Abstract
A genomic differential display method was developed that analyzes many restriction fragment length polymorphisms simultaneously. Interspersed repeat sequences were used to reduce DNA sample complexity and to target genomic subsets of interest. This work focused on trinucleotide repeats because of their importance in human inherited diseases. Immobilized repeat-containing oligonucleotides were used to capture genomic DNA fragments containing sequences complementary to the oligonucleotide. Captured fragments were amplified by PCR and fluorescently labeled using primers complementary to the repeat sequence and/or to the known sequences ligated to the ends of the restriction fragments. The labeled PCR fragments were displayed by size on a high-resolution automated fluorescent DNA sequencing instrument. Although there was a conservation in the overall pattern of displayed genome subsets, many clear and reproducible differences were detected when genomes from different individuals were compared. Fewer differences were detected within, than between, monozygotic twin pair genomes. In control experiments, the method distinguished between Huntington disease alleles with normal and expanded CAG repeat lengths.
Keywords: triplet repeats, PCR, monozygotic twins, human genetics, Huntington disease
Most molecular comparisons between cells with complex genomes focus on cDNAs. The use of cDNAs provides a sample with reduced complexity and a focus on expressed sequences. Most, if not all, of the comparative methods can be adapted to analyzing whole genomes. These approaches include subtractive hybridization (1–3), comparative genome hybridization (4), and a derivative method (5) that uses arrays of DNAs rather than metaphase chromosomes as targets. Direct cDNA sequencing has also been used when cost has not been an issue. An alternative DNA sequencing approach uses templates composed of multiple short cDNA sequence tags (6). Another widely used method is mRNA differential display (7–9), in which random cDNAs are amplified and displayed by size on an electrophoresis gel.
This paper describes a genomic differential display method. Genome complexity is reduced, and focus is provided by targeting genome subsets containing specific interspersed repeats. Here, the targeted genomic subsets contain a CAG trinucleotide repeating sequence, because of the importance of expanded CAG repeats in an increasing number of neurological diseases (for reviews, see refs. 10 and 11), and because these sequences preferentially occur in coding sequences (12). A previously described analytical approach that detects expanded trinucleotide repeats uses tandem ligation of short complementary probes (13). However, this method does not facilitate the isolation of the expanded repeat, since no surrounding unique sequences are identified.
Differential display using interspersed repeat hybridization experiments has been used in the past for genomic fingerprinting (14, 15). PCR-based genomic fingerprinting using microsatellite repeats has also been described (16, 17). The method described here uses sequence-specific capture and PCR to analyze many repeat-containing sequences simultaneously and allows the isolation of individual fragments with unique flanking sequences.
MATERIALS AND METHODS
Materials and Samples.
Nonphosphorylated oligonucleotides (Table 1) were from Operon Technologies (Alameda, CA). DNA samples were from an anonymous healthy donor, a Huntington disease (HD)-affected kindred (18), and monozygotic twins (19).
Table 1.
Oligonucleotide sequences
| No. | Description | Sequence (5′ → 3′)* |
|---|---|---|
| 1 | Sau3A I-adapter-24 | CGGGAATTCTGGCTCTGCGACATG |
| 2 | Sau3A I-adapter-10 | GATCCATGTC |
| 3 | MseI-adapter-20 | TCTCCAGCCTCTCACCGCAT |
| 4 | MseI-adapter-11 | TAATGCGGTGA |
| 5 | Sau96 I-adapter-22 | AGCACTCTCCAGCCTCACCGCG |
| 6 | Sau96 I-adapter-11 | GNCCGCGGTGA |
| 7 | CAG-12 | b-GATGATCCGACGCAT(CAG)12 |
| 8 | CCG-12 | b-GATGATCCGACGCAT(CCG)12 |
| 9 | HD2F | ATGAAGGCCTTCGAGTCCCTCAAGTCCTT |
| 10 | HD1R | GGAAGGACTTGAGGGACTCGAAGGCCTTCAT |
| 11 | HD1F | ACGGCCGCTCAGGTTCTGCTTTTAC |
| 12 | HD2R | GGCGGCTGAGGAAGCTGAGGA |
| 13 | CTG-4 | A(A/T/C)(CTG)4 |
| 14 | CAG-4 | T(A/T/C)(CAG)4 |
*b, Biotin.
