Abstract
The TAL1 (or SCL) gene, originally identified from its involvement by a recurrent chromosomal translocation, encodes a basic helix-loop-helix transcription factor essential for erythropoiesis. Although presumed to regulate transcription, its target genes are largely unknown. We show here that a nuclear complex containing TAL1, its DNA-binding partner E47, zinc finger transcription factor GATA-1, LIM domain protein LMO2, and LIM domain-binding protein Ldb1 transactivates the protein 4.2 (P4.2) gene through two E box GATA elements in its proximal promoter. Binding of this complex to DNA was dependent on the integrity of both E box and GATA sites and was demonstrated to occur on the P4.2 promoter in cells. Maximal transcription in transiently transfected cells required both E box GATA elements and expression of all five components of the complex. This complex was shown, in addition, to be capable of linking in solution double-stranded oligonucleotides corresponding to the two P4.2 E box GATA elements. This DNA-linking activity required Ldb1 and increased with dimethyl sulfoxide-induced differentiation of murine erythroleukemia (MEL) cells. In contrast, enforced expression in MEL cells of dimerization-defective mutant Ldb1, as well as wild-type Ldb1, significantly decreased E box GATA DNA-binding activities, P4.2 promoter activity, and accumulation of P4.2 and β-globin mRNAs. These studies define a physiologic target for a TAL1- and GATA-1-containing ternary complex and reveal a positive role for Ldb1 in erythroid gene expression and differentiation.
Red blood cell production is controlled by the concerted actions of signal transducers and nuclear regulators. Gene-targeting studies have demonstrated a requirement for several nuclear proteins in erythropoiesis, including the TAL1 (or SCL) and GATA-1 transcription factors and the LIM domain protein LMO2. These proteins, singly and in combination, bind to specific promoter and enhancer elements to regulate the expression of genes that execute the differentiation program.
Two of these transcriptional regulators, TAL1 and LMO2, were initially identified from their involvement by chromosomal rearrangements in T-cell acute lymphoblastic leukemia. Both are essential for embryonic erythropoiesis, with embryos bearing targeted mutations of either gene failing to progress beyond midgestation and lacking all hematopoietic cells (46, 49, 59). In addition, cells nullizygous for Tal1 or Lmo2 were not represented in any hematopoietic lineage in chimeric mice derived from mixtures of wild-type and knockout embryonic stem cells (41, 45, 61). Besides its actions in hematopoietic stem cells, TAL1 promotes the terminal differentiation of erythroid progenitors (4, 14) and was recently shown to have an essential function in postnatal erythropoiesis (17, 30, 47).
TAL1 belongs to the basic helix-loop-helix (bHLH) family of transcription factors, many of which regulate cell fate determination or differentiation. Like other tissue-restricted members of this group, it binds DNA as a heterodimer with any of several widely expressed bHLH proteins, including E47, E12, and HEB/HTF4, to a specific sequence motif, CANNTG, termed the E box. In addition to its bHLH DNA-binding partners, TAL1 can interact with two LIM domain proteins, LMO2 and LMO1, and the zinc finger transcription factor GATA-1, with LMO2 serving as a bridge between GATA-1 and TAL1/E47 dimers (37). A role for this TAL1-, LMO2-, and GATA-1-containing complex in embryonic hematopoiesis was suggested by overexpression studies in Xenopus embryos (28), and the similarity in phenotype of their respective gene knockouts is also compatible with their contribution to a common complex.
As a first step toward identification of its target genes, site selection experiments identified an E box sequence, CAGATG, that is bound preferentially by TAL1-E protein heterodimers (19). Subsequently, Wadman et al. showed that a multiprotein complex present in murine erythroleukemia (MEL) cell nuclei and composed of TAL1, E47, GATA-1, LMO2, and a LIM domain-binding protein, Ldb1, recognized a different motif with a distinct E box and GATA site separated by 9 to 12 bp from a GATA site (58). Assembly of the complex on this bipartite element required all five proteins, and their combined expression was necessary for maximal transactivation of a reporter gene linked to two copies of the consensus E box GATA element (58). In addition, chromatin immunoprecipitation (ChIP) assays revealed TAL1 association with E box GATA elements in vivo and identified a possible target gene containing such an element in its first intron (10). The E box GATA motif has been noted in the regulatory regions of several other erythroid cell-expressed genes, including those encoding the transcription factors GATA-1 (35, 57) and EKLF (3). However, evidence is either lacking or against (35) their regulation by TAL1.
We identify the gene for protein 4.2 (P4.2), an important component of the red cell membrane skeleton, as a target of this TAL1-containing protein complex in mouse erythroid progenitors. We found that TAL1, E47, GATA-1, LMO2, and Ldb1 synergistically stimulate P4.2 gene transcription, a complex containing these factors binds two E box GATA elements in the P4.2 promoter, and complexes assembled on these two elements can be linked in solution in a manner dependent on Ldb1. By using the P4.2 promoter to investigate the transcriptional properties and biological functions of this E box GATA DNA-binding complex, we establish that its constituent proteins, and Ldb1 in particular, positively regulate erythroid gene expression and differentiation. Finally, we propose a model in which Ldb1 homodimerization mediates the physical interaction of complexes bound to independent E box GATA elements.
MATERIALS AND METHODS
Plasmid constructs.
The pGL2-P4.2p1700-Luc reporter plasmid was described previously (25). Plasmids pEFIRES-P and pEFIRES-N were provided by Stephen Hobbs (Institute of Cancer Research, London, United Kingdom), pCMV-E47 was provided by Gregory Kato (Johns Hopkins University, Baltimore, Md.), pcDNA3L-NLI was provided by Gordon Gill (University of California, San Diego), and pXM-GATA-1 was provided by Leonard Zon (Boston Childrens Hospital, Boston, Mass.). pcDNA3.1-Tal1 was described recently (52), and the analogous pcDNA3.1-E47 was constructed by subcloning an XbaI fragment containing E47 coding sequence from pCMV-E47 into the XbaI site of vector pcDNA3.1. Plasmid pcDNA3.1-GATA-1 was made by similarly transferring an XhoI fragment containing the GATA-1 coding sequence from pXM-GATA-1 into the XhoI site of pcDNA3.1. A full-length LMO2 cDNA was amplified by PCR and cloned into the unique EcoRI site of vector pEFIRES-N (18) to generate pEFIRES-LMO2. cDNAs encoding full-length Ldb1 and an N-terminal truncation mutant form, Ldb1200-375, were generated by PCR with plasmid pcDNA3L-NLI serving as the template and subcloned into the EcoRI and XbaI sites of vector pEFIRES-P (18). The retroviral vector MSCV (murine stem cell virus)-IRES-GFP (green fluorescent protein)-TAL1 was described previously (21).
Site-specific mutagenesis was carried out with the GeneEditor in vitro site-directed mutagenesis system under conditions recommended by the manufacturer (Promega, Madison, Wis.). Mutations were made in the pGL2-P4.2p1700-Luc reporter in the E1 E box (CATCTG to GATCTT), the E2 E box (CATGTG to GATGTT), and the G1 and G2 GATA sites (GATA to GCTC). A P4.2 promoter-reporter construct with mutations in both E box and GATA sites (E1G1-E2G2 mutant) was generated by sequential mutation of the E1, E2, G1, and G2 sites. A DNA binding-defective Tal1 mutant was generated by substitution of Pro for Thr192 in the protein's basic region. The sequences of all DNA constructs were confirmed by nucleotide sequencing, and their ability to direct the expression of proteins of the predicted size was verified by Western blot analysis.
Cell culture and transient transfections.
MEL cells (F4-12B2 line) were cultured and transfected with DMRIE-C reagent (Life Technologies, Rockville, Md.) as described previously (21). Briefly, 1 μg of the pGL2-P4.2p1700-Luc reporter or different mutant reporters, 100 ng of a Renilla luciferase vector (used as a transfection control), and 3 μg of plasmid pCMV4 (used as filler DNA) were cotransfected into MEL cells grown in six-well plates. COS-7L cells (Life Technologies, Rockville, Md.) cultured in Dulbecco's modified Eagle medium containing 10% fetal bovine serum and 0.1 mM nonessential amino acids were transfected with Lipofectamine 2000 as recommended by the manufacturer (Invitrogen, Carlsbad, Calif.). In brief, 1.5 μg of the pGL2-P4.2p1700-Luc reporter or the E1G1-E2G2 mutant reporter was cotransfected with the indicated combinations of pcDNA3.1-TAL1 (50 ng), pcDNA3.1-E47 (1.5 ng), pcDNA3.1-GATA-1 (12.5 ng), pEFIRES-LMO2 (40 ng), pEFIRES-Ldb1 or pEFIRES-Ldb1200-375 (40 ng), and Renilla luciferase vector (5 ng) into COS-7L cells grown in poly-d-lysine treated 12-well plates (Becton Dickinson, Bedford, Mass.). All extracts were prepared 48 h after transfection, and luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Promega, Madison, Wis.), with the reporter activities normalized to Renilla luciferase activity. Each transfection was done in triplicate and repeated three or more times.
