Abstract
Sister chromatid cohesion is fundamental for the faithful transmission of chromosomes during both meiosis and mitosis. Proteins involved in this process are highly conserved from yeasts to humans. In screenings for sterile animals with abnormal vulval morphology, mutations in the Caenorhabditis elegans evl-14 and scc-3 genes were isolated. Defects in cell divisions were observed in germ line as well as in vulval and somatic gonad lineages. Through positional cloning of these genes, we have shown that EVL-14 and SCC-3 are likely the only C. elegans homologs of the yeast sister chromatid cohesion proteins Pds5 and Scc3, respectively. Both evl-14 and scc-3 mutants displayed defects in the meiotic germ line. In evl-14 mutants, synaptonemal complexes (SCs) were detectable but more than the usual six DAPI (4′,6′-diamidino-2-phenylindole)-positive structures were seen at diakinesis, suggesting that EVL-14/PDS-5 is important for the maintenance of sister chromatid cohesion in late prophase. In scc-3 mutant animals, normal SCs were not visible and ∼24 DAPI-positive structures were seen at diakinesis, indicating that SCC-3 is necessary for sister chromatid cohesion. Immunostaining revealed that localization of REC-8, a homolog of the yeast meiotic cohesin subunit Rec8, to the chromosomes depends on the presence of SCC-3 but not that of EVL-14/PDS-5. scc-3 RNA interference (RNAi)-treated embryos were 100% lethal and displayed defects in cell divisions. evl-14 RNAi caused a range of phenotypes. These results indicate that EVL-14/PDS-5 and SCC-3 have functions in both mitosis and meiosis.
In eukaryotes, the faithful transmission of chromosomes during both meiosis and mitosis is critical for species propagation and the survival of individual organisms. Sister chromatid cohesion is fundamental to this process. In mitosis and meiosis II, sister chromatid cohesion ensures proper binding of sister kinetochores to spindle pole microtubules. In meiosis I, it is also required to arrange kinetochores associated with homologous chromosomes to face the appropriate spindle poles. Sister chromatid cohesion is established during the S phase and persists until the onset of anaphase (for reviews, see references 27, 35, and 47).
A multiprotein complex called cohesin is a major effector of sister chromatid cohesion. The complex consists of at least four conserved subunits, homologs of which have been identified in all major phyla. In Saccharomyces cerevisiae they are Scc1/Mcd1, Scc3, Smc1, and Smc3, which colocalize to chromatin in an interdependent manner (18, 31, 46). Scc1 is replaced in meiosis by its paralogue Rec8 (7, 25). In Caenorhabditis elegans there are four Scc1 homologs of which one, REC-8, is the likely worm ortholog of yeast Rec8 (34). Scc3 has multiple homologs in other species as well. Fission yeast contains two homologs of Scc3: Psc3 and the predicted meiosis-specific member Rec11 (13, 26, 45). Three Scc3 homologs, SA1 to SA3 (stromal antigen 1 to 3) (of which one, SA3, joins the meiotic cohesin complex) (8, 28, 36, 42), have been found in vertebrates.
Several other highly conserved proteins are also involved in sister chromatid cohesion. Studies of budding yeast have shown that cohesion is established by Scc2, Scc4, and Eco1/Ctf7 (9, 46), maintained by Pds5 (22, 33), and released by Esp1 (also known as separase) (48). Pds5 function is necessary for viability from the S phase through mitosis, and its localization to chromosomes depends on Scc1/Mcd1 (22, 33). Since Pds5 associates with the cohesin complex in a salt-sensitive manner, it does not appear to be an integral member of the complex (22, 33, 42). Interestingly, in fission yeast, cells lacking the pds5 gene can still germinate and propagate well and Pds5 is essential for sister chromatid cohesion only after prolonged time in G2 (43, 50).
Although recent studies have made progress towards understanding the mechanism of sister chromatid cohesion, many questions still need to be addressed. Here we report positional cloning and functional characterization of evl-14 and scc-3, which encode the likely sole homologs of yeast Pds5 and Scc3 in C. elegans, respectively. Studies of PDS-5 and SCC-3 in a relatively simple multicellular system (C. elegans) will help us further understand this mechanism.
MATERIALS AND METHODS
Strains.
The genotypes of strains used in this study were as follows: wild type (N2 Bristol), unc-36(e251) evl-14(ar96)/dpy-17(e164) unc-32(e189)III, unc-36(e251) evl-14(ar97)/dpy-17(e164) unc-32(e189)III, GS387 unc-36(e251) evl-14(ar112)/dpy-17(e164) unc-32(e189)III (39), unc-93(e1500) dpy-17(e1640)III, ced-4(n1162)III, scc-3(ku263)/+V, rol-4(sc8) scc-3(ku263)/lin-25(n545ts)V, dpy-11(e224) unc-76(e911)V, sma-1(e30) unc-76(e911)V, spo-11(ok79)IV/nT1[unc(n754) let](IV;V), and kuIs29[unc-119(+) and cog-2::gfp(pWH17)]. All C. elegans strains were grown in cultures and maintained at 20°C according to standard procedures (6).
