while four do not (Fig. 2), it could be the case that the change from the MOL-present to the MOL-absent state could have a simple genetic basis. We tested this indirectly by generating hybrid males (25) from D. mauritiana fathers (a MOL-containing species), crossed to either D. yakuba or D. teissieri mothers (species containing no MOL). All 10 hybrid males analyzed from each cross contained a MOL (Fig. 1g; only the D. mauritiana × D. teissieri hybrid male is shown). These crosses provide two important pieces of information: (i) the D. mauritiana autosomes are sufficient for triggering the development of the MOL when in combination with either MOL-less genome; and (ii) it is unlikely that either D. yakuba or D. teissieri lack the MOL due to the evolution of a dominant suppressor of MOL development (assuming that such a suppressor would be capable of function in the D. mauritiana genetic background). It could be that a MOL-less genetic background is recessive to the genetic cues triggering MOL development that reside on MOL-containing species’ autosomes. This implies that within this subgroup the genetic differences between the MOL-present and the MOL-absent states are indeed minimal.
fru Effects on the MOL.
Perhaps MOL variation is influenced by evolutionary divergence of the informational content or expression pattern of fru. The interrelationship between MOL development and the fru locus is genetically defined by four fru alleles, which in varying degrees result in aberrations of at least two aspects of male courtship behavior (7). The fru1 and fru2 mutations affect the development of the MOL, leading to an incompletely developed structure in most individual males (5). The fru3 and fru4 mutations, tagged transposon insertions in the fru locus that lead to mutant fru behavior (7, 26), also completely block expression of the MOL when either is homozygous, or when in heterozygous combination (A.V., D.A.G., B. Berwald, S.O., P. T. Barnes, and J.C.H., unpublished results). The etiologies of these mutant effects are likely to be vastly different at the molecular level relative to the establishment of behavioral patterns vs. regulation of the development of a muscle. This is consistent with the notion that fru itself is a master male-behavior and MOL-determinant gene, and that fru expression is controlled in part by the transformer gene of the sex determination cascade (9).
Evolutionary addition or loss of the MOL may have directly involved the fru locus. With the recent cloning and molecular characterization of fru (6, 9), the evolutionary question just implied can be addressed experimentally. In this respect our hybrid analysis is tantalizing. Given the recent evolution of the melanogaster subgroup species it is unlikely that the change from the MOL-present to the MOL-absent state, or vice versa, involved many genetic alterations. Thus molecular comparisons of fru among Drosophila species might reveal a correlation.
CONCLUSIONS
The MOL is a dorsal abdominal muscle occurring in the males of a limited range of drosophilid species. It shows an unusual pattern of presence and absence that we have interpreted as repeated evolutionary loss. What makes the MOL a uniquely evolving structure, however, is its apparent loss even within closely related evolutionary lineages. What results is a pattern of presence that does not match accepted drosophilid phylogeny.
The utility of this muscle to the male flies possessing it has yet to be elucidated. An answer is conceivable through comparative physiological and behavioral analysis. But so far, no male behavioral difference has been noted that correlates with presence or absence of the MOL, especially within the melanogaster species subgroup (27).
Nevertheless, the fact that it is possible genetically to uncouple male-specific MOL development from that of the remaining adult musculature may allow a unique opportunity to study the developmental processes underlying MOL formation, by an approach that combines evolutionary perspectives with molecular genetics.
Acknowledgments
We thank the National Drosophila Species Resource Center, Bowling Green, OH, for complying with seemingly endless requests for Drosophila stocks. We especially appreciate the helpful and detailed criticism supplied by an anonymous reviewer, and the fine technical support from M. Eyassu, M. Macpherson, and M. Washington. This work was supported by National Institutes of Health Minority Biomedical Research Support Grant GM48135-02 (to D.A.G.), and National Institutes of Health Grants GM21473 and NS33352 (to J.C.H.).