Capture of Targeted Genome Subsets.
Genomic DNA (100 ng), digested with the restriction enzyme Sau3A I or MseI, was ligated with 50 pmol of corresponding adapters (oligonucleotides 1 and 2, and 3 and 4 for MseI, respectively; Table 1) in a 10–20 μl total reaction volume overnight at 14°C with 40 units of T4 DNA ligase (New England Biolabs). Each pair of oligonucleotides was first annealed by cooling the mixture from 70°C to 10°C in a 1-hr period. Ligase was inactivated by heating at 75°C for 10 min, and a fill-in reaction was done at 72°C for 10 min after the addition of dNTPs (100 μM each) and 0.5 unit of AmpliTaq DNA polymerase (Perkin–Elmer). DNA was phenol extracted, precipitated with ethanol, washed with 70% ethanol, dried, and dissolved in TE buffer (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA). A biotinylated oligonucleotide (10 pmol) containing a (CAG)12 or (CCG)12 sequence (oligonucleotide 7 or 8, respectively; Table 1) was mixed with 50 ng of ligation products in 50 μl of TE buffer containing 2 μM of the corresponding adapter oligonucleotides to prevent annealing of the fragment ends to each other. After the addition of mineral oil, the sample was heated to 95°C, slowly cooled to room temperature, added to 100 μg of prewashed streptavidin-coated magnetic beads M-280 [as directed by Dynal (Oslo)] using a 3-fold molar excess of biotin-binding capacity over biotinylated oligonucleotides, and incubated at room temperature for 1 hr with gentle rotation. The beads were collected with a magnet, washed twice at 55–60°C for 20 min with 3× standard saline citrate (SSC; 1× SSC = 0.15 M NaCl/15 mM sodium citrate) and 0.5% SDS and, at room temperature, twice each, with TE containing 1 M NaCl and with TE alone. Beads with captured DNA were stored in TE buffer at 4°C.
DNA Amplification and Labeling by PCR.
One-fifth of the captured DNA was amplified by PCR in a PTC-100 thermal cycler (MJ Research, Cambridge, MA) as described (1). The 50-μl reaction contained 67 mM Tris⋅HCl, pH 8.8/4 mM MgCl2/16 mM (NH4)2SO4/10 mM 2-mercaptoethanol/300 μM of each dNTP/2 units of AmpliTaq DNA polymerase/5 μM fluorescein-labeled adapter primer (i.e., in the absence of a repeat primer). The samples were incubated at 94°C for 3 min and subjected to 20–23 cycles, each consisting of 1 min at 94°C and 3 min at 72°C, and a final incubation at 72°C for 5 min (e.g., Fig. 1).
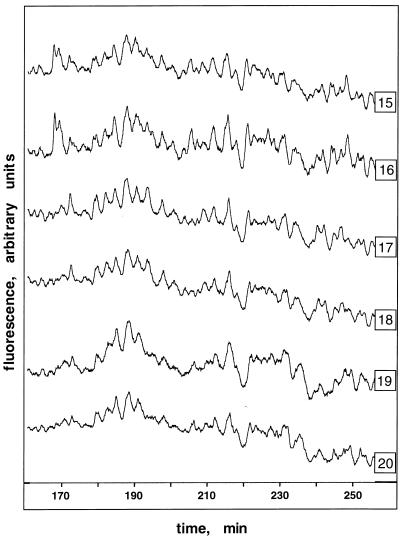
Figure 1.
Display of fluorescein-labeled MseI CAG-containing DNA fragments from three pairs of monozygotic twins. Genomic DNAs were digested with the restriction enzyme MseI, ligated to adapter oligonucleotides of known sequence, and hybridized to an immobilized single-stranded probe containing (CAG)12 repeats (see Materials and Methods). Captured DNAs were amplified by PCR with fluorescein-labeled primer 3 (Table 1) and displayed on an automated laser fluorescence (ALF) sequencing instrument. DNAs were from three pairs of monozygotic twins (pair 1, traces 15 and 16; pair 2, traces 17 and 18; pair 3, traces 19 and 20). The elution time (min) is shown on the x axis, and the fluorescence intensity is shown on the y axis (arbitrary units). The size range of the displayed fragments is ≈230 to ≈350 bp.
