Abstract
Human artificial chromosomes have been used to model requirements for human chromosome segregation and to explore the nature of sequences competent for centromere function. Normal human centromeres require specialized chromatin that consists of alpha satellite DNA complexed with epigenetically modified histones and centromere-specific proteins. While several types of alpha satellite DNA have been used to assemble de novo centromeres in artificial chromosome assays, the extent to which they fully recapitulate normal centromere function has not been explored. Here, we have used two kinds of alpha satellite DNA, DXZ1 (from the X chromosome) and D17Z1 (from chromosome 17), to generate human artificial chromosomes. Although artificial chromosomes are mitotically stable over many months in culture, when we examined their segregation in individual cell divisions using an anaphase assay, artificial chromosomes exhibited more segregation errors than natural human chromosomes (P < 0.001). Naturally occurring, but abnormal small ring chromosomes derived from chromosome 17 and the X chromosome also missegregate more than normal chromosomes, implicating overall chromosome size and/or structure in the fidelity of chromosome segregation. As different artificial chromosomes missegregate over a fivefold range, the data suggest that variable centromeric DNA content and/or epigenetic assembly can influence the mitotic behavior of artificial chromosomes.
Twentyyears have passed since the first yeast artificial chromosomes (YACs) were constructed in an effort to elucidate the minimal components necessary for eukaryotic chromosome function (37). These pioneering studies demonstrated that both centromere competence and overall chromosome organization play a role in the segregation of chromosomes (19, 36, 56). From the yeast Saccharomyces cerevisiae to humans, mitotically stable chromosomes require centromeres and other chromosomal features, including telomeres and origins of replication (10, 13, 49). While these components have been well characterized in the yeasts S. cerevisiae and Schizosaccharomyces pombe (2, 4, 32, 46, 61), tractable experimental approaches to define the corresponding elements in humans or other organisms with larger chromosomes have only recently begun to be developed (16, 26, 49, 68).
The first step in creating human artificial chromosomes was to identify the DNA sequences responsible for human centromere function. Despite the evident importance of the centromere in all eukaryotic organisms, the DNA sequences responsible for centromere activity are not conserved evolutionarily (30). Whereas the S. cerevisiae “point” centromere is only 125 bp and is shared among all yeast chromosomes (21), other eukaryotic centromeres are substantially larger, more complex, and markedly heterogeneous in sequence and composition. For example, the S. pombe centromere is composed of inner and outer repeat sequences and varies from 40 to 100 kb in size (7, 39). Arabidopsis thaliana centromeres have been localized to 500- to 1,300-kb regions of chromosomes resistant to meiotic recombination, and the sequences at these loci are composed of 180-bp tandem repeats and retrotransposon elements (9, 15, 42). Similarly, the Drosophila centromere, modeled by a 420-kb region of a stable minichromosome, appears to be made up of simple repeats and transposable elements (54, 55).
Human centromeric regions are composed of megabases of repetitive alpha satellite DNA that varies in sequence identity and organization from chromosome to chromosome (62). The only commonality between alpha satellite DNA and all of the other well-characterized centromeric DNA sequences is an increase in AT-richness compared to the genome average (5). The structure of alpha satellite DNA is based on ∼171-bp monomers repeated in tandem to make up higher-order repeat units that are in turn repeated to generate megabase size arrays (for reviews, see references 1 and 68). While monomers within each higher-order repeat are only 65 to 90% identical to each other, the multimeric higher-order repeats within the same array are highly homogeneous (44, 45, 71). Although alpha satellite DNA is located at the centromeres of all normal human chromosomes (31, 45, 71), the defining characteristics of alpha satellite required for centromere function are unknown. Notwithstanding the likely role of epigenetic influences on centromere activity (17, 49), the functional competence of alpha satellite may be due to its highly repetitive structure, its AT-richness, or the presence of critical binding sites for particular centromere proteins (5, 40).
While it is well-known that centromeres play a pivotal role in chromosome segregation, the precise mechanisms are incompletely understood and are likely to be both complex and multifactorial. First, the centromere must assemble a kinetochore, a proteinaceous structure responsible for attaching the chromosome to spindle microtubules (for a review, see reference 58). Checkpoint pathways ensure that all chromosomes have aligned properly at metaphase before proceeding into anaphase (for a review, see reference 38). Once a bipolar spindle attachment has been established, the sister chromatids separate by resolving cohesins present at the centromere until their release at anaphase (for reviews, see references 27 and 60). To segregate properly, therefore, chromosomes must contain sequences competent to assemble kinetochores, maintain cohesion, separate sister chromatids, and satisfy spindle checkpoints.
The study of human artificial chromosomes (as well as naturally occurring or engineered human chromosome rearrangements) has demonstrated that certain sequences are capable of establishing and maintaining centromeres, but the functionality of these centromeres has not been studied in detail. Studies from a number of groups have shown that alpha satellite DNA from several human chromosomes is capable of forming de novo centromeres in artificial chromosome assays (11, 14, 16, 18, 22, 24, 34, 40, 45). Some types of alpha satellite assemble artificial chromosomes that are maintained stably in culture over many cell divisions and contain centromeres that colocalize with kinetochore proteins associated specifically with active centromeres. Additional supporting evidence for the critical role of alpha satellite in human centromere function has come from detailed analysis of either natural or engineered chromosome rearrangements (20, 35, 45, 47, 59). These data suggest that, to a first approximation, artificial and rearranged chromosomes are behaving like normal chromosomes. However, the detailed segregation of these chromosomes has not been systematically evaluated or compared to that of normal chromosomes.
Here, we have investigated the extent to which human artificial chromosomes provide a model to study chromosome segregation. In this study, we examine the mitotic segregation of natural human chromosomes, human artificial chromosomes, and rearranged human chromosomes to investigate the roles of centromere and chromosome structure in proper chromosome segregation.
MATERIALS AND METHODS
Cell lines.
The near tetraploid human fibrosarcoma cell line HT1080 was grown in alpha minimal essential medium (MEM) (Gibco Inc.) supplemented with 10% fetal bovine serum (HyClone), penicillin and streptomycin, and glutamine. Human fibroblast cell line GM8148 (66) was derived from a patient with the karyotype 47,XX,del(17)(pter→p11.2::cen→qter), + der(17)(:p11.2→cen:). This cell line has a deletion in chromosome 17 (approximately 2.5 Mb of D17Z1 and several megabases of euchromatin adjacent to the centromere deleted) and also contains a ring chromosome derived from the deletion event.
A human fibroblast cell line was derived from a patient with the karyotype 46,X,r(X)/45,X. This cell line contains a small ring chromosome estimated to be approximately 10 Mb in size. The ring chromosome contains an apparently complete DXZ1 array, plus several megabases of euchromatin adjacent to DXZ1 (S. Schwartz, unpublished data). Both fibroblast lines were grown in alpha MEM supplemented with 15% fetal bovine serum, penicillin and streptomycin, and glutamine.
Large insert clones and artificial chromosome formation.