For studies of in vivo assembly of the ternary complex, expression vectors described above for TAL1 (5 μg), E47 (0.15 μg), GATA-1 (1.25 μg), LMO2 (4 μg), and Ldb1 or Ldb1200-375 (4 μg) were cotransfected into COS-7L cells cultured in 10-cm-diameter dishes. The total mass of DNA applied was adjusted to 15 μg with plasmid pCMV4. Nuclear extracts were prepared 48 h after transfection and analyzed as described.
Preparation of stably transduced cells.
Infectious retrovirus was produced by Lipofectamine-mediated transfection of φNX-Ampho cells and frozen in aliquots at −70°C. Retroviral infection of MEL cells was carried out as described previously (21). GFP-expressing transductants were isolated by fluorescence-activated cell sorting and then expanded in culture. Full-length Ldb1 and Ldb1200-375 cDNAs were introduced into MEL cells in the expression plasmid pEFIRES-P with DMRIE-C as described above. Cells were selected in culture medium containing 2 μg of puromycin per ml, beginning 48 h after transfection. The concentration of puromycin was increased to 10 μg/ml 5 days later and maintained.
EMSA.
Preparation of nuclear extracts and electrophoretic mobility shift analysis (EMSA) of DNA-binding activity were carried out as previously described (21). In brief, 5 μg of MEL cell nuclear extract was incubated with 2 nM 32P-labeled double-stranded oligonucleotide for 25 min at room temperature. Where indicated, 1 μl of the relevant antibody was also added. Rabbit polyclonal antibodies against Tal1 (24) and Ldb1 (58) have been described previously. In addition, antibodies to E47 (sc-763X), GATA-1 (sc-265X), and LMO2 (sc-10498X) and normal rabbit immunoglobulin G (IgG; sc-2027) were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Protein-DNA complexes were electrophoresed in 4% native polyacrylamide gels in Tris-glycine buffer at 60 V for 12 to 15 h at 4°C. After the gels were dried, DNA-protein complexes were visualized by autoradiography. For clarity, only the relevant portions of these autoradiograms are shown in the figures. The sense strands of the oligonucleotides used in the EMSA are as follows (the E box and GATA sites are underlined, and mutant nucleotides are in lowercase): wild-type E1G1, ATTTCCTTATCTCGTTCAAACAGATGGTTTCCT; E1 mutant, ATTTCCTTATCTCGTTCAAAaAGATcGTTTCCT; G1 mutant, ATTTCCTgAgCTCGTTCAAACAGATGGTTTCCT; E1G1 double mutant, ATTTCCTgAgCTCGTTCAAAaAGATcGTTTCCT; wild-type E2G2, CTCCCAGCAGCTGGCCTAGGAGATAGCAGCAG; E2 mutant, CTCCCAGgAGCTtGCCTAGGAGATAGCAGCAG; G2 mutant, CTCCCAGCAGCTGGCCTAGGAGcTcGCAGCAG; E2G2 double mutant, CTCCCAGgAGCTtGCCTAGGAGcTcGCAGCAG.
DNA-linking assay.
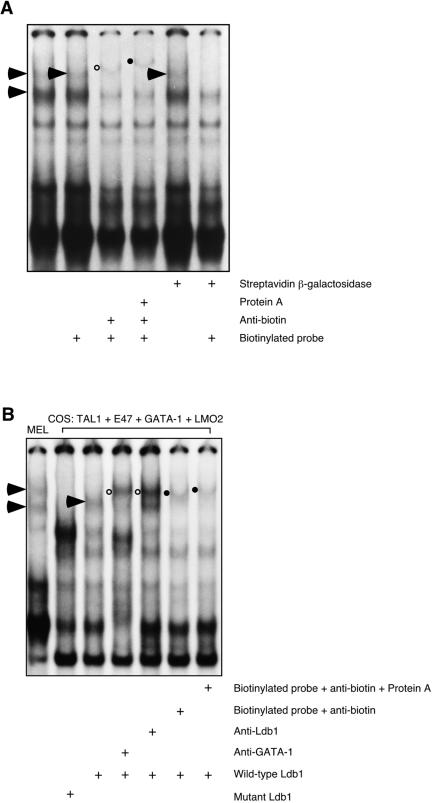
Five micrograms of nuclear extract was incubated with 2 nM 32P-labeled double-stranded E2G2 oligonucleotide at room temperature for 5 min, 5′-biotinylated double-stranded E1G1 oligonucleotide was added at a concentration of 4 nM, and the reaction was allowed to continue for an additional 15 min. Where noted, 2 μg of antibiotin antibody and protein A (Sigma) or 2.5 U of streptavidin-conjugated β-galactosidase (Roche) was added subsequently and the mixture was incubated for 10 min. Protein-DNA complexes were resolved and visualized as described above for a standard EMSA.
Western blot analysis.
Western blot analysis was performed as previously described (21), and the antibodies used are detailed above.
ChIP analysis.
ChIP analysis was carried out with a commercial kit (Upstate Biotechnology) with some modifications to the manufacturer's recommended conditions. Briefly, 1% formaldehyde was added at 37°C for 20 min to 1.0 × 108 MEL cells cultured for 3 days with or without 1.5% dimethyl sulfoxide (DMSO). Cell pellets were incubated in 10 mM HEPES (pH 7.9)-1.5 mM MgCl2-10 mM KCl-0.5 mM dithiothreitol-protease inhibitors for 10 min on ice and homogenized. The resulting nuclear pellet was suspended in 1 ml of sodium dodecyl sulfate lysis buffer, incubated for 10 min on ice, and sonicated sufficiently to shear the DNA to an average size of 500 to 1,000 bp. Following sonication chromatin was diluted 10-fold with ChIP dilution buffer and precleared with 80 μl of protein A-agarose. Precleared chromatin from 1.0 × 107 cells was then incubated with the indicated antibodies with rotation at 4°C overnight. The remaining procedures leading to purified DNA were performed in accordance with the manufacturer's protocol. Purified DNA was resuspended in 20 μl of H2O and analyzed by PCR. DNA for the input control was diluted 1:100 before PCR. Reactions were carried out in a volume of 25 μl, with initial denaturation at 94°C for 5 min, followed by 29, 31, or 33 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 1 min, and extension at 72°C for 40 s, followed by a 5-min terminal extension at 72°C. For amplification of 3′ untranslated sequences, 31 cycles were used. PCR products were resolved on 1.5% agarose gels containing ethidium bromide. The sequences of the primers used in these reaction mixtures were GCCCCAAACAATTCTAA and CCCCTTACTCCTCCTGTGA for the proximal promoter and TCCCAAACAACCCTCAACCG and CCTGGACCAACAGCACTACATAGAG for the 3′ untranslated region (UTR).
Northern blot analysis.
Northern blot analysis was performed as previously described (21). A 1.2-kb murine β-globin cDNA fragment, a 1.4-kb murine glyceraldehyde phosphate dehydrogenase cDNA, a 2.2-kb murine P4.2 cDNA, and a 1.1-kb murine Ldb1 cDNA were radiolabeled with 32P by random primer extension (Life Technologies). Purified total cellular RNA (20 μg) was fractionated in formaldehyde-agarose gels and transferred by capillary action to a nylon membrane. The blotted membranes were hybridized and washed as previously described (21).
Autoradiographic analysis.
Band intensities on photographic film were quantitated with the National Institutes of Health Image software package (version 1.5).
RESULTS
Identification and characterization of a P4.2 promoter DNA-binding complex.
Site selection (58) and chromatin immunoprecipitation (10) analyses identified a bipartite E box GATA element as the preferred binding site for a TAL1- and GATA-1-containing complex. As a sequence motif of this length should occur only infrequently by chance, a candidate gene approach was taken to define TAL1-regulated genes. From inspection of the regulatory sequences of erythroid cell-expressed genes, one potential target, the murine P4.2 gene, was selected for closer examination. We noted, first, that two E box GATA elements were present in the murine P4.2 promoter at bases −318 to −336 and −25 to −42 (25), numbered from the major transcriptional initiation site and designated here E1G1 and E2G2, respectively. The E2G2 site is identical to and the E1G1 site closely resembles the consensus sequence for the bipartite element identified by Wadman et al. (58). Further, P4.2 expression closely overlaps that of Tal1 at sites of erythropoiesis in embryonic and adult mice (63). Finally, the region of the P4.2 promoter containing these E box GATA elements was shown to be important in activating transcription of the gene during MEL cell differentiation (25).