In other experiments, captured DNA was amplified by PCR using the appropriate adapter primer (2.5 μM) and a primer complementary to the repeat (5 μM, oligonucleotides 13 or 14; Table 1). PCR conditions were as described above, except that the annealing temperature was 45°C. The primer labeled with fluorescein varied in different experiments.
Display of Targeted Genome Subsets.
Amplified PCR products (1–2 μl) were denatured for 5 min at 90°C in 4 μl of a stop solution containing 6 mg/ml of dextran blue and 0.1% SDS in deionized formamide, loaded onto a 6% denaturing polyacrylamide gel, and analyzed on an ALF DNA Sequencer (Pharmacia–Biotech). The results were displayed using fragment manager software provided with the instrument. The size standard was a fluorescein-labeled 100-bp ladder (GIBCO/BRL). The electrophoresis conditions fractionated fragments from 80 to 800 bp.
Cloning and Analysis of Captured Sequences.
The CAG-containing double-stranded fragments obtained after capture and PCR amplification with adapter primers were cloned using a TA cloning kit (Invitrogen). Randomly chosen clones were sequenced using a Sequenase 2.0 kit (Pharmacia–Biotech) and an ALF sequencer.
Quantitation of CAG Repeats in the Human Genome.
One microgram of human genomic DNA was digested to completion with HindIII (New England Biolabs). The 5′ ends were labeled with 32P by a T4 polynucleotide kinase exchange reaction as recommended by New England Biolabs. Radiolabeled DNA (5.8 pmol) was hybridized to 5 pmol of a biotinylated (CAG-12) probe (oligonucleotide 7; Table 1) and captured on streptavidin-coated magnetic beads. Hybridization and washing conditions were as described above for purification of genome subsets. The amount of radioactive DNA retained on the beads was determined by Cerenkov counting in a Beckman scintillation counter.
Detection of Specific Allelic Differences in CAG-Containing Genome Subsets.
These control experiments were done to demonstrate that known allelic differences in the HD locus could be detected in a CAG-enriched genome subset. The samples compared were from nonidentical genomes. Thus, additional experiments were also done to determine the length of the HD alleles using locus-specific PCR and to identify the HD allele sizes produced by PCR with adapters and repeat primers.
A standard HD-specific PCR amplification (see Fig. 2 for relevant HD sequence and primer and restriction site locations) with oligonucleotides 9 and 12, or 11 and 12 (Table 1) was done to determine the HD allele sizes. Each 50-μl reaction contained 100 ng DNA/0.5 μM of each primer/20 mM Tris⋅HCl, pH 8.4/50 mM KCl/200 μM dNTPs/2 mM MgCl2/3.5% formamide/15% glycerol/2.5 units of AmpliTaq DNA polymerase. Cycling conditions were 94°C for 3 min, followed by 35 cycles at 94°C for 1 min, 64°C for 1 min, and 72°C for 1 min, and a final incubation at 72°C for 7 min.
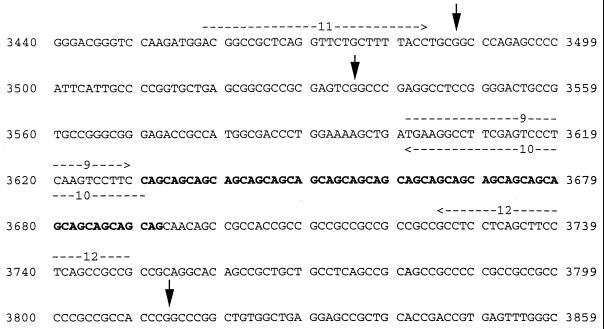
Figure 2.
Summary of HD sequence information used in these experiments. A partial sequence of the first exon of the HD gene (GenBank accession no. L34020L34020) is shown along with the locations of HD primers (oligonucleotides 9–12; Table 1; horizontal arrows) and Sau96 I restriction sites (vertical arrows) surrounding a CAG repeat (boldface type).