A ∼85-kb NotI fragment from bacterial artificial chromosome (BAC) RPCI-11 242E23 containing DXZ1 higher-order alpha satellite DNA repeats from the human X chromosome (45) was isolated and cloned into the pPAC4 vector, containing a blasticidin resistance gene (12). The construction of BAC VJ104α32, containing ∼86 kb of synthetic alpha satellite from chromosome 17 (D17Z1), has been described elsewhere (16). To generate artificial chromosomes, DXZ1- and D17Z1-based constructs were transfected in multiple independent experiments into human HT1080 fibrosarcoma cells and subjected to drug selection (14, 45).
In an alternative strategy (R. W. Mays, unpublished data), VJ104α32 and a BAC containing ∼140 kb of a genomic fragment including the human HPRT gene were both modified to contain 800 bp of human telomere repeat sequences (16) and were then cotransfected into HT1080 cells. Transfected cells were grown in medium containing both HAT (hypoxanthine-aminopterin-thymidine) to select for hypoxanthine phosphoribosyltransferase (HPRT) expression and G418 to select for βgeo expression. Two artificial chromosome-containing cell lines, PF2.6 and PF2.7, were generated using this method. Pulsed-field gel analysis (16) demonstrated that PF2.6 was linear and PF2.7 was circular (R. W. Mays and M. K. Rudd, unpublished data).
FISH and immunostaining.
After transfection, drug-resistant colonies were screened for artificial chromosomes by fluorescence in situ hybridization (FISH) as described previously (14, 16). Once artificial chromosomes were identified cytogenetically, cell lines were grown in the presence or absence of drug selection for at least 30 days to measure mitotic stability (14, 16), expressed on a per cell division basis as previously described (16). Metaphase chromosomes were prepared by standard protocols, and 50 metaphase spreads were scored per time point. Alpha satellite FISH was performed as described previously (14).
Telomere DNA probes obtained from Applied Biosystems, Inc., were hybridized to chromosome spreads fixed in 3:1 methanol-acetic acid fixative as described previously (25). Immunostaining for centromere protein E (CENP-E) was performed as described previously (16).
Alpha satellite DNA acquisition.
Cell lines containing putative artificial chromosomes were tested for acquisition of alpha satellite DNA from endogenous HT1080 chromosomes with centromere-specific FISH probes. Pancentromeric primers amplified alpha satellite DNA from the HT1080 cell line, which was subsequently labeled as a FISH probe, as described previously (22). Excess unlabeled pBamX7 (70) or p17H8 (64) DNA was denatured and preannealed with the pancentromeric FISH probe to block FISH signals from the alpha satellite DNA used to generate the artificial chromosome. Artificial chromosomes lacking pancentromeric FISH signals were then tested with individual centromeric FISH cocktails as a more rigorous test. Degenerate alpha satellite consensus primers (65) were modified (5′TCA(A/T)(C/G)T(C/A)ACAGAGTT(G/T)AAC3′/5′CACATC(A/C)CAAAG(A/T/C)AGTTTC3′) to amplify alpha satellite DNA from monochromosomal rodent/human somatic cell hybrid DNA (from Coriell Cell Repositories, mapping panel 2). PCR products from each monochromosomal hybrid were pooled into six cocktails. Alpha satellite PCR products were pooled from chromosomes 1, 6, 13, and 20 (cocktail 1); chromosomes 2, 7, 14, and Y (cocktail 2); chromosomes 3, 8, 15, and 19 (cocktail 3); chromosomes 4, 9, 16, and 21 (cocktail 4); chromosomes 5, 11, 22, and 17 or X (cocktail 5); and chromosomes 10, 12, and 18 (cocktail 6). PCR product cocktails were directly labeled as described previously (14). Cocktails of alpha satellite were hybridized to artificial chromosomes under stringent conditions (65% formamide-2× SSC washes at 42°C [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) as described previously (16) to detect alpha satellite other than the DXZ1 or D17Z1 present on the artificial chromosome. Artificial chromosomes that were negative with the pancentromere probe and all of the individual cocktails were concluded to be of de novo origin.
Anaphase segregation assay.
To measure chromosome segregation, cell lines were arrested at anaphase with nocodazole, a benzimidazole derivative that binds to tubulin dimers, inhibiting microtubule assembly. After anaphase arrest, cells were treated with dihydrocytochalasin B to prevent cytokinesis, suspending cells (and their segregating daughter chromosomes) in anaphase or telophase (52, 53). After such treatment, an estimated 20% of cells are found in anaphase or telophase configurations. Artificial chromosome, ring chromosome, and normal chromosome segregation was monitored by FISH to identify the relevant chromosome(s), using chromosome-specific alpha satellite probes. The vector probes pPAC4 (12) and VJ104 (16) were used to identify the DXZ1 and D17Z1 artificial chromosomes, respectively. Chromosome-specific centromere probes and whole chromosome paints (from Vysis Inc.) were used to measure chromosome segregation. For each cell line, 200 cells were scored in duplicate, for a total of 400 cells scored per data point. The frequency of missegregation was calculated for each chromosome as the number of missegregating chromosomes divided by the total number of chromosomes scored for each type.
RESULTS
We have used two approaches to make human artificial chromosomes. In the first approach (14, 45), we used large fragments of alpha satellite DNA cloned into BAC or P1 artificial chromosome (PAC) vectors to generate circular human artificial chromosomes. In the second approach, we used cotransfection of synthetic arrays of alpha satellite combined with large fragments of human genomic DNA and synthetic telomeres (16) (Mays et al., unpublished) to generate both linear and circular artificial chromosomes.
Type of alpha satellite DNA influences artificial chromosome formation.
Artificial chromosome studies have used a variety of chromosome-specific alpha satellite DNA sequences to generate de novo centromeres (11, 14, 16, 18, 22, 24, 34, 40, 45). Our studies focused on higher-order X chromosome (DXZ1) and chromosome 17 (D17Z1) alpha satellite. DXZ1 and D17Z1 alpha satellite are closely related; both are composed of ∼171-bp monomers arranged in a pentameric organization, and corresponding monomers within DXZ1 and D17Z1 are 85% identical on average (64).
Despite their sequence relatedness, DXZ1 and D17Z1 form artificial chromosomes at very different frequencies in HT1080 cells. (Although the DXZ1 and D17Z1 constructs used have different BAC or PAC vectors and drug resistance markers, previous studies have shown that this difference does not significantly influence artificial chromosome formation rates [14].) After transfecting either the DXZ1- or D17Z1-containing constructs into HT1080 cells, we selected blasticidin- or G418-resistant colonies and then detected artificial chromosomes by FISH with probes to either type of alpha satellite DNA. Consistent with other studies (14, 34), D17Z1 formed stable artificial chromosomes in a high proportion of drug-resistant colonies (30 to 40% in other studies; 6 of 8 colonies here). However, the artificial chromosome formation rate for DXZ1 was much lower; DXZ1 formed artificial chromosomes in only 7% (6 of 88) of drug-resistant colonies in three experiments.
Artificial chromosome composition.