Genetic mapping and molecular cloning.
evl-14(ar96) was mapped to LGIII between unc-93 and dpy-17. A total of 47 of 69 Unc-non-Dpy recombinants from unc-93 dpy-17/evl-14 heterozygotes segregated evl-14. A total of 4 of 13 Dpy-non-Evl recombinants from evl-14 dpy-17/ced-4 heterozygotes segregated ced-4. scc-3(ku263) was mapped to LGV between dpy-11 and unc-76. A total of 10 of 12 Dpy-non-Unc recombinants and 2 of 8 Unc-non-Dpy recombinants from dpy-11 unc-76/ku263 heterozygotes segregated ku263. A total of 7 of 23 Sma-non-Unc recombinants and 10 of 17 Unc-non-Sma recombinants from sma-1 unc-76/ku263 heterozygotes segregated ku263. A total of 15 of 17 Rol-non-Pvl-Ste recombinants from rol-4 ku263/lin-25 heterozygotes segregated Bag. Transgenic strains were generated by germ line transformation (30). Cosmids and fosmids were obtained from the Sanger Center. Fosmid H38K22 (10 ng/μl) was injected together with pTG96, the sur-5::gfp marker (17) (100 ng/μl), into the unc-36 evl-14(ar96)/dpy-17 unc-32 strain. Positive rescuing activity was scored by restoration of fertility to transgenic animals with the unc-36 phenotype. The minimal rescuing subclones contained one open reading frame (ORF), H38K22.1. Cosmids (15 ng/μl) covering the scc-3 region were injected together with 100 ng of the sur-5::gfp marker/μl into the rol-4 scc-3(ku263)/lin-25 strain. Positive rescuing activity was scored by restoration of fertility to transgenic animals with the rol-4 phenotype. The minimal rescuing subclone contained one ORF, F18E2.3. Molecular lesions of mutations were determined by sequencing PCR-amplified genomic DNA from mutants. PCR amplification in the region around the 11th and 12th exons of H38K22.1 from evl-14(ar97) fail to generate DNA fragments, suggesting that ar97 animals are likely to have undergone a deletion or rearrangement in this region.
The N-terminal eight exons of the predicted evl-14 gene structure were not covered by cDNA clones (WormBase). We used primers corresponding to sequences in the predicted exons to perform a reverse transcription-PCR (RT-PCR) analysis of this gene. We obtained the cDNA sequence from exons 2 to 8, but not from the first exon, possibly due to technical difficulties.
Double-stranded RNAi.
evl-14 and scc-3 RNA interference (RNAi) treatment with the N2 and unc-36(e251) evl-14 (ar96)/dpy-17(e164) unc-32(e189) strains or with other strains with genetic mutations or markers was performed by either injection or feeding. For RNAi injection, the cDNA clones yk397b1 and yk226a12 were used as a template for the generation of evl-14 double-stranded RNA and yk448f1 was used as a template for the generation of scc-3 double-stranded RNA. RNAi was carried out as described previously (15). F1 progeny were scored for RNAi effects. For RNAi feeding, evl-14 cDNAs (yk397b1 and yk226a12) and scc-3 cDNA were cloned into pPD129.36 (a gift from A. Fire) and the resulting plasmids were transformed into the Escherichia coli strain HT115. evl-14 RNAi by feeding was performed essentially as described previously (4). L4 worms were individually placed onto RNAi plates. To examine postembryonic scc-3 RNAi phenotypes, adult hermaphrodites were bleached for the collection of eggs, which were then placed onto the RNAi plates. Alternatively, adult hermaphrodites were placed onto the RNAi plates and the phenotypes of the F1 survivors were examined. cDNAs were provided by Yuji Kohara (National Institute of Genetics, Mishima, Japan), and the RNAi results for the two evl-14 cDNAs (yk397b1 and yk226a12) were identical.
Gonad preparation and immunostaining.
For all immunostaining and fluorescence in situ hybridization (FISH) procedures, whole gonads were prepared as described previously (12). Immunostaining of gonads was performed as described previously (34). Rabbit anti-REC-8 (kindly provided by J. Loidl, University of Vienna) was used at a 1:100 dilution in an antibody (Ab) buffer (1% bovine serum albumin, 0.5% Triton X-100, 0.05% sodium azide-1× phosphate-buffered saline) and anti-Rb Cy3 (Jackson ImmunoResearch) was used at a 1:250 dilution in the Ab buffer.
DAPI staining and FISH.