To investigate whether it was a bona fide TAL1 target, we first used EMSA to characterize the composition, specificity, and affinity of protein complexes in MEL cell nuclear extracts for the two E box GATA elements in the P4.2 promoter. As described by Wadman et al. for a consensus E box GATA-binding element (58), incubation of nuclear extract with 32P-labeled probes led to the appearance of at least two highly retarded protein-DNA complexes (Fig. 1A and B), although the more retarded complex was evident with the E2G2 probe only after long autoradiographic exposures (Fig. 1C; see Fig. 2A). The overall abundance of these DNA-binding complexes was significantly higher for the E1G1 element than for the E2G2 element (compare Fig. 1A and B), consistent with a greater affinity of the binding complex for the E1G1 E box GATA sequence. Addition of antibodies to Tal1, E47, GATA-1, LMO2, or Ldb1, but not normal rabbit IgG, either eliminated or supershifted both complexes (Fig. 1A to C), demonstrating a contribution from each of these proteins. For the E1G1 element, neither complex was observed when probes with mutations in either or both E box and GATA sequences were used (Fig. 1D), indicating a requirement for both sites in complex formation. In complementary studies, these same retarded complexes were competed by a 50-fold excess of unlabeled oligonucleotide of the wild-type E1G1 sequence or by oligonucleotides containing a mutation in either the E box (E box mutant) or the GATA site (GATA mutant) but not by an oligonucleotide in which both E box and GATA sites were mutated (double mutant). In contrast, another, less retarded DNA-protein complex (Fig. 1A and E) that was supershifted only by an antibody to GATA-1 was eliminated by unlabeled competitors containing an intact GATA site (wild type and E box mutant) but not by an oligonucleotide in which the E box was intact and the GATA site was mutated (GATA mutant), indicating that it contained GATA-1 alone. Quantitation of the binding activity remaining after addition of increasing concentrations of unlabeled competitor (Fig. 1F) indicated that the two ternary complexes had a greater affinity for this E box GATA element than the other, more abundant GATA-binding activity in MEL cell extracts. Together, these data demonstrate that ternary complexes containing TAL1/E47, GATA-1, and LMO2/Ldb1 bind specifically to the two E box GATA elements in the murine P4.2 promoter and that assembly of these complexes requires both DNA-binding motifs.
FIG. 1.
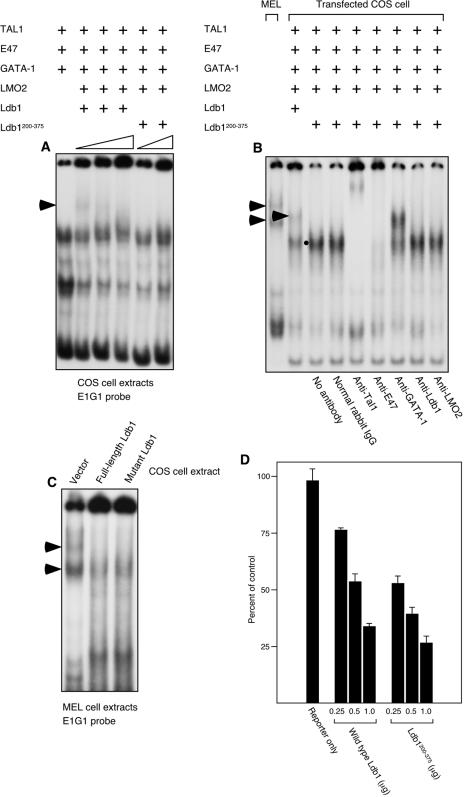
Characterization of TAL1-containing ternary complexes binding to two E box GATA elements in the murine P4.2 promoter. These elements were designated, from 5′ to 3′, E1G1 and E2G2. 32P-labeled E1G1 (A) and E2G2 (B, C) probes were incubated with 5 μg of nuclear extract from undifferentiated MEL cells. As indicated, antibodies to Tal1, GATA-1, Ldb1, LMO2, E47, or normal rabbit IgG were used in supershift analysis. Ternary complexes are marked with arrowheads. (D) Five micrograms of MEL cell nuclear extract was incubated with the radiolabeled wild-type probe or the indicated mutant E1G1 probe (labeled probe). Radiolabeled wild-type E1G1 probe was also mixed with a 50- or 100-fold molar excess of the unlabeled wild-type or mutant oligonucleotide (cold [unlabeled] competitor). (E) Stability of E1G1 DNA-binding complexes was assessed by competition with 2-, 4-, 8- and 16-fold (left to right) molar excesses of the unlabeled wild-type E1G1 oligonucleotide. (F) Quantitation of autoradiographic intensities from panel E. Symbols correspond to the denoted binding complexes in panel E. Arrowheads, TAL1-containing multiprotein complexes; filled circles, GATA-1-containing complex.
FIG. 2.
Characterization of Ldb1-mediated DNA-linking activity. (A) Five micrograms of MEL cell nuclear extract was incubated with 2 nM 32P-labeled double-stranded E2G2 oligonucleotide at room temperature for 5 min. Biotin end-labeled E1G1 oligonucleotide (4 nM) was then added, and the mixture was incubated for 15 min. Finally, an antibody to biotin, singly or with protein A, or streptavidin-conjugated β-galactosidase was added and the mixture was incubated for an additional 10 min. A retarded protein-DNA complex supershifted by the biotin antibody (open circle) and supershifted further by infusion of protein A with the biotin antibody (filled circle) and abolished by streptavidin-conjugated β-galactosidase is predicted to contain radiolabeled and biotin-labeled oligonucleotides that became bridged in solution. (B) Results of DNA-linking assay carried out with 5 μg of nuclear extract from COS-7L cells transfected with expression vectors for Tal1, E47, GATA-1, LMO2, and either wild-type Ldb1 or the Ldb1200-375 truncation mutant. A retarded protein-DNA complex supershifted by antibodies to two of its constituent proteins (open circle), GATA-1 and Ldb1, and to biotin (filled circles) is predicted to contain radiolabeled and biotin-labeled oligonucleotides that became bridged in solution.
The TAL1- and Ldb1-1-containing ternary complex can link independent protein-DNA complexes in solution.
Given the ability of Ldb1 to homodimerize in vitro (7) and genetic data demonstrating a requirement for dimerization in the mediation of promoter-enhancer interactions by Drosophila Ldb1 (or Chip) (33, 54, 55), we considered the possibility that the more retarded ternary complex observed in EMSA resulted from interaction of two protein-DNA complexes in solution. To investigate, a novel electrophoretic binding method that we termed a DNA-linking assay was developed. In practice, MEL cell nuclear extract was incubated with a mixture of double-stranded, 32P-labeled E2G2 oligonucleotide and double-stranded, biotin-labeled E1G1 oligonucleotide. We reasoned that if the E1G1 and E2G2 probes became linked in solution, a retarded complex would form with a mobility similar to that of the upper complex in a standard EMSA but containing both radioactive and biotinylated oligonucleotides. As shown in Fig. 2A, incubation of MEL cell nuclear extract with an empirically determined mixture of radiolabeled E2G2 and biotinylated E1G1 oligonucleotides led to the formation of a single E box GATA-binding complex that was retarded by antibody to biotin (open circle) and supershifted to an even greater extent by inclusion of protein A with the antibody (filled circle). This protein-DNA complex was, in addition, ablated by incubation of extracts with streptavidin conjugated to β-galactosidase (Fig. 2A) or beads (data not shown). In contrast, the mobility of the lower complex was unaltered by these reagents. From these results we concluded that the upper complex included both biotinylated E1G1 oligonucleotide and radiolabeled E2G2 oligonucleotide (E1G1-E2G2 complex), while the less retarded of the two E box GATA-binding complexes contained only radiolabeled E2G2 probe. The absence of any radiolabeled E2G2-E2G2 complex after addition of E1G1 oligonucleotide, seen experimentally as the complete supershifting of the upper complex, was due, we surmise, to the formation of E1G1-E2G2 complexes being favored over E2G2-E2G2 complexes. Importantly, if biotinylated E1G1 oligonucleotide were not included in the probe mixture, the mobility of the upper complex was unaffected by addition of either streptavidin-β-galactosidase (Fig. 2A) or biotin antibody (data not shown), demonstrating the specificity of these results. Together, these data show that a multiprotein DNA-binding complex in MEL cell extracts is capable of interacting with individual E box GATA elements in the P4.2 promoter and linking them physically in solution. As the two E box GATA DNA-binding complexes detected in a standard EMSA increased in parallel with DMSO-induced differentiation (see below), at least at early times, an equilibrium between these structures may exist in cells.
To determine if the five proteins present in the complex were sufficient to bridge independent E box GATA elements in solution, we transiently transfected COS-7L cells with expression vectors for Tal1, E47, GATA-1, LMO2, and Ldb1 and prepared nuclear extracts for the DNA-linking assay as described. As observed previously for a consensus E box GATA-binding element (58), a retarded complex could not be detected unless all of the proteins were present (Fig. 2B). While at least two E box GATA-binding complexes were detected with MEL cell extracts, extracts from COS-7L cell transfectants gave rise to a single DNA-protein complex with intermediate mobility (single arrowhead, Fig. 2B). Nonetheless, this retarded complex was supershifted by antibodies to its component proteins, GATA-1 and Ldb1, for example (open circles, Fig. 2B), or by antibody to biotin (filled circles, Fig. 2B), confirming that it contained both radiolabeled and biotinylated probes. A complex containing E1G1 and E2G2 probes failed to form, however, when a dimerization-defective truncation mutant form of Ldb1 was expressed in place of the full-length protein (Fig. 2B), and this dimerization-defective form of Ldb1 also significantly decreased E box GATA DNA-binding activity when stably expressed in MEL cells (see below). These data thus account for the presence of two E box GATA DNA-binding complexes with apparently identical protein compositions but different electrophoretic mobilities in EMSA (Fig. 1) (58) and are compatible with the notion of Ldb1 dimerization acting to link or loop DNA.