Previous studies have demonstrated that some artificial chromosomes may acquire DNA sequences other than the transfected input DNA (14, 16). Because we wished to address the centromeric competence of DXZ1 and D17Z1 sequences, it was necessary to rule out that any other alpha satellite DNA had assembled into the artificial chromosome that might confer centromere activity. To this end, we used centromeric FISH assays to detect alpha satellite besides that of the input DNA on each artificial chromosome (Materials and Methods). While it is formally possible that the artificial chromosomes could have acquired DXZ1 or D17Z1 from endogenous chromosomes, this possibility would not be detected by these methods. However, on the basis of previous experiments (16), we expect such highly specific recombination-mediated acquisition to be unlikely. Of the five DXZ1-containing chromosomes tested, three had acquired alpha satellite DNA other than DXZ1; only two, therefore, could be validated as containing de novo centromeres (45). In contrast, none of the four D17Z1-based artificial chromosomes tested was positive for non-D17Z1 alpha satellite DNA by FISH; thus, all were of de novo origin (Table 1). The difference between the two types of input alpha satellite is likely related to the formation efficiency of the two constructs (14); poorly or moderately competent constructs are thus more likely to show acquisition of endogenous alpha satellite sequences. Interestingly, the two validated de novo DXZ1-based artificial chromosomes (X-4 and X-5) appear to be cytologically larger than the three acquisition DXZ1-containing artificial chromosomes (Fig. 1). This could reflect a greater size requirement for DXZ1-based artificial chromosomes. We assayed the artificial chromosomes in this study only for the acquisition of alpha satellite, as no other sequences have been implicated in centromere function in humans (43, 45).
TABLE 1.
Characteristics of human artificial chromosomes
| Input DNA | Cell line | Characteristic
|
||||
|---|---|---|---|---|---|---|
| Relative sizea | CENP-Eb | Mitotic stabilityc | Centromered | Telomeree | ||
| Strategy 1 | ||||||
| DXZ1 | X-1 | Small | + | + | Acquisition | − |
| X-2 | Small | + | − | ND | ND | |
| X-3 | Small | + | + | Acquisition | − | |
| X-4 | Medium | + | + | De novo | − | |
| X-5 | Medium | + | + | De novo | − | |
| X-6 | Small | + | ND | Acquisition | − | |
| D17Z1 | 17-1 | Small | + | + | De novo | − |
| 17-2 | Small | + | + | De novo | − | |
| 17-3 | Medium | + | ND | De novo | − | |
| 17-4 | Medium | + | ND | De novo | − | |
| Strategy 2 | ||||||
| D17Z1 | PF2.6 | Small | + | + | De novo | + |
| PF2.7 | Small | + | + | De novo | − | |
| 17-15 | Large | + | + | De novo | + | |
Relative size was determined by DAPI staining, and artificial chromosomes were grouped into small, medium, and large categories. Small artificial chromosomes were estimated to be ∼15 Mb or less, medium artificial chromosomes were estimated to be between 15 and 30 Mb, and large artificial chromosomes were estimated to be larger than 30 Mb.
Immunostaining for the presence of anti-CENP-E antibodies on the artificial chromosome (16).
Mitotic stability was measured as described in Materials and Methods. Mitotically stable artificial chromosomes were retained at a rate of 98.6 to 100% per cell division, calculated as described previously (16). ND, not done.
Artificial chromosomes were tested for the acquisition of non-input alpha satellite DNA sequences (Materials and Methods).
Artificial chromosomes were assayed for the presence of functional telomeres. Artificial chromosomes constructed from circular input DNA (in cell lines X-1 to 17-4) did not acquire telomere sequences as demonstrated by FISH. The presence of functional telomeres in artificial chromosomes in which telomere sequences were included in the input constructs (PF2.6 and PF2.7) was confirmed by gamma irradiation and pulsed-field gel analysis.
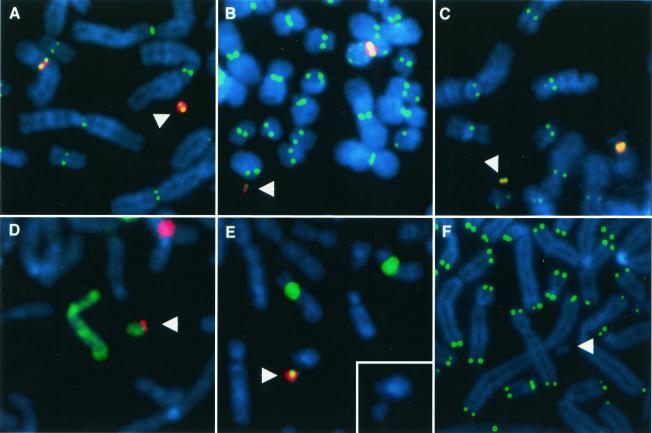
FIG. 1.
FISH analysis of artificial chromosomes. Arrowheads denote artificial chromosomes. DAPI (4′,6′-diamidino-3-phenylindole) stains chromosomes in blue. (A to C) CENP-E immunostaining (green) stains active centromeres, while alpha satellite FISH (red) hybridizes to the artificial chromosome and the relevant endogenous centromere. Cell lines X-5 (A) and X-6 (B) contain DXZ1-based artificial chromosomes probed with DXZ1. (C) Cell line 17-1 contains a D17Z1-based artificial chromosome probed with D17Z1. (D) Cell line 17-15. A green chromosome 8 paint probe hybridizes to the endogenous chromosome 8 as well as the artificial chromosome. The D17Z1 FISH probe in red hybridizes to the artificial chromosome as well as the endogenous chromosome 17. (E) Cell line PF2.6. The green D17Z1 probe hybridizes to the artificial chromosome and to the centromeres of the endogenous chromosomes 17. An HPRT probe in red hybridizes to the artificial chromosome. The insert shows a DAPI image of the artificial chromosome. (F) Cell line 17-1. A telomere probe in green hybridizes to the ends of all chromosomes except for the artificial chromosome.
Artificial chromosome centromere function.
While both DXZ1 and D17Z1 can form de novo centromeres on an artificial chromosome, the functionality of these centromeres remained to be determined. CENP-E, a motor protein, is part of the kinetochore (72, 73) and localizes specifically to active centromeres (50), making it a useful marker of functional centromeres. We immunostained cell lines containing human artificial chromosomes with antibodies to CENP-E. Like endogenous chromosomes, all artificial chromosomes were positive for the characteristic pattern of CENP-E staining (Fig. 1 and Table 1). To further evaluate the artificial chromosome centromeres, we examined the mitotic stability of several artificial chromosomes in the presence and absence of drug selection (Table 1, strategy 1). Of the seven artificial chromosomes tested in this way, six were mitotically stable both in the presence and absence of drug selection and were retained at a calculated rate of 98.6 to 100% per cell division. The fact that the artificial chromosomes in this study are retained after many cell divisions indicates that, like previously described artificial chromosomes (11, 14, 16, 18, 22, 24, 34, 40, 45), they segregate well, even in the absence of selection.
Artificial chromosome segregation in anaphase.