In other experiments, gel-purified HD fragments (≈150 ng), obtained from PCR amplification of genomic DNA with primers 11 and 12 (Table 1), were digested with Sau96 I and ligated to Sau96 I adapters (oligonucleotides 5 and 6; Table 1). The resulting HD fragments were then amplified by PCR using adapter primer 5 and fluorescein-labeled CTG-containing repeat primer 13 (Table 1). PCR conditions and analysis were as described above.
CAG repeat-containing genomic subsets, captured from Sau96 I-digested and tagged DNAs, were amplified by PCR using fluorescein-labeled primer 5 and a CAG-containing repeat primer (primer 14) or CTG-containing repeat primer 13 (Table 1). Two-microliter aliquots were displayed as described above. The remaining PCR product (≈100 ng) was hybridized overnight at 37°C to an immobilized HD-specific capture probe in 6× SSC/5× Denhardt’s solution/0.5% SDS/100 μg/ml of herring sperm DNA/100 pmol each of oligonucleotides (CAG)6 and (CTG)6. The capture probe, generated by PCR using oligonucleotide 10 and biotinylated oligonucleotide 11 (Table 1), was a 173-bp fragment upstream from the CAG repeat in the first exon of the HD gene. The gel-purified PCR product (175 ng, 1.5 pmol) was immobilized on streptavidin-coated magnetic beads and treated with alkali, as recommended by the manufacturer (Dynal). After hybridization, the beads were washed once in 1× SSC/0.5% SDS at 65°C for 2 hr and rinsed twice with TE at room temperature. Captured fragments were released from the beads by boiling for 5 min in stop solution and displayed as described above.
RESULTS
Principles of the Method.
The goal of this work was the development of a genomic differential display method targeting interspersed repeat-containing sequences. Major considerations were simplicity and the use of a minimal amount of DNA (200 ng/experiment). The procedure takes advantage of previously developed sequence-specific capture methods (20–25). CAG- (and CGG-) trinucleotide repeating sequences were targeted because of the importance of these repeats in neurodegenerative diseases (10, 11). It should be noted that only 10 capture probes are needed to profile all trinucleotide repeat sequences.
In brief, genomic DNA is cleaved with a restriction enzyme which cuts outside of the targeted repeat sequence. The restriction fragments are tagged at their ends by ligation to adapters—i.e., oligonucleotides of known sequence. The adapters permit subsequent amplification and labeling of the fragments by PCR. The fragments are denatured, hybridized to a biotinylated single-stranded oligonucleotide probe containing a sequence, (CAG)12 [or (CCG)12], complementary to the targeted repeat sequence, and captured on streptavidin-coated magnetic beads. Captured fragments are then amplified by PCR using an adapter primer alone or in combination with a primer complementary to the targeted repeat. Either the adapter or the repeat primer was fluorescein-labeled to allow detection of the products on a high-resolution automated fluorescent DNA sequencing instrument. Others have also used an automated DNA sequencing instrument for analysis for differential display (9, 26).
Efficacy of the Procedure.
Captured fragments containing CAG- (or CCG-) repeating sequences were amplified by PCR using an adapter primer and cloned. The clones were hybridized to the targeted repeat sequence. As expected, most (≈90% and ≈60% of the putative CAG and CCG clones, respectively) hybridized to the corresponding probes. Sequencing of four randomly selected CAG-containing clones revealed the presence of four different CAG repeated sequences—i.e., (CAG)3, (CAG)4, (CAG)6CCAGAGCCAG, and (CAG)2ACAGCA. These results showed that the capture procedure generated genome subsets enriched for targeted repeat-containing sequences.
The completeness of the capture CAG-containing genome subset was evaluated by determining the total number of 32P end-labeled HindIII restriction fragments captured by hybridization to an immobilized oligonucleotide containing (CAG)12 (oligonucleotide 7; Table 1). A total of ≈0.5% of the fragments (or ≈5 × 103 fragments) were captured. A similar number (≈2.5 × 103) of CAG repeats were predicted when human DNA sequences in GenBank were analyzed (12). This suggests that the capture procedure enriches for a particular genome subset with little loss of targeted sequences.
Reproducibility and Sensitivity of the Method.