Intact worms were fixed with Carnoy's solution (41) for 2 h and then incubated for 5 min with 0.1 μg of DAPI (4′,6′-diamidino-2-phenylindole)/ml. A BioNick labeling system (Invitrogen) was used to label the 5S ribosomal gene (rDNA) probe (11) with biotin-14-dATP. FISH was performed as described previously (12), with the following modifications. Briefly, ethanol-fixed preparations were washed twice for 10 min each in 2× SSCT (0.3 M NaCl, 0.03 M sodium citrate, 0.1% Tween 20) and subsequently RNase treated (30 μg per slide) for 30 min at 37°C. Slides were washed in 2× SSCT-20% formamide, 2× SSCT-40% formamide, and 2× SSCT-50% formamide for 10 min each and then transferred into fresh 2× SSCT-50% formamide and incubated at 37°C for 30 min. Labeled probe DNA (100 ng) mixed with salmon sperm DNA (20 μg) was vacuum dried and resuspended in hybridization mix (3× SSC-50% formamide-10% dextran sulfate). The probe was denatured at 95°C, placed on ice, and dropped onto the slide. A coverslip was placed, and the sample was sealed using rubber cement. Slides were heated to 95°C for 3 min and incubated overnight at 37°C. Coverslips were removed by incubating slides in 2× SSCT-50% formamide in a Coplin jar at 42°C. Slides were washed successively in 1× SSC, 0.2× SSC, and 0.1× SSC at 42°C. Blocking solution (3% bovine serum albumin-0.1% Tween 20-4× SSCT) was added for 30 min at 37°C. Slides were stained with rhodamine B-conjugated streptavidin (Molecular Probes) for 40 min at 37°C and washed three times for 5 min in 4× SSCT-0.1% Tween 20. Finally, slides were mounted in Vectashield supplemented with 0.1 μg of DAPI/ml.
Microscopy.
Images were taken with a Hamamatsu digital camera on a Zeiss Axioplan 2 microscope or with a Diagnostic camera on a Leica DMRXA microscope. Deconvolution was performed with the use of OpenLab 2.0 software (Improvision). For time-lapsed images, a Leica DMRXE microscope equipped with a COHU camera was used. These images were taken using Scion Image 1.62c software (Scion Corporation), processed using Turnaround 3.24 software, and analyzed using 4D Viewer 4.12 software (44).
RESULTS
Mutations in evl-14 and scc-3 disrupted the development of the vulva and gonad.
evl-14 was defined by three recessive alleles, ar96, ar97, and ar112, that were isolated in a screening for mutations causing abnormal eversion of the vulva (39). We performed a similar EMS-induced F1 clonal screening to generate a collection of mutants that are both sterile and abnormal in vulval structure (14, 51). One recessive allele, ku263, was further analyzed in this study. A number of other mutations that produce both sterility and abnormality in vulval structure define genes involved in cell division and cell cycle regulation processes (e.g., see reference 14). Postembryonic cell division defects are also commonly seen in sterile uncoordinated mutants (2).
ku263 and the evl-14 mutants shared several postembryonic phenotypes (Fig. 1) that were indicative of common defects in cell division. The vulva is normally derived from three vulval precursor cells, each of which undergoes three rounds of precisely defined divisions to generate 22 vulva-specific cells (23). evl-14 mutants generated 18 ± 3 (n = 15) vulval cells, while scc-3 mutant animals produced 15 ± 3 (n = 12) vulval cells. These defects in cell division were likely to have been responsible for the asymmetric vulval invaginations observed at the L4 stage (Fig. 1B and C). Lineage analysis of six evl-14 mutant animals is shown in Fig. 1F.
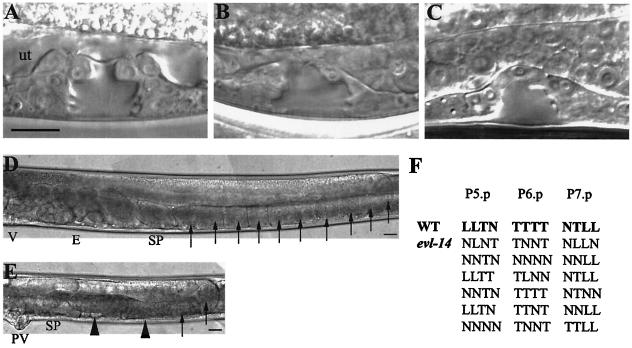
FIG. 1.
Vulval and gonadal abnormalities caused by evl-14(ar96) and scc-3(ku263). All images are oriented with the anterior portion to the left and the dorsal portion to the top. (A to C) Mid-L4-stage vulva in a wild-type and two mutant animals. (A) The wild-type vulva (containing 22 nuclei) displayed a symmetric “Christmas tree” shape of the vulval lumen. “ut” indicates the location of the uterine space. (B and C) The irregular and asymmetric shape of the vulval lumen displayed in evl-14 (B) and scc-3 (C) mutants resulted from fewer vulval cells. (D) Posterior arm of the gonad in a young adult wild type. V, vulva; E, embryos; SP, spermatheca. Arrows indicate oocytes. (E) Posterior arm of the gonad in a young adult evl-14 mutant animal. Note lack of embryos and displayed endomitotic oocytes. PV, protruding vulva. Arrowheads indicate endomitotic oocytes. Similar gonad phenotypes were seen in scc-3 mutants. (F) Lineage analysis of the evl-14 mutant vulva. Each line represents either a wild-type (first line) or evl-14 mutant pattern of final divisions of P5.p to P7.p (23). T, cell division along the left-right (transverse) axis; L, cell divided along the anterior-posterior (longitudinal) axis; N, cell did not divide. The genotypes of the two mutant strains used for the experiments whose results are shown here and in Fig. 4 to 6 are as follows: unc-36(e251) evl-14(ar96) and rol-4(sc8) scc-3(ku263). Bars, 10 μm.