Mitotic stability in the drug selection assay reflects the proportion of cells containing one or more artificial chromosomes over time; as such, it is only an indirect measure of segregation events in individual cell divisions. To investigate the segregation of the artificial chromosomes more directly, we implemented an assay based on use of nocodozole to trap cells in anaphase and thus monitor chromosome segregation directly at each cell division (52, 53). We monitored the segregation of seven human artificial chromosomes (see Fig. 3B), three formed with DXZ1 and four formed with D17Z1. Six of the seven artificial chromosomes tested were fully validated and of de novo origin (see above), while one of the DXZ1 artificial chromosomes, X-3, had acquired endogenous chromosome 7 alpha satellite and thus contained multiple types of alpha satellite. After the cells were suspended in anaphase, we performed FISH with probes to identify the artificial chromosome and endogenous X chromosomes or chromosomes 17 in each cell line, providing a direct comparison of the segregation of artificial chromosomes to the corresponding endogenous chromosomes in the same cell (Fig. 2). In addition to the endogenous chromosomes 17 and X, we also examined the segregation of natural human chromosomes 7, 18, and Y as controls.
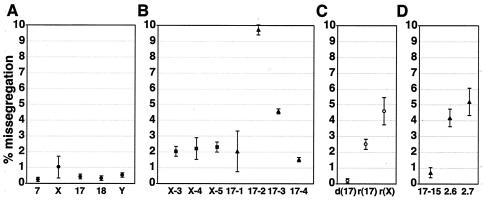
FIG. 3.
Missegregation of natural, artificial, and variant chromosomes. Missegregation rates were calculated as the number of missegregating chromosomes divided by the total number of chromosomes present of each type. Error bars represent one standard deviation from the mean of two segregation experiments for each chromosome. The total number of cells scored for each data point was >400. (A) Missegregation rates for natural chromosomes 7, X, 17, 18, and Y. (B) Missegregation rates for artificial chromosomes in cell lines X-3, X-4, X-5, 17-1, 17-2, 17-3, and 17-4. DXZ1-based artificial chromosomes (squares) and D17Z1-based artificial chromosomes (triangles) are indicated. (C) Missegregation frequencies for chromosome 17 with a deletion [d(17)], the ring chromosome 17 [r(17)], and the ring X chromosome [r(X)]. (D) Missegregation rates for D17Z1-based artificial chromosomes in cell lines 17-15, PF2.6, and PF2.7.
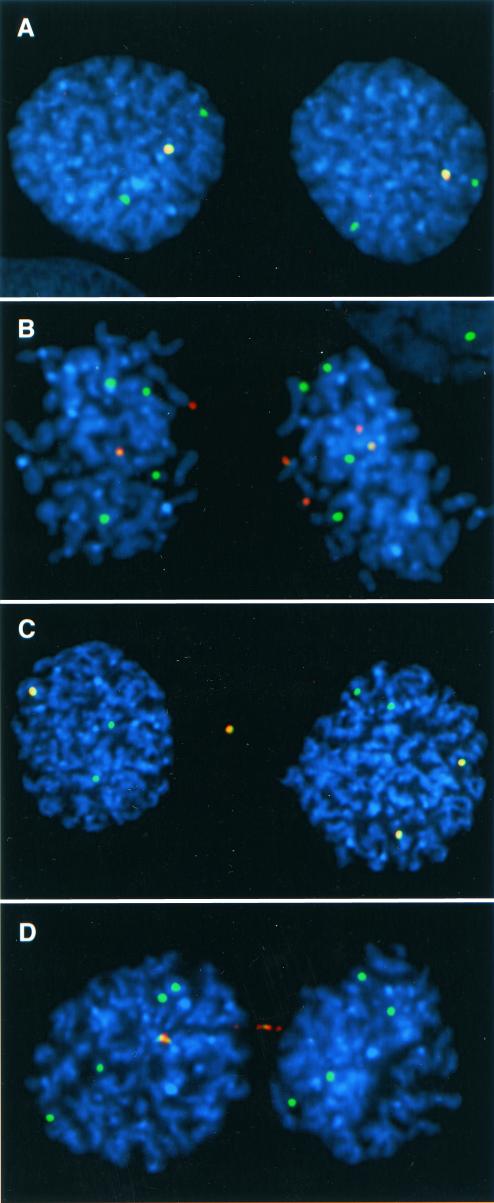
FIG. 2.
Anaphase segregation assay. Anaphase nuclei were stained blue with DAPI and hybridized with FISH probes to detect artificial and natural chromosomes. In each example, the green alpha satellite FISH probe hybridized to the artificial chromosome as well as natural chromosomes with the same alpha satellite DNA as the artificial chromosome. A red vector probe hybridized to the artificial chromosomes, but not the natural chromosomes. Thus, the natural chromosomes appear as green dots, while the artificial chromosomes appear asmerged green and red signals. (A) Cell line X-5. The natural X chromosomes segregated 2:2, and the artificial chromosomes segregated 1:1. (B) Cell line 17-2. The natural chromosomes 17 segregated 4:4, and the artificial chromosomes segregated 2:4, reflecting a likely nondisjunction event. (C) Cell line X-5. The natural X chromosomes segregated 2:2, and the artificial chromosomes segregated 1:2 with a lagging artificial chromosome in between the other segregating chromosomes. (D) Cell line 17-3. The natural chromosomes 17 segregated 4:4, and one of the artificial chromosomes was in an anaphase bridge.
Missegregation of the natural human chromosomes tested ranged from 0.33 to 1.1% per chromosome per cell division (Fig. 3A). These data are consistent with previous studies of chromosome segregation in other human cell lines using this or similar assays (6, 53). In contrast, the human artificial chromosomes tested missegregated between 1.6 and 9.7% per chromosome per cell division (Fig. 3B). Notably, the artificial chromosome in cell line X-3 missegregated at a rate of 2.0% ± 0.4%, similar to the rates of other artificial chromosomes studied. Although this artificial chromosome had acquired endogenous alpha satellite that may or may not be acting as the functional centromere, it behaved like the other artificial chromosomes. The wide range of artificial chromosome missegregation rates is due largely to the artificial chromosomes in cell lines 17-3 and 17-2, with missegregation rates of 4.3% ± 0.05% and 9.7% ± 0.2%, respectively. Even without including these two artificial chromosomes, however, the missegregation rates of artificial chromosomes and natural chromosomes were significantly different from each other (P < 0.001).
Variant chromosome segregation.
The differences in segregation between natural chromosomes and artificial chromosomes could be explained by a number of factors relevant to centromere and/or chromosome function. To shed light on the possible differences in missegregation rates between artificial and natural chromosomes, we next examined a number of variant human chromosomes. To control for possible variation between diploid human cell lines and the hyperdiploid HT1080 line, we first examined the segregation of the normal 17 and X chromosomes in the diploid lines. The frequencies of missegregation detected (0.6 and 0.1%, respectively) were indistinguishable from those shown in Fig. 3A for HT1080, indicating that such possible cell line effects are negligible.