E box GATA DNA-binding activity increases with DMSO-induced MEL cell differentiation.
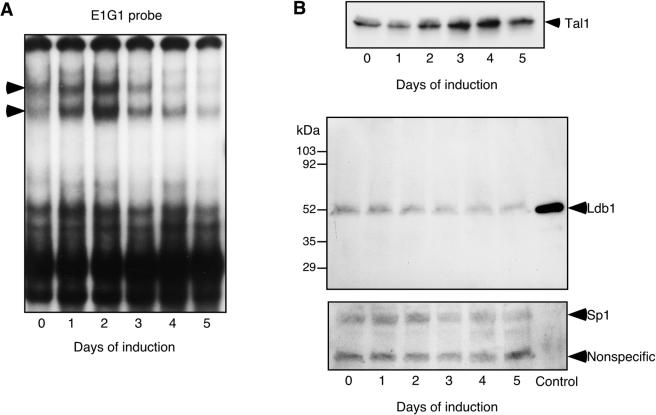
Expression of the mouse P4.2 gene is tightly regulated during erythroid differentiation (25) (see below). To investigate the relationship between activation of P4.2 transcription and E box GATA DNA-binding activity, an EMSA was carried out with the E1G1 oligonucleotide in MEL cells induced to differentiate by DMSO. DNA-binding activity was present in extracts from uninduced MEL cells, increased in the first 2 days after addition of DMSO, but ultimately declined with terminal differentiation (Fig. 3A). While E box GATA DNA-binding activity was detectable even before the gene was expressed, it increased in parallel with accumulation of P4.2 mRNA. To examine whether specific components of the ternary complex might be limiting for its assembly, Tal1 and Ldb1 protein levels were determined in differentiating MEL cells by immunoblot analysis. Although Ldb1 mRNA was reported to decrease with differentiation of murine erythroid cell lines (56), expression of Ldb1, like that of the transcription factor Sp1, was maintained in DMSO-treated MEL cells (Fig. 3B) and Friend virus-induced proerythroblasts (data not shown) until relatively late stages of differentiation. At early time points, the kinetics of neither Tal1 nor Ldb1 protein accumulation overlapped P4.2 promoter E box GATA DNA-binding activity. However, the late decrease in Ldb1 concentration paralleled and could potentially have contributed to the terminal decline in DNA-binding activity.
FIG. 3.
Time course of induction of P4.2 promoter E box GATA DNA-binding activity in DMSO-treated MEL cells. (A) E1G1 probe was incubated with 5 μg of nuclear extract from MEL cells treated with 1.5% DMSO for the indicated times. Ternary complexes are indicated by arrowheads. (B) Western blot analysis of Tal1 and Ldb1 protein expression in nuclear extracts from MEL cells treated with 1.5% DMSO for the indicated times. Western blot analysis of Sp1 expression was carried out on the stripped membrane as a control for protein loading. A much smaller amount of extract from Ldb1-transfected COS-7L cells (control) was used.
The E box GATA-binding complex occupies the P4.2 promoter in cells.
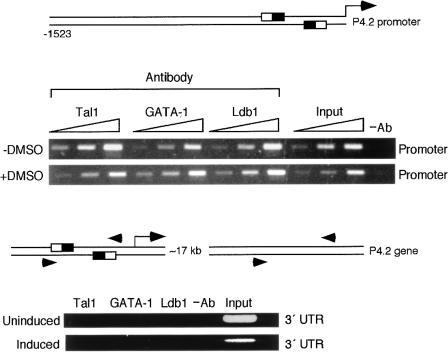
The above-described studies show only the potential for the TAL1-containing complex to bind E box GATA elements in the P4.2 promoter. To verify that the P4.2 promoter was actually occupied by the ternary complex in cells, ChIP analysis was carried out. To that end, MEL cells were cultured for 3 days in the presence or absence of 1.5% DMSO and treated with 1% formaldehyde to cross-link DNA and associated proteins. Their chromatin was then sonicated and the resulting fragments were subjected to immunoprecipitation with specific antibodies. After reversal of cross-links, DNA was extracted from the immunoprecipitated material and a PCR was performed with primers encompassing the two E box GATA motifs in the P4.2 promoter or a region 17 kb downstream of the P4.2 coding region (arrowheads). As demonstrated in Fig. 4, antibodies to Tal1, GATA-1, and Ldb1, but not buffer or normal rabbit immunoglobulin (data not shown), selected P4.2 promoter fragments from both uninduced and induced MEL cells. In contrast, chromatin derived from the gene's 3′ UTR was not immunoprecipitated by any antibody, indicating that only the promoter was occupied by the ternary complex in vivo. Since P4.2 is not expressed in uninduced MEL cells, at least at the sensitivity of Northern blot analysis, these data suggest that the ternary complex is assembled before the gene is transcribed and that its presence, while necessary, is not sufficient for transcription to proceed. This raises the possibility that additional factors are recruited to the promoter with differentiation, conceivably by the complex itself, or that constituents of the complex undergo posttranslational modification (see Discussion).
FIG. 4.
ChIP analysis of TAL1, GATA-1, and Ldb1 occupancy of the P4.2 promoter in cells. MEL cells were incubated with or without 1.5% DMSO for 3 days, and nuclear proteins were cross-linked to DNA by addition of formaldehyde to the culture. Chromatin fragments generated by sonication were immunoprecipitated with antibodies to Tal1, GATA-1, or Ldb1 or with normal rabbit IgG (data not shown) and then used for amplification of the indicated regions in the promoter encompassing the two E box GATA elements (top) or the gene's 3′ UTR (bottom). Ethidium bromide staining of the PCR product was carried out following 29, 31, and 33 cycles of amplification (left to right). Arrowheads denote the locations of the primers used in the PCR.
The TAL1- and GATA-1-containing complex transactivates the P4.2 promoter.
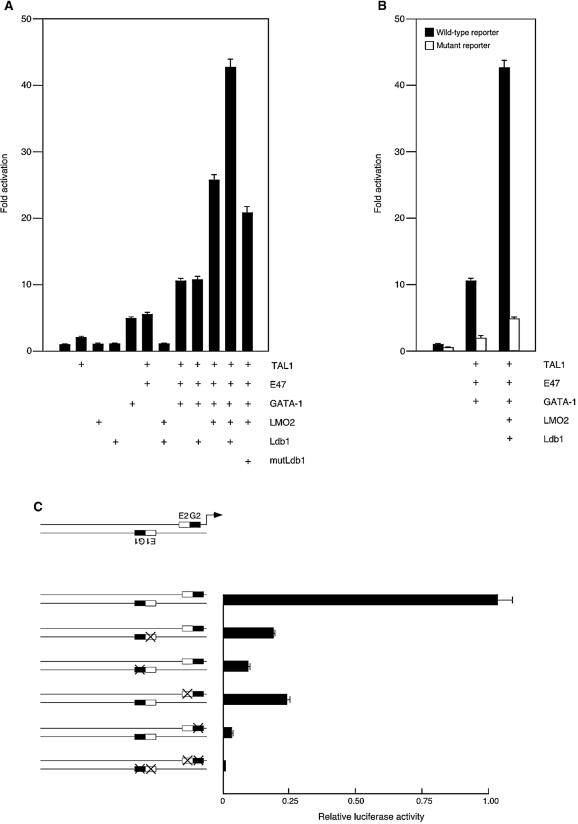
Having established that the multiprotein complex bound the P4.2 promoter in vitro and in vivo, we investigated whether it regulated the gene's transcription. To that end, a 1.7-kb fragment of the mouse P4.2 promoter linked to a luciferase reporter gene was cotransfected into COS-7L cells with expression plasmids for Tal1, E47, GATA-1, LMO2, and Ldb1. As shown in Fig. 5A, reporter activity was stimulated five-fold by GATA-1 or Tal1 plus E47, while LMO2 and Ldb1, singly or in combination, were without effect. When GATA-1 was coexpressed with Tal1 and E47, reporter activity increased 10-fold, for an approximately additive effect on reporter gene expression. Addition of LMO2 to Tal1, E47, and GATA-1 resulted in a 26-fold increase in reporter activity, likely reflecting LMO2-promoted stabilization of the bHLH heterodimer- and GATA-1-containing complexes. Maximal promoter activation, or a 42-fold induction of reporter activity, was achieved with cotransfection of all five members of the ternary complex. Importantly, Ldb1 did not enhance Tal1-, E47-, and GATA-1-stimulated transcription in the absence of cotransfected LMO2, supporting the notion that Ldb1's actions, and presumably its recruitment into the complex, require its interaction with the LIM domain of LMO2. Additionally, transfection of the dimerization-defective mutant form Ldb1200-375 (mutLdb1 in Fig. 5A) in place of full-length Ldb1 reduced reporter activity to a level below that observed with Tal1, E47, GATA-1, and LMO2. As Ldb1 is expressed in COS-7L cells (see Fig. 8B) and Ldb1200-375 retains the full capacity to interact with LIM domain proteins (22), this mutant form likely interfered with endogenously expressed Ldb1 (see below). Together, these results establish that TAL1, E47, GATA-1, LMO2, and Ldb1 act synergistically to promote P4.2 gene transcription.