evl-14 and scc-3 mutants also showed gonadal defects and never produced embryos (Fig. 1E). Two gonad arms with normal turns were observed in both mutants, but the size of the gonad was about one-half of that seen in wild-type animals. In wild-type animals, embryos are contained in the uterus, which is located at the most proximal end of the gonad (Fig. 1D). In evl-14 and scc-3 mutant animals, uteri were abnormal and small (Fig. 1E and data not shown). This may in part have been due to the presence of fewer uterine cells (see below and Fig. 2). Additionally, 100% of evl-14 and scc-3 mutants also displayed an endomitotic oocyte (Emo) phenotype (Fig. 1E), in which chromosomes have gone through endoreduplication without cytokinesis and karyokinesis (24). The Emo phenotype often results from the lack of somatic gonad cells (5, 29). Spermatheca in young adult mutants did not display obvious morphological defects (data not shown).
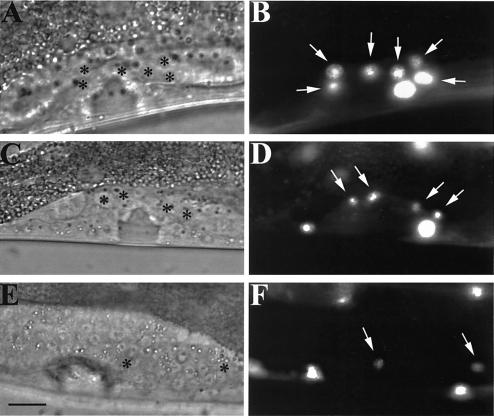
FIG. 2.
scc-3 and evl-14/pds-5 mutants have abnormal π cell linage. (A, C, and E) Differential interference contrast images of L4 hermaphrodites carrying an integrated cog-2::gfp fusion construct. (B, D, and F) Fluorescence images corresponding to images in panels A, C, and E. Arrows, π cells. (A and B) Wild-type worm; (C and D) evl-14 RNAi-treated worm; (E and F) scc-3 RNAi-treated worm. In the fluorescence images, cog-2::gfp expressed in π cells and body wall muscle cells is depicted. Asterisks in panels A, C, and E indicate π cells. Six π cells (ventral uterine cells) are visible in the wild-type image (B). Fewer π cells appeared in the animals treated with either evl-14 RNAi or scc-3 RNAi. Bar, 10 μm.
To further examine the somatic gonad defects in scc-3 and evl-14 mutants, we performed RNAi on wild-type animals carrying an integrated cog-2::gfp fusion array (see Materials and Methods). cog-2::gfp is a π-cell-specific marker (21). π precursor cells are 6 of the 12 granddaughters of the ventral uterine cells. Following induction by a signal from the gonadal anchor cell, the π precursor cells divide to form 12 π cells (32). In wild-type animals, the six π precursor cells express cog-2::gfp at the early- to mid-L4 stage (Fig. 2B). RNAi with both scc-3 and evl-14 strains resulted in fewer π cells at equivalent stages of development (Fig. 2D and F). RNAi-treated worms still had anchor cells (data not shown); therefore, the reduction in the number of π cells was likely to have been the result of failed cell divisions or cell cycle arrest or quiescence which would have prevented further divisions. Therefore, the somatic gonad defects in scc-3 and evl-14 mutants were at least partially due to cell division defects.
evl-14 and scc-3 encode the likely homologs of the yeast Pds5 and Scc3, respectively.
evl-14 had been previously mapped to chromosome III between unc-93 and dpy-17 (39). Using ced-4 and dpy-17, we placed evl-14 in a small genetic region (see Materials and Methods). DNA-mediated microinjection transformation experiments were performed to identify the affected genetic locus. The mutant phenotypes were rescued by a single fosmid clone, H38K22. Coinjecting, but not individually injecting, two subclones that contained overlapping DNA fragments of H38K22 also rescued the evl-14 mutant phenotypes. These two clones together contained a single predicted ORF, H38K22.1. Sequencing this ORF in ar96 mutant animals revealed a single nucleotide substitution (C to T) which created a premature stop codon at residue 372 in the seventh exon. In ar112 animals, a single nucleotide substitution was found to alter the 5′ splice site (GT to AT) at the end of the 11th exon. An in-frame stop codon is present in the following intron; therefore, splicing failure at the site leads to a truncated protein or a rapidly degraded message. The molecular nature of the lesions identified in these alleles and the similarity of the phenotypes that they produced strongly suggest that ar96 and ar112 are null or strong loss-of-function (lf) alleles (Fig. 3C). Protein sequence comparison using the NCBI BLAST analysis revealed that EVL-14 is 18% identical and 34% similar to the S. cerevisiae cohesin-associated protein Pds5 throughout the entire length of the protein. The similarity of EVL-14 to human AS3 (androgen-induced proliferation inhibitor) is higher than to the other family members examined (Fig. 3A). In addition, evl-14 appears to be the only gene in the C. elegans genome with significant similarity to the yeast pds5.