As one hypothesis, artificial chromosomes may missegregate more than natural chromosomes due to the presence of de novo centromeres, which may be compromised in some as yet unidentified way relative to endogenous centromeres. If de novo centromeres were the only factor causing the observed increase in missegregation, then we would expect chromosomes of a given size with those with natural centromeres to segregate better than those with artificial chromosomes. To test this, we measured the segregation of two naturally occurring, patient-derived ring chromosomes, one derived from the X chromosome and one derived from chromosome 17. These ring chromosomes are cytologically similar in size to artificial chromosomes and contain either DXZ1 or D17Z1 alpha satellite at their natural centromeres. When analyzed in the anaphase assay, the chromosome 17-derived ring chromosome missegregated with a frequency of 2.5% ± 0.3%, while the X-derived ring missegregated with a frequency of 4.5% ± 0.8% (Fig. 3C). Notably, these rates are within the range of artificial chromosome missegregation, rather than the range of natural chromosome missegregation. These data argue, therefore, that the presence of de novo centromeres alone does not account for an increase in segregation defects.
The amount of alpha satellite DNA could also play a role in chromosome segregation. With the exception of the Y centromere, natural human centromeres are composed of several megabases of alpha satellite (29, 67), although the minimum amount of alpha satellite capable of centromere function is unknown. To investigate the possible effect of alpha satellite array size on segregation, we next analyzed the segregation of a patient-derived chromosome 17 with a deletion in the alpha satellite array. This chromosome contains less than a quarter of the alpha satellite present at the normal chromosome 17 centromere (700 kb versus 3.2 Mb) (66); however, it segregates normally in the anaphase segregation assay, with a missegregation rate of 0.12% ± 0.1% (Fig. 3C). Thus, at least above 700 kb of alpha satellite, alpha satellite array size does not determine chromosome segregation. On the basis of the relative intensity of FISH signals, we estimate that the artificial chromosomes in this study contain more than 700 kb of alpha satellite. Therefore, it is unlikely that the segregation errors we observe in artificial chromosomes are due to a lack of alpha satellite DNA.
Since both artificial chromosomes with de novo centromeres and ring chromosomes with natural centromeres missegregate more than normal chromosomes, the data suggest that some characteristic of chromosome structure other than the origin of the centromere plays a role in chromosome segregation. Both artificial chromosomes and ring chromosomes are smaller than natural chromosomes. If chromosome size is involved in segregation, then we would expect larger artificial chromosomes to segregate better than smaller artificial chromosomes, as is the case with YACs (36, 37). To determine whether the size of human artificial chromosomes affects segregation, we examined an artificial chromosome with a de novo D17Z1 centromere that, by translocation, had acquired a large region of chromosome 8q (16). This chromosome (in clone 17-15) is much larger than any of the other artificial chromosomes or ring chromosomes in this study (Fig. 1) (Table 1, strategy 2) and is the only artificial chromosome with a normal euchromatic chromosome arm. Unlike the artificial chromosomes and ring chromosomes, it is linear and is capped by telomere repeats at each end of the chromosome (16). This large linear artificial chromosome missegregates at a frequency of 0.75% ± 0.35%, within the range of natural human chromosomes (Fig. 3D). These data argue that chromosome size and/or organization is involved in ensuring proper segregation.
If linear structure alone contributes to proper chromosome segregation, then linear artificial chromosomes should segregate better than circular ones. Indeed, studies in S. cerevisiae have shown that circular YACs missegregate more frequently than linear YACs of a comparable size (37). Small human artificial chromosomes could also be affected by circular structure, since the seven artificial chromosomes studied here in detail were formed from constructs without telomere sequences and, when assayed for the acquisition of telomere sequences using telomere probes (Materials and Methods), contain no detectable telomere sequences (Table 1 and Fig. 1).
To evaluate the effect of linearity on chromosome segregation, we next studied the segregation of two additional artificial chromosomes, one linear (PF2.6) and one circular (PF2.7) (Table 1, strategy 2), containing copies of the human HPRT gene (Mays and Rudd, unpublished). Both of these artificial chromosomes contain de novo D17Z1 alpha satellite centromeres, a fragment of genomic DNA containing HPRT gene sequences, and telomere repeats (Materials and Methods) (Mays et al., unpublished). The missegregation frequencies for PF2.6 and PF2.7 are 4.1% ± 0.35% and 5.2% ± 0.55%, respectively, both within the range of artificial chromosome missegregation rates (Fig. 3D). The similarity in segregation between the circular and linear artificial chromosomes refutes the possibility that circular structure alone causes an increase in segregation errors.
Types of missegregation errors.
In addition to the frequency of missegregation, the types of segregation errors differed between artificial chromosomes and natural chromosomes (Table 2). We analyzed each type of chromosome for the number of detected nondisjunction and anaphase lag events, as these types of error could easily be distinguished cytogenetically (Fig. 2). (We excluded from this analysis other errors in which the mechanism could not be inferred, i.e., apparent 1:0 segregation. These uncharacterized errors were equally represented among natural and artificial chromosomes, making up 12 to 18% of the total segregation errors, and could reflect absence of replication, chromosome loss, or hybridization failure.)
TABLE 2.
Segregation errors
| Chromosome type | Total no. of chromosomes scored | No. (%) of missegregating chromosomesa
|
||
|---|---|---|---|---|
| Total | NDJ | Lag | ||
| Normal | 31,867 | 153 | 124 (81) | 29 (19) |
| Artificial | 13,193 | 447 | 268 (60) | 179 (40) |
| Ring | 947 | 13 | 11 (85) | 2 (15) |
Missegregating chromosomes exhibiting nondisjunction (NDJ) or anaphase lag.
Natural chromosomes are more likely to exhibit nondisjunction than lag during anaphase (Table 2); of the 153 missegregating natural chromosomes detected in this study, 81% exhibited nondisjunction, while only 19% exhibited anaphase lag. This trend was also apparent in the limited number of ring chromosome missegregation events detected, among which 85% exhibited nondisjunction and 15% exhibited anaphase lag. In contrast, missegregating artificial chromosomes (n = 447) exhibited nondisjunction 60% of the time and exhibited anaphase lag 40% of the time. The frequencies of nondisjunction and anaphase lag exhibited by artificial chromosomes and natural chromosomes are statistically different (P < 0.001 by the chi-square test), suggesting that the mechanism of segregation error varies between natural and artificial chromosomes. Even among artificial chromosomes, missegregation rates and type of missegregation vary from clone to clone (data not shown), likely reflecting the different de novo centromeres. For example, the artificial chromosome in cell line X-5 exhibited anaphase lag 30% of the time (n = 13) and nondisjunction 70% of the time (n = 30), while the artificial chromosome in cell line 17-3 had a higher incidence of lag (54% of missegregating chromosomes [n = 27] versus 46% nondisjunction [n = 23]). The propensity for anaphase lag in artificial chromosomes compared to natural chromosomes and ring chromosomes suggests that de novo centromeres are structurally and functionally compromised in some manner and/or may be missing critical factors required for normal chromosome segregation.
DISCUSSION
Chromosome segregation is a complicated process involving factors specific to centromere structure and function, as well as more general aspects of chromosome organization (49). In S. cerevisiae, the requirements for proper chromosome segregation have been well defined using YACs as a model. Here, we have used a human artificial chromosome system as a similar model for addressing the critical DNA elements required for proper segregation of human chromosomes in mitosis. By studying the segregation of natural chromosomes, artificial chromosomes, and patient-derived variant chromosomes, we have begun to examine the requirements for proper chromosome segregation.