Display of CAG repeat-containing fragments from three pairs of monozygotic twins is shown in Fig. 1. In this experiment genomic DNAs were digested with MseI, ligated to an adapter consisting of oligonucleotides 3 and 4 (Table 1), and hybridized to an immobilized single-stranded oligonucleotide probe containing (CAG)12 (oligonucleotide 7; Table 1). The captured DNA was amplified by PCR using a fluorescein-labeled adapter primer (oligonucleotide 3; Table 1). The fluorescence intensity versus elution time profiles, shown in Fig. 1, represents size-fractionated fluorescein-labeled fragments.
Although it was shown that the capture step is very efficient (see above), it is difficult to estimate the total number of displayed fragments. It is clear that the patterns are very complex, and many peaks are detected. Many peaks likely contain multiple fragments. Given the expected complexity of the captured genome subset and the expected average size of genomic MseI fragments (i.e., ≈160 bp), it is not likely that the entire CAG-containing MseI genome subset is shown. Large fragments will not be efficiently amplified by PCR or, even if amplified, they may be outside the size range analyzed. The annealing of the inverted terminal repeats at the ends of the fragments may prevent PCR amplification of some fragments (27, 28). The hairpin structures formed by long CAG repeats (29, 30) are likely to interfere with annealing to complementary repeat capture probes or PCR primers (see below and refs. 31 and 32). Nevertheless, the results show many clear differences between monozygotic twin pairs (Fig. 1). As expected, differences between pairs were much greater than differences within pairs. Fewer differences between pairs were observed when a fluorescein-labeled adapter primer was used in combination with a repeat primer (data not shown). Differences between samples were enhanced when the complexity of the captured fragments was reduced further by selective PCR using adapter primers anchored at their 3′ end to unique sequences flanking the repeat sequence, or by cleaving the captured sample with a second restriction enzyme before PCR labeling (data not shown). These results are reproducible in replicated experiments.
Detection of Specific Allelic Differences in the CAG-Containing Genome Subset.
The genomic differential display method was tested for its ability to distinguish different HD alleles (Fig. 3). The HD sequence relevant to these experiments is shown in Fig. 2. Many differences were expected between the samples used in these experiments. Thus, a number of control experiments were used to identify the HD-containing fragments.
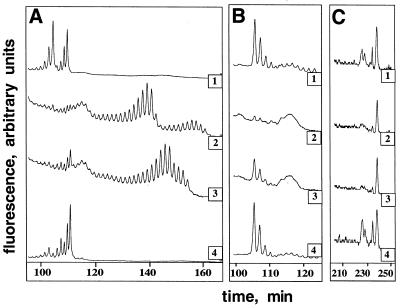
Figure 3.
Display of different HD alleles from three related individuals (traces: 1, HD-A; 2, HD-B; and 3, HD-C) and an anonymous unrelated control (trace 4) in a Sau96 I CAG-containing genome subset. Because there were many differences between samples, a number of control experiments was done to determine the size of the HD alleles, to identify HD-containing fragments, and to understand the primary structure of the PCR products using different primers. A subset of these control experiments is shown in A and B, whereas the true HD genomic display is shown in C. (A) HD-specific amplification of genomic DNA using primers 9 (fluorescein-labeled) and 12 (Table 1) measured the size of the HD alleles. (B) The HD fragments, generated from genomic DNA using primers 11 and 12 (Table 1 and Fig. 2A), were digested with Sau96 I, tagged with Sau96 I adapters, and amplified by PCR using primer 5 and fluorescein-labeled CTG-repeat primer 13 (Table 1). This experiment measured the size of the HD-containing fragments when the PCR used adapter and repeat primers. (C) Genomic DNAs were digested with the restriction enzyme Sau96 I, ligated to adapter oligonucleotides of known sequence, and hybridized to a (CAG)12-containing oligonucleotide probe. The captured fragments were amplified by PCR using CAG repeat-containing primer 14 and fluorescein-labeled primer 5 (Table 1). This experiment displays Sau96 I CAG-containing genome subsets en masse. The differentially displayed fragments eluting at ≈227 min hybridized to an HD-specific capture probe (data not shown). Also not shown are the results of experiments using adapter primers only. These experiments detected the same differentially displayed fragments eluting at ≈227 min as shown in C.