FIG. 5.
Synergistic transactivation of the P4.2 promoter by ternary DNA-binding complex. (A) A luciferase reporter construct containing 1.7 kb of the P4.2 promoter was cotransfected into COS-7L cells with different combinations of expression plasmids encoding Tal1, E47, GATA-1, LMO2, and full-length Ldb1 or the Ldb1200-375 mutant form. Cell lysates were prepared 48 h after transfection and assayed for luciferase activity. Plotted is the mean induction in luciferase activity ± the standard error from three independent experiments. (B) Transactivation by the indicated expression vectors of the wild-type promoter and a 1.7-kb promoter construct with mutations in both E box and GATA sites (E1G1-E2G2) were compared by using the same conditions as for panel A. (C) Reporter constructs containing the wild-type P4.2 promoter reporter or the indicated promoter mutants were transfected into MEL cells that had been incubated with 1.5% DMSO for 24 h. Following transfection, cells were cultured in DMSO for an additional 48 h, cellular lysates were prepared, and luciferase activities were determined.
FIG. 8.
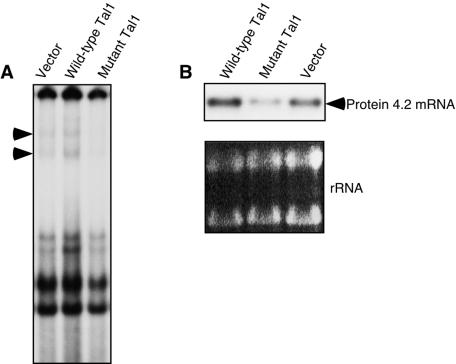
Effects of wild-type Ldb1 and a dimerization-defective mutant form on endogenous P4.2 gene expression and erythroid differentiation. (A) Northern blot analysis of 20 μg of total cellular RNA from corresponding MEL cell transductants. Membranes were sequentially hybridized with P4.2, β-globin, and Ldb1 cDNA probes. Ethidium bromide-stained 18S and 28S rRNAs served as loading controls. Coincidentally, the introduced RNAs migrated with the same mobility as two endogenous Ldb1 transcripts. (B) Western blot analysis of Ldb1 (upper) and Ldb1200-375 (lower) protein expression in nuclear extracts from transduced cells. Extracts from COS-7L cells transiently transfected with Ldb1 and Ldb1200-375 expression vectors were used as controls (transfected control). (C) EMSA with E1G1 probe and nuclear extract (5 μg) from the indicated MEL cell transductants. Both E box GATA DNA-binding complexes (arrowheads) were supershifted by antibody to GATA-1 (shown) and Ldb1 (not shown).
To investigate the requirement of specific DNA-binding sequences for the ternary complex's activity, we made mutations in the two E box GATA elements in the context of the full 1.7-kb promoter fragment. As shown in Fig. 5B, this compound mutant (E1G1-E2G2) was almost totally impaired in the ability to be transactivated by Tal1, E47, and GATA-1, with or without expression of LMO2 and Ldb1.
Finally, to examine in a more relevant cellular environment whether the E box GATA elements were required for P4.2 promoter activity, promoter-luciferase reporter constructs containing individual or compound site mutations were transiently transfected into MEL cells, which were then cultured for 2 additional days in the presence or absence of 1.5% DMSO. Confirming the previous report (25), 1.7 kb of proximal promoter conferred high-level, differentiation-dependent reporter activity in DMSO-treated cells, with little to no activity detected in cells cultured without DMSO (data not shown). Compared to the wild-type construct, mutation of either E box (E1 or E2) decreased promoter activity approximately 75%, while mutation of the G1 or G2 GATA site reduced activity 90 or 95%, respectively (Fig. 5C). In accord with the data obtained with COS-7L cells, reporter activity was essentially abolished by mutation of all E box and GATA sites. In sum, these data show that the inducibility of the P4.2 promoter by the TAL1-containing ternary complex is dependent on the integrity of both E box GATA elements. Further, the greater-than-additive contribution made by individual elements to promoter activity (Fig. 5C) and the supra-additive stimulation of reporter activity with expression of individual activators (Fig. 5A) provide evidence for transcriptional synergy.
TAL1 DNA-binding activity is required for P4.2 gene expression.
Although dispensable for its actions in hematopoietic stem cell development (40), DNA binding is required for TAL1 function in erythroid differentiation (4, 40). Given the requirement for the E box sequences in both formation of the E box GATA-binding complex and transactivation of the 1.7-kb P4.2 promoter-reporter construct, we hypothesized that TAL1 DNA-binding activity would be important for P4.2 gene expression in cells. To investigate, cDNAs encoding full-length Tal1 or a mutant protein, Tal1T192P, containing a proline substitution in its basic region and established previously as a potent trans-dominant inhibitor of wild-type TAL1 DNA binding (S. Huang and S. J. Brandt, unpublished data), were introduced into MEL cells with the MSCV-IRES-GFP retroviral vector (38). Polyclonal populations of cells transduced with these cDNAs or the parental vector were selected by fluorescence-activated cell sorting, and GFP-expressing cells were cultured for 5 days in the presence or absence of 1.5% DMSO. Nuclear extracts and total cellular RNA were then prepared, and DNA-binding activity and endogenous P4.2 gene expression were examined. When normalized to an anonymous complex whose abundance did not differ between conditions, enforced expression of wild-type Tal1 increased the two E box GATA DNA-binding complexes 33 and 74% relative to that of the vector control, while the dominant negative Tal1 mutant decreased these complexes by 64 and 56% (Fig. 6A). In association, steady-state P4.2 mRNA levels increased slightly in Tal1-overexpressing cells while being reduced almost 70% in cells transduced with the binding-defective mutant (Fig. 6B). These data indicate that DNA binding is required for TAL1 regulation of P4.2 gene expression.
FIG. 6.
Effects of wild-type Tal1 or the DNA binding-defective Tal1T192P mutant form on P4.2 promoter E box GATA DNA-binding activity and endogenous P4.2 gene expression. (A) EMSA was performed with a 32P-labeled E1G1 probe and 5 μg of nuclear extract from transduced MEL cells as indicated. Arrowheads, TAL1-containing multiprotein complexes. (B) Northern blot analysis of 20 μg of total cellular RNA from corresponding MEL cell transductants. Ethidium bromide-stained 18S and 28S rRNAs served as loading controls.
Overexpression of Ldb1 inhibits E box GATA DNA-binding activity and P4.2 promoter function.
Enforced expression of Ldb1 and Lmo2 was shown previously to inhibit the differentiation of erythroid cell lines (56), implying a negative role for these proteins in terminal differentiation. For Chip, however, enforced expression of wild-type protein or a dimerization-defective mutant form analogous to the one used in our studies phenocopied a loss-of-function allele (15, 31, 54). This raised the possibility of Ldb1 acting physiologically to promote erythroid differentiation. To investigate the effect of overexpressed Ldb1 on E box GATA DNA-binding activity, both the dimerization-defective mutant, Ldb1200-375, and wild-type Ldb1 were transfected into COS-7L cells together with cDNAs for the other components of the binding complex. As described above, only a single DNA-binding complex with a mobility intermediate between those observed with MEL cell nuclear extract was apparent in an EMSA. Although LMO2 and Ldb1 were clearly necessary for this retarded complex to form (compare lanes 1 and 2, Fig. 7A), both Ldb1 and the dominant negative mutant form inhibited complex formation in a dose-related fashion when overexpressed (Fig. 7A). The effect of Ldb1200-375 on E box GATA DNA-binding activity was also investigated with extracts from transfected COS-7L cells. Transfection of an Ldb1200-375 cDNA inhibited formation of this complex while enhancing that of a less retarded complex (filled circle, Fig. 7B), which, from supershift analysis, comprised only TAL1/E47 heterodimers. Significantly, no DNA-binding complex was recognized that appeared to incorporate the truncated Ldb1 protein. Further, preincubation of MEL cell nuclear extracts with Ldb1 or Ldb1200-375 prepared in COS-7L cells inhibited the formation of both of the retarded complexes (Fig. 7C). Finally, overexpression of Ldb1 or Ldb1200-375 in MEL cells effected a dose-related inhibition of DMSO-induced P4.2 promoter activity (Fig. 7D). Together, these results show that overexpression of Ldb1 inhibits E box GATA DNA binding and, as a consequence, P4.2 promoter function.
FIG. 7.