Chromosome size and structure affect segregation.
Chromosome size is a likely factor in chromosome segregation, as revealed initially with YACs (37). Our data with human natural and artificial chromosomes support this notion; human artificial chromosomes and ring chromosomes analyzed in this study are smaller than normal chromosomes, and they missegregate more often than normal chromosomes (Fig. 3). Thus, there may be a minimum chromosome size threshold (presumably smaller than the normal Y chromosome) below which chromosomes are more prone to mitotic error.
Overall chromosome structure and organization may also affect segregation. Ring chromosomes and the majority of human artificial chromosomes in this study are circular, and both of these types of chromosome missegregate more often than normal chromosomes. However, a linear artificial chromosome in this study, PF2.6, missegregates at a rate similar to those of circular artificial chromosomes, arguing that circular structure alone does not account for the observed increased frequency of segregation errors. In contrast, linear artificial chromosome 17-15 missegregates within the range of normal chromosomes (Fig. 3); however, this chromosome has acquired a region of a normal chromosome arm (16) and consequently is much larger than the other artificial chromosomes examined in this study. This artificial chromosome may segregate properly due to its large size or to the presence of a bona fide chromosome arm. This conclusion has implications for the design of next-generation human artificial chromosomes.
The type of segregation error exhibited by chromosomes is indicative of the mechanism of centromere dysfunction.
The fact that normal chromosomes and ring chromosomes exhibit relatively less chromosome lag than artificial chromosomes suggests that de novo centromeres are compromised in a way that increases the frequency of anaphase lag. Artificial chromosomes are composed of alpha satellite DNA plus BAC or PAC vector sequences concatamerized into chromosomes larger than the original 100-kb input DNA. Multiple stretches of alpha satellite separated by vector sequences on an artificial chromosome may be assembling dicentric or multicentric chromosomes. Dicentric chromosomes in S. cerevisiae and humans are prone to segregation defects. In S. cerevisiae, dicentric minichromosomes are mitotically unstable and have a higher copy number than monocentric minichromosomes (23). Similarly, human dicentric chromosomes are more prone to lag than normal chromosomes (53). In both of these previous studies, the distance between centromeres affects segregation; when centromeres are close to each other, they may function as a monocentric chromosome. At the resolution of metaphase chromosomes, artificial chromosomes appear to bind centromere proteins in one discrete kinetochore; however, the precise organization of DNA and proteins is not currently known. Further analysis of the structure of artificial chromosomes may prove that some are functionally di- or multicentric and/or others lack proteins required for centromere function.
Factors affecting chromosome segregation.
Among the artificial chromosomes examined in this study, we observe a wide range of missegregation rates (Fig. 3). Furthermore, the frequency of anaphase lag is greater in artificial chromosomes than in normal chromosomes (Table 2). Unlike natural human chromosomes, artificial chromosomes must form de novo centromeres in culture, a process requiring specific DNA sequences (40, 69), centromeric proteins, and epigenetic chromatin modifications (8). As each artificial chromosome centromere is created independently, some may establish better centromeric configurations than others.
Segregation errors may arise from impaired assembly of trans-acting centromeric factors on the artificial chromosome. For example, CENP-A, a histone H3 variant found at the centromere, plays a critical role in centromere function and is conserved from S. cerevisiae to humans (41, 48). While we did not directly address whether the artificial chromosomes in this study contain CENP-A, its presence is suggested by the localization of the centromere protein CENP-E (Table 1) (14, 16, 22, 33). Furthermore, CENP-E associates with only a subset of the total mass of alpha satellite DNA on both normal and artificial chromosomes (Fig. 1) (14, 45, 51, 63). Whether the remaining alpha satellite is truly redundant or whether it serves a more general function as pericentric heterochromatin remains unknown (49). Thus, at the current level of analysis, there are no indications that interactions between kinetochore proteins and centromeric DNA are compromised.
Examination of centromere function in model organisms demonstrates that, in addition to the CENPs, other proteins are required at the centromere to ensure proper chromosome segregation. For example, deletion of the heterochromatin protein HP1 homolog, Swi6, in fission yeast results in chromosome lag (3). Mouse cells depleted for HP1 exhibit micronuclei, an indicator of missegregation (57). Checkpoint proteins monitor bipolar kinetochore attachment to spindle microtubules and initiate the metaphase-to-anaphase transition. The absence of checkpoint proteins necessary for the anaphase-promoting complex causes a delay in the onset of anaphase (38). Centromeric cohesion is also critical for chromosome segregation. The absence of Drosophila cohesion protein MEI-S332 results in chromosome segregation defects, as well as reduced transmission frequency of minichromosomes (28). Similarly, when Rad21, a member of the fission yeast cohesion complex, is depleted from fission yeast chromosomes, there is an increase in the incidence of chromosome lag (3). Therefore, the presence of heterochromatin, checkpoint, cohesion, or other as yet unidentified proteins may be necessary for proper segregation of artificial chromosomes, and one or more of these aspects may be suboptimal in the human artificial chromosomes examined here.
This study suggests that both chromosome and centromere structure play important roles in chromosome segregation. From our analysis of artificial, variant, and normal chromosomes, it appears that chromosome size and/or composition is involved in chromosome segregation. Future investigations into the organization of a range of different human artificial chromosomes may help to identify the factor(s) responsible for impaired de novo centromeres and give us more insight into the requirements (both genetic and epigenetic) for human chromosome segregation.
Acknowledgments
We thank Brenda Grimes, Bala Balakumaran, Kristin Scott, Mary Schueler, Beth Sullivan, Gil Van Bokkelen, and John Harrington for useful discussions.
This work was supported in part by a Franklin Delano Roosevelt research grant from the March of Dimes Birth Defects Foundation and by a Sponsored Research Agreement from Athersys, Inc. Both R.W.M. and H.F.W. have a financial interest in Athersys, and this potential conflict of interest has been disclosed to and managed by Case Western Reserve University. M.K.R. was supported in part by a training grant from the National Institutes of Health.