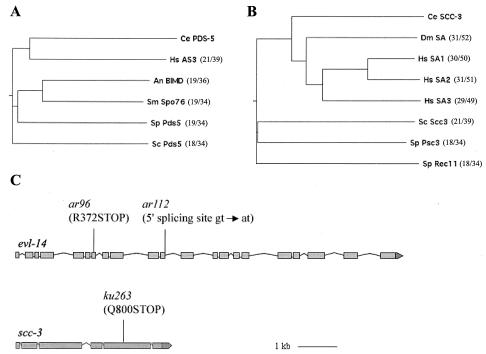
FIG. 3.
evl-14 and scc-3 encode cohesion proteins. (A and B) Phylogenetic trees of PDS-5 and SCC-3 family members in different species. The trees were computed using MacVector 6.5.3 software. Ce, C. elegans; Hs, Homo sapiens; An, Aspergillus nidulans; Sm, Sordaria macrospora; Sp, Schizosaccharomyces pombe; Sc, S. cerevisiae; Dm, Drosophila melanogaster. The overall degrees of relatedness between the C. elegans proteins and those in other species are indicated in parentheses (percent identity/percent similarity). (C) evl-14 and scc-3 gene structures. Gene structures were determined on the basis of assessment of WORMBASE and cDNA clones. For evl-14, there are no existing cDNA clones that cover the N-terminal region (exons 1 to 8). We have performed RT-PCR analysis of the gene and obtained cDNA sequences from exons 2 to 8. The locations and natures of the molecular lesions of three alleles are indicated.
We mapped ku263 to chromosome V between dpy-11 and unc-76. Further mapping placed ku263 just to the right of lin-25 (see Materials and Methods). We were able to rescue ku263 phenotypes with either the C15C8 or the F18E2 cosmid, indicating that the rescuing ORF is on both C15C8 and F18E2. A minimal rescuing subclone was created which contained a single predicted ORF, F18E2.3. This ORF encodes the C. elegans homolog of the budding yeast cohesin subunit Scc3 and was previously named scc-3 by curators at the Caenorhabditis Genetics Center. To confirm that the mutant phenotypes were caused by a mutation in this gene, the entire scc-3 ORF from ku263 mutant animals was sequenced. A single nucleotide substitution (C to T) (found in the fifth exon) created a premature stop codon at residue 800 (Fig. 3C). Therefore, ku263 is also likely to be a null or a strong loss-of-function (lf) allele. An NCBI Blast search with SCC-3 found homologs from yeast to human with 18 to 31% identity throughout the entire length of the protein (Fig. 3B). Like EVL-14, SCC-3 is likely to be the only homolog of the cohesin subunit Scc3/SA in C. elegans.
Loss of evl-14 and scc-3 gene functions disrupt sister chromatid cohesion during meiosis.
As determined on the basis of morphological changes, meiotic prophase is traditionally divided into five sequential stages: leptotene, zygotene, pachytene, diplotene, and diakinesis (1). In the C. elegans germ line, mitotic proliferation, meiotic prophase progression, and gametogenesis are present sequentially (38). From the distal to the proximal ends, the hermaphrodite gonad can be divided into several zones: the distal mitotic zone, the transition (Trans) zone, the pachytene zone, and the zone containing oocytes at diplotene and diakinesis stages. Within the Trans zone, nuclei progress from the mitotic cell cycle to the leptotene and zygotene stages of meiosis. The distinct morphologies of meiotic nuclei within the C. elegans gonad facilitate analyses of mutant defects in chromosome morphology, homolog pairing, and sister chromatid cohesion during the meiotic prophase.
In whole-mount gonads stained with DAPI for visualizing DNA, wild-type, evl-14, and scc-3 mutant animals showed similar densities of nuclei in the distal arm of the gonad (data not shown). In the proximal arm of the gonad, the number of oocytes in both scc-3 and evl-14 mutants was significantly reduced compared to the number seen with wild-type animals. The germ line zones of mitosis and meiosis appeared normal in evl-14 mutants, but oocytes displayed defects (Fig. 4 and data not shown). scc-3 mutant animals had the mitotic zone, but a clear Trans zone was not seen, and abnormal morphologies were observed in both pachytene nuclei and oocytes (Fig. 4 and data not shown). Additionally, DAPI staining also revealed that a DNA condensation event had occurred in many nuclei of the gonads of adult evl-14 and scc-3 mutants (data not shown), suggesting an increase in the number of apoptotic nuclei (19).