First, the length of the HD CAG repeat was determined in three members of an HD-affected kindred (HD-A, -B, and -C) and an unrelated (control) individual (Fig. 3A). The control sample had two normal alleles (i.e., both alleles had <30 repeats) as did the HD-A sample. Both the HD-B and HD-C samples had two expanded alleles (>40 repeats); these HD homozygotes were reported previously (18). The HD-B and HD-C samples also contained small amounts of fragments with shorter repeats. This type of HD mosaicism has been seen by others (33).
The next experiments (Fig. 3B) tested the ability of the method to distinguish between normal and expanded HD alleles in the absence of other genomic restriction fragments. The experiments took advantage of known Sau96 I restriction enzyme recognition sites located near the HD CAG repeat sequence. First, HD-specific PCR with primers (oligonucleotides 11 and 12; Table 1) flanking the repeat was used to generate HD-containing fragments with Sau96 I recognition sites proximal to the repeat. The PCR products were digested with Sau96 I, ligated to Sau96 I adapters (oligonucleotides 5 and 6; Table 1), and amplified by PCR using a Sau96 I adapter primer and a fluorescein-labeled repeat primer (oligonucleotides 5 and 13, respectively; Table 1). Clear differences were detected in the samples containing normal and expanded alleles (Fig. 3B). A heterogeneous ≈130-bp product, eluting at ≈105 min (Fig. 3B), was present in samples with normal HD alleles, but this fragment was absent in HD-B, and its amount was substantially reduced in HD-C. The expected size of the normal HD allele-containing fragments was ≥175 bases (the distance from the Sau96 I site to the distal 3′ end of the CAG repeat). An HD-containing fragment about 130 bp long could contain 4–6 CAG repeats. This result suggests that during PCR variable length CAG repeats with a low number of repeats (<40) were converted to a constant length equal to that contained in the repeat primer (16).
Because the repeat primer does not amplify across the repeat, one must explain why there are no HD fragments in the samples with expanded alleles (Fig. 3B). CAG repeat-containing fragments are known to form hairpin structures with stabilities that increase with repeat length (29, 30). These structures appear to inhibit PCR amplification of expanded alleles as seen in these experiments and as reported before (31, 32).
A broad faint peak eluting at ≈115 min (≈150 bp) was seen in the samples with expanded HD alleles and in control experiments in the absence of templates. This peak is apparently generated by primer interactions when there is no competing complementary sequence, supporting the notion that the long HD repeats were not available for annealing to the repeat primers.
Finally, experiments similar to those shown in Fig. 1 were done to demonstrate that the genomic differential display procedure can distinguish between normal and expanded HD lleles even when multiple templates were present. These experiments analyzed Sau96 I CAG-containing genome subsets expected to include the HD alleles (see Materials and Methods and Figs. 2 and 3C). The same basic procedure was used as that described for the experiments shown in Fig. 1, except for the change in restriction enzyme and the inclusion of the repeat primer. As expected, many differences were detected when the genomic DNAs from different individuals were compared. Because the fluorescent label was on the Sau96 I adaptor primer, two different HD-containing PCR products could be displayed. The longer fragment (≈330 bp, elution time ≈227 min) should be the PCR product formed by the adapter primer only. The shorter fragment (≈155 bp, elution time 115 min) should be the PCR product formed by the adapter and repeat primer. Fragments eluting at ≈227 min in HD-A and control samples were absent in the HD-B and HD-C samples (Fig. 3C). Small peaks at ≈115 min were detected in the HD-A and control samples but not in the HD-B and HD-C samples (data not shown). As expected, PCR with adapter primers alone also amplified the ≈330 bp fragments in the HD-A and control samples but not in the HD-B and HD-C samples (data not shown).
Additional experiments were done to prove that the ≈330-bp fragments contained the normal HD allele. A 173-base sequence adjacent to, but not including, the CAG-repeat region within the HD gene (see Materials and Methods and Fig. 2) was used as an HD-specific capture probe. The CAG-containing genome subsets shown in Fig. 3C were hybridized to the HD-specific capture probe and then displayed (data not shown). As expected, the fragments captured and displayed from the control and HD-A samples eluting at ≈227 min were not present in the HD-B and HD-C samples. Thus, the results in Fig. 3 show that our genomic differential display method is effective in distinguishing normal versus expanded CAG allele lengths.