REFERENCES
- 1.Alexandrov, I., A. Kazakov, I. Tumeneva, V. Shepelev, and Y. Yurov. 2001. Alpha-satellite DNA of primates: old and new families. Chromosoma 110:253-266. [DOI] [PubMed] [Google Scholar]
- 2.Beach, D., M. Piper, and S. Shall. 1980. Isolation of chromosomal origins of replication in yeast. Nature 284:185-187. [DOI] [PubMed] [Google Scholar]
- 3.Bernard, P., J. F. Maure, J. F. Partridge, S. Genier, J. P. Javerzat, and R. C. Allshire. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294:2539-2542. [DOI] [PubMed] [Google Scholar]
- 4.Bloom, K. S., and J. Carbon. 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29:305-317. [DOI] [PubMed] [Google Scholar]
- 5.Choo, K. H. 2001. Domain organization at the centromere and neocentromere. Dev. Cell 1:165-177. [DOI] [PubMed] [Google Scholar]
- 6.Cimini, D., D. Fioravanti, E. D. Salmon, and F. Degrassi. 2002. Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J. Cell Sci. 115:507-515. [DOI] [PubMed] [Google Scholar]
- 7.Clarke, L., H. Amstutz, B. Fishel, and J. Carbon. 1986. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 83:8253-8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 9.Copenhaver, G. P., K. Nickel, T. Kuromori, M. I. Benito, S. Kaul, X. Lin, M. Bevan, G. Murphy, B. Harris, L. D. Parnell, W. R. McCombie, R. A. Martienssen, M. Marra, and D. Preuss. 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286:2468-2474. [DOI] [PubMed] [Google Scholar]
- 10.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 11.Ebersole, T. A., A. Ross, E. Clark, N. McGill, D. Schindelhauer, H. Cooke, and B. Grimes. 2000. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum. Mol. Genet. 9:1623-1631. [DOI] [PubMed] [Google Scholar]
- 12.Frengen, E., B. Zhao, S. Howe, D. Weichenhan, K. Osoegawa, E. Gjernes, J. Jessee, H. Prydz, C. Huxley, and P. J. de Jong. 2000. Modular bacterial artificial chromosome vectors for transfer of large inserts into mammalian cells. Genomics 68:118-126. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimes, B. R., A. A. Rhoades, and H. F. Willard. 2002. Alpha-satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol. Ther. 5:798-805. [DOI] [PubMed] [Google Scholar]
- 15.Hall, S. E., G. Kettler, and D. Preuss. 2003. Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Res. 13:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrington, J. J., G. Van Bokkelen, R. W. Mays, K. Gustashaw, and H. F. Willard. 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 15:345-355. [DOI] [PubMed] [Google Scholar]
- 17.Henikoff, S., K. Ahmad, and H. S. Malik. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098-1102. [DOI] [PubMed] [Google Scholar]
- 18.Henning, K. A., E. A. Novotny, S. T. Compton, X. Y. Guan, P. P. Liu, and M. A. Ashlock. 1999. Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human alpha satellite and single-copy DNA sequences. Proc. Natl. Acad. Sci. USA 96:592-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hieter, P., D. Pridmore, J. H. Hegemann, M. Thomas, R. W. Davis, and P. Philippsen. 1985. Functional selection and analysis of yeast centromeric DNA. Cell 42:913-921. [DOI] [PubMed] [Google Scholar]
- 20.Higgins, A. W., M. G. Schueler, and H. F. Willard. 1999. Chromosome engineering: generation of mono- and dicentric isochromosomes in a somatic cell hybrid system. Chromosoma 108:256-265. [DOI] [PubMed] [Google Scholar]
- 21.Hyman, A. A., and P. K. Sorger. 1995. Structure and function of kinetochores in budding yeast. Annu. Rev. Cell Dev. Biol. 11:471-495. [DOI] [PubMed] [Google Scholar]
- 22.Ikeno, M., B. Grimes, T. Okazaki, M. Nakano, K. Saitoh, H. Hoshino, N. I. McGill, H. Cooke, and H. Masumoto. 1998. Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 16:431-439. [DOI] [PubMed] [Google Scholar]
- 23.Koshland, D., L. Rutledge, M. Fitzgerald-Hayes, and L. H. Hartwell. 1987. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell 48:801-812. [DOI] [PubMed] [Google Scholar]
- 24.Kouprina, N., T. Ebersole, M. Koriabine, E. Pak, I. B. Rogozin, M. Katoh, M. Oshimura, K. Ogi, M. Peredelchuk, G. Solomon, W. Brown, J. C. Barrett, and V. Larionov. 2003. Cloning of human centromeres by transformation-associated recombination in yeast and generation of functional human artificial chromosomes. Nucleic Acids Res. 31:922-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lansdorp, P. M., N. P. Verwoerd, F. M. van de Rijke, V. Dragowska, M. T. Little, R. W. Dirks, A. K. Raap, and H. J. Tanke. 1996. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 5:685-691. [DOI] [PubMed] [Google Scholar]
- 26.Larin, Z., and J. E. Mejia. 2002. Advances in human artificial chromosome technology. Trends Genet. 18:313-319. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J. Y., and T. L. Orr-Weaver. 2001. The molecular basis of sister-chromatid cohesion. Annu. Rev. Cell Dev. Biol. 17:753-777. [DOI] [PubMed] [Google Scholar]
- 28.Lopez, J. M., G. H. Karpen, and T. L. Orr-Weaver. 2000. Sister-chromatid cohesion via MEI-S332 and kinetochore assembly are separable functions of the Drosophila centromere. Curr. Biol. 10:997-1000. [DOI] [PubMed] [Google Scholar]
- 29.Mahtani, M. M., and H. F. Willard. 1990. Pulsed-field gel analysis of alpha satellite DNA at the human X chromosome centromere: high frequency polymorphisms and array size estimate. Genomics 7:607-613. [DOI] [PubMed] [Google Scholar]
- 30.Malik, H. S., and S. Henikoff. 2002. Conflict begets complexity: the evolution of centromeres. Curr. Opin. Genet. Dev. 12:711-718. [DOI] [PubMed] [Google Scholar]
- 31.Manuelidis, L. 1978. Chromosomal localization of complex and simple repeated human DNAs. Chromosoma 66:23-32. [DOI] [PubMed] [Google Scholar]
- 32.Marahrens, Y., and B. Stillman. 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255:817-823. [DOI] [PubMed] [Google Scholar]
- 33.Masumoto, H., M. Ikeno, M. Nakano, T. Okazaki, B. Grimes, H. Cooke, and N. Suzuki. 1998. Assay of centromere function using a human artificial chromosome. Chromosoma 107:406-416. [DOI] [PubMed] [Google Scholar]
- 34.Mejia, J. E., A. Alazami, A. Willmott, P. Marschall, E. Levy, W. C. Earnshaw, and Z. Larin. 2002. Efficiency of de novo centromere formation in human artificial chromosomes. Genomics 79:297-304. [DOI] [PubMed] [Google Scholar]
- 35.Mills, W., R. Critcher, C. Lee, and C. J. Farr. 1999. Generation of an approximately 2.4 Mb human X centromere-based minichromosome by targeted telomere-associated chromosome fragmentation in DT40. Hum. Mol. Genet. 8:751-761. [DOI] [PubMed] [Google Scholar]
- 36.Murray, A. W., N. P. Schultes, and J. W. Szostak. 1986. Chromosome length controls mitotic chromosome segregation in yeast. Cell 45:529-536. [DOI] [PubMed] [Google Scholar]
- 37.Murray, A. W., and J. W. Szostak. 1983. Construction of artificial chromosomes in yeast. Nature 305:189-193. [DOI] [PubMed] [Google Scholar]
- 38.Musacchio, A., and K. G. Hardwick. 2002. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 3:731-741. [DOI] [PubMed] [Google Scholar]