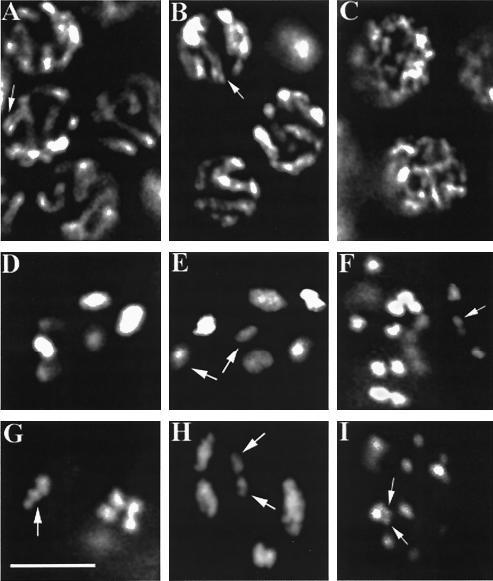
FIG. 4.
evl-14 and scc-3 mutants show cytological defects in meiotic germ line. (A to C) Nuclei in the pachytene zone of the germ line from a wild-type and two mutant animals. The evl-14 mutant nuclei (B) did not exhibit significant differences from wild-type nuclei (A). Both panels A and B display normal SCs (indicated by arrows). (C) The normal SCs are not visible in the scc-3 mutant nuclei. (D to I) Oocytes at the diakinesis stage. (D) The wild-type oocyte displays six symmetrical bivalents (three of them slightly out of focus), representing the six pairs of homologous chromosomes. (E) In the evl-14 mutant, seven DAPI-positive structures are visible, suggesting that homologous chromosomes in one pair were separated or that one sister chromatid drifted away from its partner. As determined on the basis of the size of the DAPI-stained area, two spots (indicated by arrows) likely represent the separated partners. (G) In an evl-14 mutant, one bivalent showed aberrant morphology (indicated by the arrow), suggesting a loosening of sister chromatid cohesion or a defect in chromosome condensation. (The other three bivalents were not in this focus plane.) (H) In an evl-14 mutant, one pair of homologs is slightly separated (indicated by arrows), suggesting a defect in sister chromatid cohesion. On the right side of this image, two bivalents overlap. In two scc-3 mutants (F and I), 25 and 26 DAPI structures are visible (only some of the spots are seen in one plane of focus), indicating that the sister chromatids were completely separated and that fragmentation occurred to at least one chromosome. Arrows indicate likely chromosomal fragments. Bar, 5 μm.
No obvious cytological differences were seen in mitotic germ cell and leptotene, zygotene (data not shown), and pachytene (n > 50 worms) nuclei between wild-type and evl-14 mutant animals. Synaptonemal complexes (SCs) appeared normal in pachytene nuclei of evl-14 mutants (Fig. 4A and B). However, defects in diakinesis of these mutants were observed. In wild-type diakinesis, six bivalents with a consistently symmetrical, compact shape were clearly observed (Fig. 4D). When we examined 37 oocytes from 25 evl-14 mutant animals in diakinesis, 29 of them displayed abnormal morphology: 18 nuclei exhibited seven or eight DAPI-positive structures (Fig. 4E), 5 nuclei exhibited slightly separated spots within bivalent-like DAPI structures (Fig. 4H), and 6 nuclei exhibited aberrant bivalents (Fig. 4G). The extra DAPI-positive spots suggest that homologous chromosomes or sister chromatids had become separated, while abnormal bivalents suggest that sister chromatid cohesion was weakened or impaired or that there were defects in chromosome condensation.
A very different DAPI-staining pattern was observed with scc-3(ku263) mutant animals from that observed with the wild type. We did not detect normal SCs in the pachytene zone of the scc-3 mutant gonad (as determined on the basis of the chromosome morphology) (Fig. 4C). The most striking feature was that at the diakinesis stage, 24.5 ± 1.6 (n = 35 nuclei) DAPI-positive structures were present in the 20 observed scc-3 mutant animals, indicating that sister chromatids were completely separated. Additionally, chromosome fragmentation may have occurred since up to four small DAPI-positive structures were present in addition to the 24 chromatids in some nuclei (Fig. 4F and I). Fragmentation was also observed in scc-3 RNAi-treated animals (data not shown). Such chromosome fragments may have originated from unrepaired meiotic double-stranded DNA breaks (DSBs). To address this question, we performed scc-3 RNAi in spo-11 mutant animals (see Materials and Methods). SPO-11 is an enzyme that creates DSBs during meiotic recombination (11). A total of 23.4 ± 0.7 (n = 20 nuclei) DAPI spots were seen in the scc-3 RNAi-treated spo-11 mutant worms, without additional small spots which would indicate the presence of extra chromosome fragments, suggesting that the extra fragments in scc-3 mutant or RNAi animals were generated by DSBs.