Effects of wild-type Ldb1 and a dimerization-defective mutant form on P4.2 promoter E box GATA DNA-binding activity and P4.2 promoter activity. (A) An EMSA was carried out with a 32P-labeled E1G1 probe and 5 μg of nuclear extract from COS-7L cells transfected with expression vectors for Tal1, E47, GATA-1, and LMO2 and increasing concentrations of wild-type Ldb1 or Ldb1200-375. The ternary complex is marked with an arrowhead. (B) An EMSA was carried out with a 32P-labeled E1G1 probe and 5 μg of nuclear extract from MEL and COS-7L cells transfected with expression vectors for Tal1, E47, GATA-1, and LMO2 and wild-type Ldb1 or Ldb1200-375. As indicated, antibodies to Tal1, GATA-1, Ldb1, LMO2, or E47 or normal rabbit IgG were added to the reaction mixtures. Ternary complexes are marked with arrowheads, and the Tal1/E47 complex deduced from supershift analysis is marked with a filled circle. (C) An EMSA was carried out with a 32P-labeled E1G1 probeand 5 μg of MEL cell nuclear extract that had been preincubated with 5 μg of nuclear extract from COS-7L cells transfected with an expression vector for Ldb1 or Ldb1200-375. Ternary complexes are marked with arrowheads. (D) A luciferase reporter construct (0.25 μg) containing 1.7 kb of P4.2 promoter was cotransfected with the indicated amounts of expression plasmids into MEL cells that had been incubated with 1.5% DMSO for 24 h. Following transfection, cells were cultured in DMSO for an additional 48 h, cellular lysates were prepared, and luciferase activities were determined. Plotted is luciferase activity relative to the reporter control ± the standard error from three independent experiments.
Overexpression of Ldb1 inhibits P4.2 gene expression and erythroid differentiation.
To investigate the effects of enforced expression of Ldb1 and Ldb1200-375 on endogenous P4.2 gene expression, a plasmid vector containing the strong EF1α promoter (18) was used to introduce their respective cDNAs into MEL cells. Polyclonal populations of transduced cells were then incubated with or without 1.5% DMSO for 5 days, and benzidine staining for hemoglobin expression, Northern blot analysis of P4.2 and β-globin expression, and EMSA of E box GATA DNA-binding activity were carried out. To maximize expression of Ldb1 protein, which was encoded with a drug resistance gene from a bicistronic transcript, high concentrations of the selecting agent, puromycin, were used (18). Despite an abundance of message (Fig. 8A), however, the transfected proteins were expressed at very low levels in independently prepared populations (Fig. 8B). While use of polyclonal populations could have led to underrepresentation of high expressers, more likely these cells were unable to tolerate increased levels of Ldb1 and Ldb1200-375. Nevertheless, both the proportion of benzidine-stained cells and the extent of morphological differentiation were significantly reduced in wild-type Ldb1 and to an even greater extent in Ldb1200-375 transductants (data not shown). Overexpression of these Ldb1 proteins also dramatically reduced steady-state levels of P4.2 and β-globin mRNAs in DMSO-induced cells (Fig. 8A). Finally, in accord with the in vitro results described above, stable expression of wild-type Ldb1 and the Ldb1200-375 mutant form decreased endogenous E box GATA DNA-binding activities (Fig. 8C). Thus, analogous to the effects of Chip on wing development (15, 31, 44, 54), overexpression of Ldb1 phenocopied a loss-of-function mutation.
DISCUSSION
The importance of the bHLH transcription factor TAL1 has been clearly established for primitive and definitive erythropoiesis. The identities of the genes under its control and the specific mechanisms by which it regulates transcription, however, remain largely unknown. We establish here that the murine P4.2 gene is a target of a multiprotein complex in erythroid progenitors containing TAL1, GATA-1, LMO2, and Ldb1. Our studies show that this complex synergistically activates P4.2 transcription and positively regulates erythroid gene expression and differentiation.
A major limitation to the analysis of TAL1's transcriptional properties has been an incomplete knowledge of its target genes. While site selection assays have defined a bipartite E box GATA element as the preferred binding site for a TAL1- and GATA-1-containing ternary complex in erythroid cells (58) and a gene containing this motif was found to be occupied by TAL1 in cells (10), TAL1 DNA binding was dispensable for transactivation of the c-kit promoter by a TAL1-, E47-, LMO2-, GATA-1/2-, and Ldb1-containing complex (26) and a cryptic promoter in the retinaldehyde dehydrogenase 2 gene by a TAL1-, LMO1-, and GATA-3-containing complex (36). In contrast, we found that two E box GATA elements in the P4.2 promoter bound the pentameric complex in vitro, were occupied by at least three members of the complex in vivo, and were required for synergistic transactivation of the promoter in transfected cells. Furthermore, E box GATA DNA-binding activity increased with DMSO-induced differentiation of MEL cells, paralleling P4.2 gene expression. Finally, overexpression of a full-length Tal1 cDNA enhanced endogenous P4.2 expression in differentiating MEL cells, while a DNA binding-defective mutant significantly inhibited its expression. Together, these studies provide compelling evidence for the P4.2 gene as a direct target of TAL1 in differentiating erythroid progenitors.
P4.2 is present in a complex with band 3 and ankyrin that is essential for the integrity of the red blood cell membrane (9). Humans with inherited mutations in this gene and mice homozygous for a targeted mutation of the locus exhibit defective erythrocyte ion transport and spherocytic anemia (11, 16, 39). P4.2 is an abundant protein, with an estimated 200,000 copies per red cell (6), and this requirement for high-level expression may explain its regulation by multiple transcription factors. The strict order of the two binding sites in the E box GATA element (58), the greater stability of the pentameric protein complex compared to the major GATA-binding activity in the cell (Fig. 1), and the ability of its component proteins to synergistically stimulate transcription are all consistent with this nuclear complex acting as an enhanceosome (8). The presence of Ldb1, with its apparent ability to alter higher-order DNA structure, is particularly reminiscent of the contribution of high-molecular-weight group proteins to enhanceosome function (13, 62). Similar to the T-cell receptor α enhanceosome (50), this E box GATA-binding complex appears to assemble on the P4.2 promoter prior to initiation of transcription. As the complex is necessary but not sufficient for P4.2 transcription, additional proteins are likely recruited to a specific activation surface formed with its assembly. These could include histone acetyltransferases p300 (29) and P/CAF (1), which we showed interact with TAL1 in differentiating MEL cells (20, 21), ATP-dependent chromatin remodeling proteins, or components of the basal transcriptional machinery.
Identification of the P4.2 promoter as a target of a multimeric TAL1-containing complex led us to investigate the actions of its other constituent proteins, and Ldb1 in particular. First isolated as a LIM domain-interacting protein (2, 5, 23), Ldb1, also known as NLI and CLIM-2, binds all nuclear LIM proteins, including LIM homeodomain (33, 54) and LIM-only (LMO) (32, 51) proteins, and several homeodomain transcription factors lacking a LIM domain (53). Although not capable itself of binding DNA, Ldb1 potentiates the activity of a number of sequence-specific DNA-binding proteins (5, 7, 53), most notably the LIM homeobox protein Apterous (54). A self-dimerization domain at its amino terminus and a carboxy-terminal LIM-interacting domain (7, 22) are essential for protein function (55), and a requirement for homodimerization exists for both its transcriptional (7) and biological (54, 55) actions. Ldb1 homodimers have been proposed to mediate physical interaction, sometimes over a very long distance, between promoters and enhancers (12, 33, 34).
Ldb1 was found by Rabbitts and colleagues to contribute to a TAL1- and GATA-1-containing complex capable of stimulating transcription from a model promoter with multiple copies of its consensus DNA-binding site (58). In an analogous manner, Chip activates the achaete-scute proneural complex by bridging the GATA transcription factor Pannier to a heterodimer of the HLH proteins Achaete (Ac)/Scute (Sc) and Daughterless (43). Through the use of a novel modification of the gel mobility shift assay, we have established that Ldb1 can mediate the interaction in solution of two independent protein-DNA complexes, potentially simulating its action in cells in which elements colinear in DNA might be brought into association (Fig. 9). Ldb1-mediated looping of the P4.2 promoter is not established by these data, however, as no technique short of electron or atomic force microscopy (27, 48) is capable of demonstrating looping over the relatively short distance involved, and considerable effort to apply our assay to circular substrates proved unsuccessful. Further, while the functional interaction between the E1G1 and E2G2 sites in the P4.2 promoter (Fig. 5C) is compatible with their physical interaction, since a specific inhibitor of Ldb1 dimerization is not available and the dimerization-defective Ldb1 mutant form Ldb1200-375 was apparently not incorporated into the TAL1- and GATA-1-containing complex (Fig. 7), a requirement for Ldb1 dimerization in the transactivation of the P4.2 promoter remains to be proven.
FIG. 9.