ABBREVIATIONS
- MOL
muscle of Lawrence
- A5
fifth abdominal segment
- fru
fruitless
References
- 1.Lawrence P A, Johnston P. Cell. 1984;36:775–782. doi: 10.1016/0092-8674(84)90357-x. [DOI] [PubMed] [Google Scholar]
- 2.Miller A. In: Biology of Drosophila. Demerec M, editor. New York: Wiley; 1950. pp. 420–534. [Google Scholar]
- 3.Lawrence P A, Johnston P. Cell. 1986;45:505–513. doi: 10.1016/0092-8674(86)90282-5. [DOI] [PubMed] [Google Scholar]
- 4.Currie D A, Bate M. Development. 1995;121:2549–2557. doi: 10.1242/dev.121.8.2549. [DOI] [PubMed] [Google Scholar]
- 5.Gailey D A, Taylor B J, Hall J C. Development. 1991;113:879–890. doi: 10.1242/dev.113.3.879. [DOI] [PubMed] [Google Scholar]
- 6.Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D. Proc Natl Acad Sci USA. 1996;93:9687–9692. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall J C. Science. 1994;264:1702–1714. doi: 10.1126/science.8209251. [DOI] [PubMed] [Google Scholar]
- 8.Taylor B J, Villella A, Ryner L C, Baker B S, Hall J C. Dev Genet. 1994;15:275–296. doi: 10.1002/dvg.1020150309. [DOI] [PubMed] [Google Scholar]
- 9.Ryner L C, Goodwin S F, Castrillon D H, Anand A, Villella A, Baker B S, Hall J C, Taylor B J, Wasserman S A. Cell. 1996;87:1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 10.Courchesne-Smith C L, Tobin S L. Dev Biol. 1989;133:313–321. doi: 10.1016/0012-1606(89)90036-5. [DOI] [PubMed] [Google Scholar]
- 11.Cariou M L. Genet Res. 1987;50:181–185. doi: 10.1017/s0016672300023673. [DOI] [PubMed] [Google Scholar]
- 12.Lachaise D, Cariou M L, David J R, Lemeunier F, Tsacas L, Ashburner M. Evol Biol. 1988;22:159–225. [Google Scholar]
- 13.Caccone A, Amato G, Powell J R. Genetics. 1988;118:671–683. doi: 10.1093/genetics/118.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemeunier F, David J R, Tsacas L, Ashburner M. In: The Genetics and Biology of Drosophila. Ashburner M, Novitski E, editors. 3e. London: Academic; 1986. pp. 147–256. [Google Scholar]
- 15.Bicudo H E M C. Genetica. 1973;44:313–329. [Google Scholar]
- 16.Ehrman L, Powell J R. In: The Genetics and Biology of Drosophila. Ashburner M, Novitski E, editors. 3b. London: Academic; 1982. pp. 193–225. [Google Scholar]
- 17.Cariou M L, Lachaise D, Tsacas L, Sourdis J, Krimbas C, Ashburner M. Heredity. 1988;61:73–84. [Google Scholar]
- 18.Lakovaara S, Saura A. In: The Genetics and Biology of Drosophila. Ashburner M, Novitski E, editors. 3b. London: Academic; 1982. pp. 1–59. [Google Scholar]
- 19.Loukas M, Krimbas C B, Vergini Y. Heredity. 1984;53:483–493. [Google Scholar]
- 20.Throckmorton L H. In: Handbook of Genetics: Invertebrates of Genetic Interest. King R C, editor. Vol. 3. New York: Plenum; 1975. pp. 421–469. [Google Scholar]
- 21.Ashburner M. Drosophila, A Laboratory Handbook. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Wheeler M R. In: The Genetics and Biology of Drosophila. Ashburner M, Novitski E, editors. 3e. London: Academic; 1986. pp. 395–409. [Google Scholar]
- 23.Grimaldi D A. Bull Am Mus Nat His. 1990;197:1–139. [Google Scholar]
- 24.Kwiatowski J, Skarecky D, Bailey K, Ayala F J. J Mol Evol. 1994;38:443–454. doi: 10.1007/BF00178844. [DOI] [PubMed] [Google Scholar]
- 25.Lee W H, Watanabe T K. Jpn J Genet. 1987;62:225–239. [Google Scholar]
- 26.Castrillon D H, Gönczy P, Alexander S, Rawson R, Eberhart C G, Viswanathan S, DiNardo S, Wasserman S A. Genetics. 1993;135:489–505. doi: 10.1093/genetics/135.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cobb M, Connolly K, Burnet B. Behaviour. 1985;95:203–231. [Google Scholar]