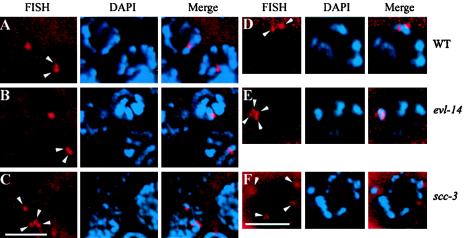
Using FISH, we further examined homologous chromosome pairing and sister chromatid cohesion in wild-type and mutant worms. A probe for the 5S rDNA, a repetitive sequence that maps close to the genetically defined pairing center on chromosome V (11), was generated and hybridized to fixed gonads from wild-type and mutant animals. In wild-type mitotic germ line nuclei, two randomly placed FISH signals were seen, indicating that the homologs were unpaired. The signals started approaching each other within the Trans zone of the gonad (reference 11 and data not shown). Throughout the pachytene zone, signals usually appeared as closely spaced doublets (n > 40) (Fig. 5A) (11). At diakinesis, 2.5 ± 0.7 (n = 20 nuclei) signals appeared on the same bivalent in wild-type oocytes (Fig. 5D).
FIG. 5.
Detection of homologous pairing and sister chromatid cohesion by FISH and DAPI staining. (A to C) Nuclei in the pachytene zone of the germ line. Arrowheads indicate the FISH signals. In the wild type (A) and the evl-14 mutant (B), two closely located FISH signals in each nucleus were detected, representing paired homologs and normal sister chromatid cohesion. (C) In contrast, four separated signals were detected in the scc-3 mutant, indicating the lack of sister chromatid cohesion. (D to F) Oocytes at the diakinesis stage. (D) In the wild type, two FISH signals were observed. (E) In the evl-14 mutant, four closely located FISH signals are visible, suggesting that the sister chromatid cohesion is not as stable as that in the wild type. (F) In the scc-3 mutant, four completely separated FISH signals are visible (two of them slightly out of focus), which is consistent with the data presented for panel C. Bar, 5 μm.
scc-3(ku263) animals showed 3.5 ± 0.5 (n = 45 nuclei from eight worms) separated FISH signals in the nuclei from the pachytene zone and four completely separated signals in oocytes at diakinesis (n = 9) (Fig. 5C and F), further suggesting that homologous chromosome pairing and sister chromatid cohesion were severely disrupted in the scc-3 mutants.
In evl-14 mutants 1.9 ± 0.3 (n > 40 nuclei from nine worms) FISH signals were seen in mitotic germ cells, and 1.6 ± 0.5 (n > 40 from nine worms) signals were seen in pachytene nuclei (Fig. 5B). These numbers are similar to those observed for the wild type, suggesting that it is unlikely that the extra DAPI-staining spots in the diakinesis oocytes in evl-14 mutant animals originated from the earlier aneuploidy events that occurred during mitosis. In evl-14 mutant oocytes, we observed an average of 3.5 ± 0.5 (n = 15 from nine worms) FISH signals on the same DAPI spot (Fig. 5). This number is slightly higher than that observed in the wild-type animals (2.5 ± 0.7), which might suggest that sister chromatid cohesion in evl-14 mutants was unstable. This result was consistent with the observation of the abnormal bivalents in evl-14 mutant oocytes (DAPI-staining images). However, we cannot exclude the possibility that the observed difference in FISH numbers in oocytes reflects subtle differences in the progression of diakinesis for the individual animals scored. Therefore, our results suggest that during C. elegans meiosis, sister chromatid cohesion cannot be established or maintained without SCC-3 and that EVL-4/PDS-5 is important for maintaining cohesion in late prophase.
The localization of REC-8 to chromosomes is completely disrupted in scc-3 mutants.
We next examined the relationships between REC-8 and EVL-14/PDS-5 or SCC-3. In budding yeast, Rec8 is a meiotic version of Scc1. The cleavage of Scc1/Rec8 by separase at the onset of anaphase promotes the separation of sister chromatids and the subsequent dissociation of the other cohesin subunits from chromosomes (37, 49). In C. elegans, SEP-1 (separase) has also been shown to be essential for homologous chromosome disjunction during meiosis I, and REC-8 might be its cleavage target (40). C. elegans REC-8 is the likely worm ortholog of yeast Rec8. REC-8 localized to the SCs at pachytene and chromosomal axes at diakinesis (34).