DNA-looping model of Ldb1 action on the P4.2 promoter. GATA-1 binds to the GATA sites in both E box GATA elements of the promoter and initiates DNA bending. Simultaneous or subsequent binding of TAL1/E47 heterodimers to the adjacent E boxes leads to recruitment of the LIM domain protein LMO2 and LIM domain-binding protein Ldb1. A higher-order DNA structure would then be formed through Ldb1 dimerization. This would enhance the stability of the multiprotein complex on DNA, mediate physical interaction of the two E box GATA elements, and, ultimately, promote transcription. Although a molecule of LMO2 is shown simultaneously contacting one TAL1/E47 heterodimer and one molecule of GATA-1, the exact stoichiometry of the proteins in this complex has not been established.
These limitations notwithstanding, the results of our DNA-linking analysis have specific implications for the function of bHLH-GATA-LMO DNA-binding complexes. While one molecule of Ldb1 interacts with a single LIM homeoprotein molecule (22), some models of its function in the erythroid E box GATA-binding complex have Ldb1 dimerization occurring within rather than between such complexes (42). Although attempts to define the stoichiometry of this complex in solution by chemical cross-linking proved unsuccessful, likely because it only assembles on DNA, our data are at least compatible with Ldb1 homodimerization mediating looping between two E box GATA elements. For other promoters containing a single such element, Ldb1 might link a promoter-associated E box GATA-binding complex to complexes on distal enhancers containing an LMO protein or other Ldb1-interacting partners. Indeed, Chip has been proposed to link Pannier bound to GATA sites in the dorsocentral enhancer to Ac/Sc-Da heterodimers on the achaete and scute promoters located 4 and 30 kb away, respectively (43).
Current understanding of the biological functions of Ldb1 comes largely from studies of the Drosophila protein Chip, which promotes embryonic segmentation (33), neuronal differentiation (55), and wing morphogenesis (54). Unexpectedly, given the well-documented actions of TAL1 and GATA-1 in erythropoiesis, Orkin and colleagues suggested that Ldb1 and LMO2 are negative regulators of erythroid differentiation (56). This conclusion was based in large part on the inhibition of erythroid differentiation by overexpressed Ldb1 or LMO2. Although Ldb1 and LMO2 transcripts were noted to decrease in differentiating G1ER cells (56), the abundance of their protein products and the DNA-binding complexes to which they contribute were not examined. In our studies, in contrast, Ldb1 protein did not decline until late stages of erythroid differentiation and P4.2 promoter E box GATA DNA-binding activity actually increased with MEL cell differentiation. Although it can act as a repressor (22), we believe that a more likely explanation for the inhibition of erythroid differentiation by overexpressed wild-type Ldb1, which was confirmed with an expression vector very similar to that of Visvader et al. (56), is titration of LMO2, for which Ldb1 has significant affinity in solution (22). Whatever the mechanism, overexpression of wild-type Ldb1 and Ldb1200-375 effected similar reductions in E box GATA DNA-binding activity (Fig. 7), P4.2 promoter activity (Fig. 7), and endogenous P4.2 gene expression (Fig. 8). Overexpression of wild-type Ldb1 thus mimicked the actions of a bona fide loss-of-function mutant, similar to what was described for Chip in the developing wing (15, 31, 44, 54). It may not be necessary, then, to have different, indeed opposing, functions for this Ldb1-containing complex at different stages of erythroid differentiation, and we propose that Ldb1 acts physiologically to stimulate the expression of genes that are required for or, like P4.2, are associated with terminal differentiation.
Although not examined directly, this model could also explain how LMO2 might promote differentiation at physiological levels and yet inhibit the process when overexpressed. In fact, enforced expression of the Drosophila LMO protein dLMO is thought to inhibit Apterous function through competition for limiting amounts of Chip (31, 32, 60), and a similar mechanism has been suggested for the effects of overexpressed LMO proteins on wing development and T-cell leukemogenesis (32). Cells contain low levels of Ldb1 and appear not to tolerate its overexpression, implying that it is regulated over a narrow range of concentrations, if at all, and underscoring the importance of proper stoichiometry for productive protein-protein interactions and, ultimately, biological activity.
Acknowledgments
We thank Stephen Hobbs, Gregory Kato, Gordon Gill, and Leonard Zon for materials.
This work was supported in part by National Institutes of Health grants R01 HL49118 (to S.J.B.) and R03 MH61406 (to L.-S.C.), a Merit Review Award from the Department of Veterans Affairs (to S.J.B.), and a Hope Street Kids Foundation fellowship award (to Z.X.).
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and T. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Agulnick, A. D., M. Taira, J. J. Breen, T. Tanaka, I. B. Dawid, and H. Westphal. 1996. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 384:270-272. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, K. P., S. C. Crable, and J. B. Lingrel. 1998. Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Krüppel-like factor (EKLF) gene. J. Biol. Chem. 273:14347-14354. [DOI] [PubMed] [Google Scholar]
- 4.Aplan, P. D., K. Nakahara, S. H. Orkin, and I. R. Kirsch. 1992. The SCL gene product: a positive regulator of erythroid differentiation. EMBO J. 11:4073-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bach, I., C. Carrière, H. P. Ostendorff, B. Andersen, and M. G. Rosenfeld. 1997. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 11:1370-1380. [DOI] [PubMed] [Google Scholar]
- 6.Branton, D., C. M. Cohen, and J. Tyler. 1981. Interaction of cytoskeletal proteins on the human erythrocyte membrane. Cell 24:24-32. [DOI] [PubMed] [Google Scholar]
- 7.Breen, J. J., A. D. Agulnick, H. Westphal, and I. B. Dawid. 1998. Interactions between LIM domains and the LIM domain-binding protein Ldb1. J. Biol. Chem. 273:4712-4717. [DOI] [PubMed] [Google Scholar]
- 8.Carey, M. 1998. The enhanceosome and transcriptional synergy. Cell 92:5-8. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, C. M., E. Dotimas, and C. Korsgren. 1993. Human erythrocyte membrane protein band 4.2 (pallidin). Semin. Hematol. 30:119-137. [PubMed] [Google Scholar]
- 10.Cohen-Kaminsky, S., L. Maouche-Chrétien, L. Vitelli, M. A. Vinit, I. Blanchard, M. Yamamoto, C. Peschle, and P.-H. Roméo. 1998. Chromatin immunoselection defines a TAL-1 target gene. EMBO J. 17:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Franceschi, L., O. Olivieri, E. Miraglia del Giudice, S. Perrotta, V. Sabato, R. Corrocher, and A. Iolascon. 1997. Membrane cation and anion transport activities in erythrocytes of hereditary spherocytosis: effects of different membrane protein defects. Am. J. Hematol. 55:121-128. [DOI] [PubMed] [Google Scholar]
- 12.Dorsett, D. 1999. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 9:505-514. [DOI] [PubMed] [Google Scholar]
- 13.Ellwood, K. B., Y. M. Yen, R. C. Johnson, and M. Carey. 2000. Mechanism for specificity by HMG-1 in enhanceosome assembly. Mol. Cell. Biol. 20:4359-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elwood, N. J., H. Zogos, D. S. Pereira, J. E. Dick, and C. G. Begley. 1998. Enhanced megakaryocyte and erythroid development from normal human CD34+ cells: consequence of enforced expression of SCL. Blood 91:3756-3765. [PubMed] [Google Scholar]
- 15.Fernández-Fúnez, P., C. H. Lu, D. E. Rincón-Limas, A. García-Bellido, and J. Botas. 1998. The relative expression amounts of apterous and its co-factor dLdb/Chip are critical for dorso-ventral compartmentalization in the Drosophila wing. EMBO J. 17:6846-6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallagher, P. G., and B. G. Forget. 1997. Hematologically important mutations: band 3 and protein 4.2 variants in hereditary spherocytosis. Blood Cells Mol. Dis. 23:417-421. [DOI] [PubMed] [Google Scholar]
- 17.Hall, M. A., D. J. Curtis, D. Metcalf, A. G. Elefanty, K. Sourris, L. Robb, J. R. Göthert, S. M. Jane, and C. G. Begley. 2003. The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis, erythropoiesis, and lineage choice in CFU-S12. Proc. Natl. Acad. Sci. USA 100:992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbs, S., S. Jitrapakdee, and J. C. Wallace. 1998. Development of a bicistronic vector driven by the human polypeptide chain elongation factor 1α promoter for creation of stable mammalian cell lines that express very high levels of recombinant proteins. Biochem. Biophys. Res. Commun. 252:368-372. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, H.-L., L. Huang, J. T. Tsan, W. Funk, W. E. Wright, J.-S. Hu, R. E. Kingston, and R. Baer. 1994. Preferred sequences for DNA recognition by the TAL1 helix-loop-helix proteins. Mol. Cell. Biol. 14:1256-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, S., and S. J. Brandt. 2000. mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol. Cell. Biol. 20:2248-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, S., Y. Qiu, Y. Shi, Z. Xu, and S. J. Brandt. 2000. P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL. EMBO J. 19:6792-6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurata, L. W., and G. N. Gill. 1997. Functional analysis of the nuclear LIM domain interactor NLI. Mol. Cell. Biol. 17:5688-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurata, L. W., D. A. Kenny, and G. N. Gill. 1996. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc. Natl. Acad. Sci. USA 93:11693-11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kallianpur, A. R., J. E. Jordan, and S. J. Brandt. 1994. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood 83:1200-1208. [PubMed] [Google Scholar]