- 39.Nakaseko, Y., N. Kinoshita, and M. Yanagida. 1987. A novel sequence common to the centromere regions of Schizosaccharomyces pombe chromosomes. Nucleic Acids Res. 15:4705-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohzeki, J., M. Nakano, T. Okada, and H. Masumoto. 2002. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 159:765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer, D. K., K. O'Day, M. H. Wener, B. S. Andrews, and R. L. Margolis. 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Round, E. K., S. K. Flowers, and E. J. Richards. 1997. Arabidopsis thaliana centromere regions: genetic map positions and repetitive DNA structure. Genome Res. 7:1045-1053. [DOI] [PubMed] [Google Scholar]
- 43.Saffery, R., L. H. Wong, D. V. Irvine, M. A. Bateman, B. Griffiths, S. M. Cutts, M. R. Cancilla, A. C. Cendron, A. J. Stafford, and K. H. Choo. 2001. Construction of neocentromere-based human minichromosomes by telomere-associated chromosomal truncation. Proc. Natl. Acad. Sci. USA 98:5705-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindelhauer, D., and T. Schwarz. 2002. Evidence for a fast, intrachromosomal conversion mechanism from mapping of nucleotide variants within a homogeneous alpha-satellite DNA array. Genome Res. 12:1815-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schueler, M. G., A. W. Higgins, M. K. Rudd, K. Gustashaw, and H. F. Willard. 2001. Genomic and genetic definition of a functional human centromere. Science 294:109-115. [DOI] [PubMed] [Google Scholar]
- 46.Shampay, J., J. W. Szostak, and E. H. Blackburn. 1984. DNA sequences of telomeres maintained in yeast. Nature 310:154-157. [DOI] [PubMed] [Google Scholar]
- 47.Shen, M. H., J. W. Yang, J. Yang, C. Pendon, and W. R. Brown. 2001. The accuracy of segregation of human mini-chromosomes varies in different vertebrate cell lines, correlates with the extent of centromere formation and provides evidence for a trans-acting centromere maintenance activity. Chromosoma 109:524-535. [DOI] [PubMed] [Google Scholar]
- 48.Stoler, S., K. C. Keith, K. E. Curnick, and M. Fitzgerald-Hayes. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573-586. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan, B. A., M. D. Blower, and G. H. Karpen. 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2:584-596. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan, B. A., and S. Schwartz. 1995. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet. 5:2189-2198. [DOI] [PubMed] [Google Scholar]
- 51.Sullivan, B. A., A. D. Skora, H. D. Le, and G. H. Karpen. 2002. CENP-A chromatin occupies “euchromatic” histone modifications. Am. J. Hum. Genet. 71:218. [Google Scholar]
- 52.Sullivan, B. A., and P. E. Warburton. 1999. Studying progression of vertebrate chromosomes through mitosis by immunofluorescence and FISH, p. 81-101. In W. A. Bickmore (ed.), Chromosome structural analysis: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 53.Sullivan, B. A., and H. F. Willard. 1998. Stable dicentric X chromosomes with two functional centromeres. Nat. Genet. 20:227-228. [DOI] [PubMed] [Google Scholar]
- 54.Sun, X., H. D. Le, J. M. Wahlstrom, and G. H. Karpen. 2003. Sequence analysis of a functional Drosophila centromere. Genome Res. 13:182-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun, X., J. Wahlstrom, and G. Karpen. 1997. Molecular structure of a functional Drosophila centromere. Cell 91:1007-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Surosky, R. T., C. S. Newlon, and B. K. Tye. 1986. The mitotic stability of deletion derivatives of chromosome III in yeast. Proc. Natl. Acad. Sci. USA 83:414-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114-120. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka, T. U. 2002. Bi-orienting chromosomes on the mitotic spindle. Curr. Opin. Cell Biol. 14:365-371. [DOI] [PubMed] [Google Scholar]
- 59.Tyler-Smith, C., R. J. Oakey, Z. Larin, R. B. Fisher, M. Crocker, N. A. Affara, M. A. Ferguson-Smith, M. Muenke, O. Zuffardi, and M. A. Jobling. 1993. Localization of DNA sequences required for human centromere function through an analysis of rearranged Y chromosomes. Nat. Genet. 5:368-375. [DOI] [PubMed] [Google Scholar]
- 60.Uhlmann, F. 2003. Chromosome cohesion and separation: from men and molecules. Curr. Biol. 13:R104-R114. [DOI] [PubMed] [Google Scholar]
- 61.Walmsley, R. W., C. S. Chan, B. K. Tye, and T. D. Petes. 1984. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature 310:157-160. [DOI] [PubMed] [Google Scholar]
- 62.Warburton, P., and H. Willard. 1996. Evolution of centromeric alpha satellite DNA: molecular organization within and between human and primate chromosomes, p. 121-145. In M. Jackson, T. Strachan, and G. Dover (ed.), Human genome evolution. BIOS Scientific Publishers, Oxford, United Kingdom.
- 63.Warburton, P. E., C. A. Cooke, S. Bourassa, O. Vafa, B. A. Sullivan, G. Stetten, G. Gimelli, D. Warburton, C. Tyler-Smith, K. F. Sullivan, G. G. Poirier, and W. C. Earnshaw. 1997. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7:901-904. [DOI] [PubMed] [Google Scholar]
- 64.Waye, J. S., and H. F. Willard. 1986. Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol. Cell. Biol. 6:3156-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weier, H. U., J. N. Lucas, M. Poggensee, R. Segraves, D. Pinkel, and J. W. Gray. 1991. Two-color hybridization with high complexity chromosome-specific probes and a degenerate alpha satellite probe DNA allows unambiguous discrimination between symmetrical and asymmetrical translocations. Chromosoma 100:371-376. [DOI] [PubMed] [Google Scholar]
- 66.Wevrick, R., W. C. Earnshaw, P. N. Howard-Peebles, and H. F. Willard. 1990. Partial deletion of alpha satellite DNA associated with reduced amounts of the centromere protein CENP-B in a mitotically stable human chromosome rearrangement. Mol. Cell. Biol. 10:6374-6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wevrick, R., and H. F. Willard. 1989. Long-range organization of tandem arrays of alpha satellite DNA at the centromeres of human chromosomes: high frequency array-length polymorphism and meiotic stability. Proc. Natl. Acad. Sci. USA 86:9394-9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Willard, H. F. 1998. Centromeres: the missing link in the development of human artificial chromosomes. Curr. Opin. Genet. Dev. 8:219-225. [DOI] [PubMed] [Google Scholar]
- 69.Willard, H. F. 2001. Neocentromeres and human artificial chromosomes: an unnatural act. Proc. Natl. Acad. Sci. USA 98:5374-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willard, H. F., K. D. Smith, and J. Sutherland. 1983. Isolation and characterization of a major tandem repeat family from the human X chromosome. Nucleic Acids Res. 11:2017-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willard, H. F., and J. S. Waye. 1987. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J. Mol. Evol. 25:207-214. [DOI] [PubMed] [Google Scholar]
- 72.Yen, T. J., D. A. Compton, D. Wise, R. P. Zinkowski, B. R. Brinkley, W. C. Earnshaw, and D. W. Cleveland. 1991. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 10:1245-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yen, T. J., G. Li, B. T. Schaar, I. Szilak, and D. W. Cleveland. 1992. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature 359:536-539. [DOI] [PubMed] [Google Scholar]