This work characterized the different products that were generated when PCRs used adapter primers alone or adapter primers in combination with repeat primers. The greatest number of peaks was displayed when fluorescein-labeled adapter primers were used in combination with a repeat primer. In this case each genomic fragment can be potentially displayed twice; once as a PCR product of the adapter primers alone and second as a fragment amplified between the repeat and the adapter primer. The use of the adapter primers alone allows the simultaneous assessment of many short CAG-repeat lengths, whereas the use of adapter primers in combination with a repeat primer permits studies to focus on sets of unique sequences flanking trinucleotide repeats.
DISCUSSION
We have developed a genomic differential display method that reduces genome complexity by capturing a genomic restriction fragment subset containing a targeted interspersed repeat. Here, the focus was on trinucleotide repeating sequences. Other interspersed repeat-containing fragments could be targeted—e.g., fragments containing SINEs (short interspersed repeated DNA elements), LINEs (long interspersed repeats), LTRs (long terminal repeats), and sequences coding for particular protein motifs or cis-acting sequence elements. Differential display of cDNAs could also be enhanced by a similar use of interspersed repeats to target interesting cDNA subsets.
The HD control experiments described above took advantage of known differences in the CAG-repeat length of the HD gene. In most experiments, the cause of a specific restriction fragment length polymorphism would not be known a priori and the display method will also detect polymorphisms that may arise in the unique sequences surrounding the repeat. Thus, each displayed polymorphism must be characterized individually to understand its origin.
The informativeness of these and other differential display methods will be maximized when large amounts of data can by analyzed automatically. Thus, ongoing work is focused on high throughput automated analysis using signal processing methods (Y. H. Graber, N.E.B., C.L.S., and C. R. Cantor, unpublished results). The display method is also being used to assess the variation within monozygotic twin pairs (N.E.B., S. A. Lukyanov, Y. H. Graber, E. D. Sverdlov, and C.L.S., unpublished data). At a minimum, such experiments should provide quantitative information on genome stability, and they have the potential to reveal interesting facets of twin biology.
Acknowledgments
We thank C. R. Cantor, T. Sano, E. Sverdlov, V. Gindilis, J. Graber, and N. Bukanov for valuable discussions and critical reading of the manuscript, and the Venezuelan Huntington’s Disease Collaborative Group and H. Fuller Torrey for human samples. This work was supported by U.S. Department of Army Grant DAM17-94-J4414, U.S. Department of Energy Grant DE-F602-95-ER62040 to C.L.S., and U.S. Department of Energy Grant DE-FG02-93ER61609 to C. R. Cantor.
ABBREVIATIONS
- HD
Huntington disease
- ALF
automated laser fluorescence
References
- 1.Lisitzyn N, Lisitzyn N, Wigler M. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 2.Lisitzyn N, Launer G, Wagner L, Akopyanz N, Martynov V, Lelikova G, Limborska S, Sverdlov E. Biomed Sci. 1990;1:513–516. [PubMed] [Google Scholar]
- 3.Lisitzyn N, Rosenberg M, Launer G, Wagner L, Potapov V, Kolesnik T, Sverdlov E. Mol Genet Microbiol Virol. 1993;3:26–29. [PubMed] [Google Scholar]
- 4.Kallioniemi A, Kallioniemi O-P, Sudar D, Rutovitz D, Gray J W, Waldman F. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 5.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–471. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 6.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 7.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 8.McClelland M, Mathieu-Daude F, Welsh J. Trends Genet. 1995;11:242–246. doi: 10.1016/s0168-9525(00)89058-7. [DOI] [PubMed] [Google Scholar]