- 25.Karacay, B., and L.-S. Chang. 1999. Induction of erythrocyte protein 4.2 gene expression during differentiation of murine erythroleukemia cells. Genomics 59:6-17. [DOI] [PubMed] [Google Scholar]
- 26.Lécuyer, E., S. Herblot, M. Saint-Denis, R. Martin, C. G. Begley, C. Porcher, S. H. Orkin, and T. Hoang. 2002. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood 100:2430-2440. [DOI] [PubMed] [Google Scholar]
- 27.Matthews, K. S. 1992. DNA looping. Microbiol. Rev. 56:123-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mead, P. E., A. E. Deconinck, T. L. Huber, S. H. Orkin, and L. I. Zon. 2001. Primitive erythropoiesis in the Xenopus embryo: the synergistic role of LMO-2, SCL and GATA-binding proteins. Development 128:2301-2308. [DOI] [PubMed] [Google Scholar]
- 29.Merika, M., A. J. Williams, G. Chen, T. Collins, and D. Thanos. 1998. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol. Cell 1:277-287. [DOI] [PubMed] [Google Scholar]
- 30.Mikkola, H. K. A., J. Klintman, H. Yang, H. Hock, T. M. Schlaeger, Y. Fujiwara, and S. H. Orkin. 2003. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421:547-551. [DOI] [PubMed] [Google Scholar]
- 31.Milán, M., and S. M. Cohen. 1999. Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol. Cell 4:267-273. [DOI] [PubMed] [Google Scholar]
- 32.Milán, M., F. J. Diaz-Benjumea, and S. M. Cohen. 1998. Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev. 12:2912-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morcillo, P., C. Rosen, M. K. Baylies, and D. Dorsett. 1997. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 11:2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morcillo, P., C. Rosen, and D. Dorsett. 1996. Genes regulating the remote wing margin enhancer in the Drosophila cut locus. Genetics 144:1143-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimura, S., S. Takahashi, T. Kuroha, N. Suwabe, T. Nagasawa, C. Trainor, and M. Yamamoto. 2000. A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene. Mol. Cell. Biol. 20:713-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono, Y., N. Fukuhara, and O. Yoshie. 1998. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol. Cell. Biol. 18:6939-6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osada, H., G. Grutz, H. Axelson, A. Forster, and T. H. Rabbitts. 1995. Association of erythroid transcription factors: complexes involving the LIM protein RBTN2 and the zinc-finger protein GATA1. Proc. Natl. Acad. Sci. USA 92:9585-9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Persons, D. A., J. A. Allay, E. R. Allay, R. J. Smeyne, R. A. Ashmun, B. P. Sorrentino, and A. W. Nienhuis. 1997. Retroviral-mediated transfer of the green fluorescent protein gene into murine hematopoietic cells facilitates scoring and selection of transduced progenitors in vitro and identification of genetically modified cells in vivo. Blood 90:1777-1786. [PubMed] [Google Scholar]
- 39.Peters, L. L., H. K. Jindel, B. Gwynn, C. Korsgren, K. M. John, S. E. Lux, N. Mohandas, C. M. Cohen, M. R. Cho, D. E. Golan, and C. Brugnara. 1999. Mild spherocytosis and altered red cell ion transport in protein 4.2-null mice. J. Clin. Investig. 103:1527-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porcher, C., E. C. Liao, Y. Fujiwara, L. I. Zon, and S. H. Orkin. 1999. Specification of hematopoietic and vascular development by the bHLH transcription factor SCL without direct DNA binding. Development 126:4603-4615. [DOI] [PubMed] [Google Scholar]
- 41.Porcher, C., W. Swat, K. Rockwell, Y. Fujiwara, F. W. Alt, and S. H. Orkin. 1996. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86:47-57. [DOI] [PubMed] [Google Scholar]
- 42.Rabbitts, T. H. 1998. LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev. 12:2651-2657. [DOI] [PubMed] [Google Scholar]
- 43.Ramain, P., R. Khechumian, K. Khechumian, N. Arbogast, C. Ackermann, and P. Heitzler. 2000. Interactions between Chip and the Achaete/Scute-Daughterless heterodimers are required for Pannier-driven proneural patterning. Mol. Cell 6:781-790. [DOI] [PubMed] [Google Scholar]
- 44.Rincón-Limas, D. E., C. H. Lu, I. Canal, and J. Botas. 2000. The level of DLDB/CHIP controls the activity of the LIM homeodomain protein apterous: evidence for a functional tetramer complex in vivo. EMBO J. 19:2602-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robb, L., N. J. Elwood, A. G. Elefanty, F. Köntgen, R. Li, L. D. Barnett, and C. G. Begley. 1996. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 15:4123-4129. [PMC free article] [PubMed] [Google Scholar]
- 46.Robb, L., I. Lyons, R. Li, L. Hartley, F. Köntgen, R. P. Harvey, D. Metcalf, and C. G. Begley. 1995. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl. Acad. Sci. USA 92:7075-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sánchez, M.-J., E.-O. Bockamp, J. Miller, L. Gambardella, and A. R. Green. 2001. Selective rescue of early haematopoietic progenitors in Scl−/− mice by expressing Scl under the control of a stem cell enhancer. Development 128:4815-4827. [DOI] [PubMed] [Google Scholar]
- 48.Schleif, R. 1992. DNA looping. Annu. Rev. Biochem. 61:199-223. [DOI] [PubMed] [Google Scholar]
- 49.Shivdasani, R. A., E. L. Mayer, and S. H. Orkin. 1995. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373:432-434. [DOI] [PubMed] [Google Scholar]
- 50.Spicuglia, S., D. Payet, R. K. Tripathi, P. Rameil, C. Verthuy, J. Imbert, P. Ferrier, and W. M. Hempel. 2000. TCRα enhancer activation occurs via a conformational change of a pre-assembled nucleo-protein complex. EMBO J. 19:2034-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugihara, T. M., I. Bach, C. Kioussi, M. G. Rosenfeld, and B. Andersen. 1998. Mouse deformed epidermal autoregulatory factor 1 recruits a LIM domain factor, LMO-4, and CLIM coregulators. Proc. Natl. Acad. Sci. USA 95:15418-15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang, T., K. S. Prasad, M. J. Koury, and S. J. Brandt. 1999. Mitogen-activated protein kinase mediates erythropoietin-induced phosphorylation of the TAL1/SCL transcription factor in murine proerythroblasts. Biochem. J. 343:615-620. [PMC free article] [PubMed] [Google Scholar]
- 53.Torigoi, E., I. M. Bennani-Baiti, C. Rosen, K. Gonzalez, P. Morcillo, M. Ptashne, and D. Dorsett. 2000. Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc. Natl. Acad. Sci. USA 97:2686-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Meyel, D. J., D. D. O'Keefe, L. W. Jurata, S. Thor, G. N. Gill, and J. B. Thomas. 1999. Chip and Apterous physically interact to form a functional complex during Drosophila development. Mol. Cell 4:259-265. [DOI] [PubMed] [Google Scholar]
- 55.van Meyel, D. J., D. D. O'Keefe, S. Thor, L. W. Jurata, G. N. Gill, and J. B. Thomas. 2000. Chip is an essential cofactor for Apterous in the regulation of axon guidance in Drosophila. Development 127:1823-1831. [DOI] [PubMed] [Google Scholar]
- 56.Visvader, J. E., X. Mao, Y. Fujiwara, K. Hahm, and S. H. Orkin. 1997. The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc. Natl. Acad. Sci. USA 94:13707-13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vyas, P., M. A. McDevitt, A. B. Cantor, S. G. Katz, Y. Fujiwara, and S. H. Orkin. 1999. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development 126:2799-2811. [DOI] [PubMed] [Google Scholar]
- 58.Wadman, I. A., H. Osada, G. G. Grütz, A. D. Agulnick, H. Westphal, A. Forster, and T. H. Rabbitts. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warren, A. J., W. H. Colledge, M. B. Carlton, M. J. Evans, A. J. Smith, and T. H. Rabbitts. 1994. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell 78:45-57. [DOI] [PubMed] [Google Scholar]
- 60.Weihe, U., M. Milán, and S. M. Cohen. 2001. Regulation of Apterous activity in Drosophila wing development. Development 128:4615-4622. [DOI] [PubMed] [Google Scholar]
- 61.Yamada, Y., A. J. Warren, C. Dobson, A. Forster, R. Pannell, and T. H. Rabbitts. 1998. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl. Acad. Sci. USA 95:3890-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yie, J., M. Merika, N. Munshi, G. Chen, and D. Thanos. 1999. The role of HMG I(Y) in the assembly and function of the IFNβ enhanceosome. EMBO J. 18:3074-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, L., S. B. Kahwash, and L.-S. Chang. 1998. Developmental expression of mouse erythrocyte protein 4.2 mRNA: evidence for specific expression in erythroid cells. Blood 91:695-705. [PubMed] [Google Scholar]