- 9.Ito T, Kito K, Adati N, Mitsui Y, Hagiwara H, Sakaki Y. FEBS Lett. 1994;351:231–236. doi: 10.1016/0014-5793(94)00867-1. [DOI] [PubMed] [Google Scholar]
- 10.Ross C A, McInnis M G, Margolis R L, Li S-H. Trends Neurosci. 1993;16:254–260. doi: 10.1016/0166-2236(93)90175-l. [DOI] [PubMed] [Google Scholar]
- 11.Sutherland G R, Richards R I. Proc Natl Acad Sci USA. 1995;92:3636–3641. doi: 10.1073/pnas.92.9.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stallings R L. Genomics. 1994;21:116–121. doi: 10.1006/geno.1994.1232. [DOI] [PubMed] [Google Scholar]
- 13.Schalling M, Hudson T J, Buetow K H, Housman D E. Nat Genet. 1993;4:135–139. doi: 10.1038/ng0693-135. [DOI] [PubMed] [Google Scholar]
- 14.Jeffreys A J, Wilson V, Thein S L. Nature (London) 1985;314:67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- 15.Uitterlinden A G, Slagboom P E, Knook D L, Vijg J. Proc Natl Acad Sci USA. 1989;86:2742–2746. doi: 10.1073/pnas.86.8.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weising K, Atkinson R G, Gardner R C. PCR Methods Appl. 1995;4:249–255. doi: 10.1101/gr.4.5.249. [DOI] [PubMed] [Google Scholar]
- 17.Zietkiewicz E, Rafalski A, Labuda D. Genomics. 1994;20:176–183. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]
- 18.Wexler N S, Young A B, Tanzi R E, Travers H, Starosta-Rubinstein S, Penney J B, Snodgrass S R, Shoulson I, Gomez F, Ramos Arroyo M A, Penchaszadeh G D, Moreno H, Gibbons K, Faryniarz A, Hobbs W, Anderson M A, Bonilla E, Conneally P M, Gusella J F. Nature (London) 1987;326:194–197. doi: 10.1038/326194a0. [DOI] [PubMed] [Google Scholar]
- 19.Torrey E F, Bowler A E, Taylor E H, Gottesman I I. Schizophrenia and Manic Depressive Disorder. New York: Harper Collins; 1994. , Appendix, pp. 1–2. [Google Scholar]
- 20.Ito T, Smith C L, Cantor C R. Nucleic Acids Res. 1992;20:3624. doi: 10.1093/nar/20.13.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broude N E, Sano T, Smith C L, Cantor C R. Proc Natl Acad Sci USA. 1994;91:3072–3076. doi: 10.1073/pnas.91.8.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kandpal R P, Kandpal G, Weissman S M. Proc Natl Acad Sci USA. 1994;91:88–92. doi: 10.1073/pnas.91.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown J, Hardwick L J, Wright A F. Mol Cell Probes. 1995;9:53–58. doi: 10.1016/s0890-8508(95)91022-0. [DOI] [PubMed] [Google Scholar]
- 24.Rothuizen J, van Raak M. Nucleic Acids Res. 1994;22:5512–5513. doi: 10.1093/nar/22.24.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parimoo S, Patanjali S R, Kolluri R, Xu H, Wei H, Weissman S M. Anal Biochem. 1995;228:1–17. doi: 10.1006/abio.1995.1308. [DOI] [PubMed] [Google Scholar]
- 26.Kato K. Nucleic Acids Res. 1995;23:3685–3690. doi: 10.1093/nar/23.18.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siebert P D A, Chenchik A, Kellogg D E, Lukyanov K A, Lukyanov S A. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukyanov K A, Launer G A, Tarabykin V S, Zaraisky A G, Lukyanov S A. Anal Biochem. 1995;229:198–202. doi: 10.1006/abio.1995.1402. [DOI] [PubMed] [Google Scholar]
- 29.Mariappan S V S, Garcia A E, Gupta G. Nucleic Acids Res. 1996;24:775–783. doi: 10.1093/nar/24.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gacy A M, Goellner G, Juranic N, Macura S, McMurray C T. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 31.Walsh P S, Erlich H A, Higuchi R. PCR Methods Appl. 1992;1:241–250. doi: 10.1101/gr.1.4.241. [DOI] [PubMed] [Google Scholar]
- 32.Demers D B, Curry E T, Egholm M, Sozer A C. Nucleic Acids Res. 1995;23:3050–3055. doi: 10.1093/nar/23.15.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldberg Y P, Kremer B, Andrew S E, Theilman J, Graham R K, Squitieri F, Telenius H, Adam S, Sajoo A, Starr E, Heiberg A, Wolff G, Hayden M R. Nat Genet. 1993;5:174–179. doi: 10.1038/ng1093-174. [DOI] [PubMed] [Google Scholar]