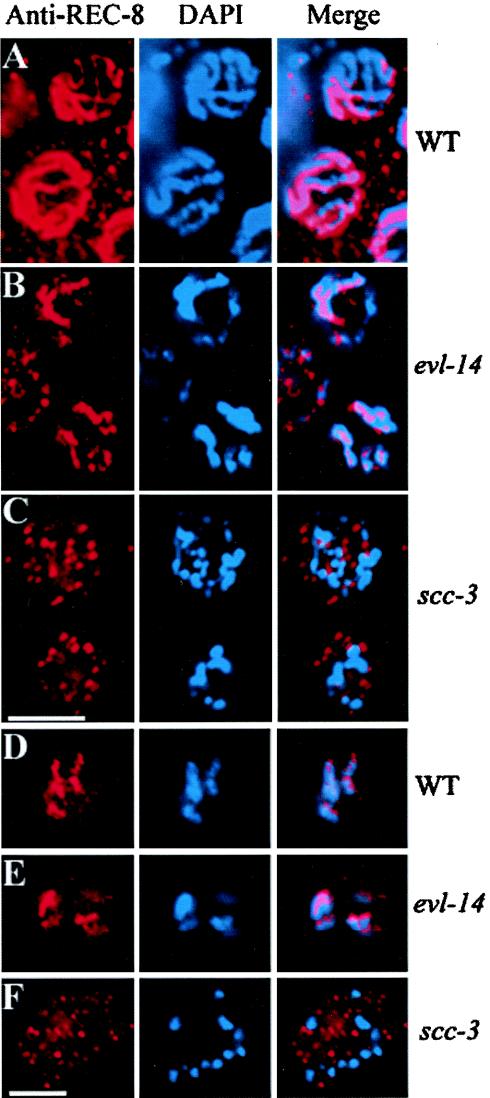
Immunostaining with anti-REC-8 antibodies was performed in wild-type, evl-14(ar96), and scc-3(ku263) mutant animals. The localization of REC-8 was not obviously affected in evl-14(ar96) (n > 20) animals, as determined by its colocalization to the DAPI signal. However, in scc-3 mutant gonads, REC-8 did not colocalize to DAPI but was seen scattered as small particles throughout the nuclei in the pachytene zone and at diakinesis (n > 40) (Fig. 6). Therefore, localization of REC-8 to the chromosomes in C. elegans depends on the presence of SCC-3.
FIG. 6.
Localization of REC-8 in evl-14(ar96) and scc-3(ku263) animals. (A to C) Nuclei in the pachytene zone of the germ line. (D to F) Oocytes at the diakinesis stage. In both wild-type (A and D) and evl-14 mutant (B and E) nuclei, the anti-REC-8 Ab staining (left panels) matches well with the DAPI staining (center panels) of the chromosomes, as shown in the right (Merge) panels. In contrast, the localization of REC-8 to the chromosomes was severely disrupted in scc-3 mutant nuclei (C and F). Bar, 5 μm.
scc-3 RNAi results in embryonic lethality and evl-14/pds-5 RNAi causes a range of phenotypes.
Animals heterozygous for evl-14 or scc-3 mutations display no observable phenotype, while homozygotes are sterile. It is possible that evl-14 and scc-3 are required for early development and that the lack of an obvious embryonic phenotype is due to the contribution of maternal gene products. To address this possibility, we performed RNAi, which can abolish the activities of transcripts from both zygotic and maternal sources. scc-3 RNAi performed by injection produced 100% embryonic lethality. RNAi with the evl-14 gene by both feeding and injection caused a range of phenotypes that included embryonic lethality, larval lethality, a high incidence of males (the Him phenotype), and sterility coupled with a protruding vulva (the Pvl-Ste phenotype) (Table 1). The Him phenotype is the result of the X chromosome nondisjunction during meiosis. Variations in phenotypes associated with evl-14 RNAi may be due to gene-dependent inefficiency in RNAi or due to the fact that evl-14 is not as essential a component of cohesion as is SCC-3. Examination of the gonads of Pvl-Ste worms after the RNAi treatment revealed cytological phenotypes similar to those of evl-14 mutants. We also performed double (scc-3 and evl-14) RNAi analysis and observed phenotypes similar to those seen with single scc-3 RNAi.
TABLE 1.
Phenotypes caused by evl-14 and scc-3 RNAi
| Gene and RNAi method | Genotypea | % of study population exhibiting indicated phenotype
|
n | ||||
|---|---|---|---|---|---|---|---|
| Embryo lethality | Larva lethalb | Pvl-Stec | Himd | WTe | |||
| scc-3 | |||||||
| Injection | WT | 100 | 0 | 0 | 0 | 0 | >100 |
| Feedingf | WT | NAg | 23.1 | 76.9 | 0 | 0 | 217 |
| evl-14 | |||||||
| Feeding | WT | 3.7 | 2.8 | 83.5 | 0.9 | 9.2 | 218 |
| evl-14(ar96)/ evl-14(+) | 7.2 | 3.9 | 86.5 | 2.4 | 0 | 207 | |
| Injection | WT | 13.8 | 4.6 | 79.4 | 2.3 | 0 | 218 |
| evl-14(ar96)/ evl-14(+) | 20.5 | 7.9 | 66.9 | 4.6 | 0 | 239 | |
Genotype of the worm strain being treated with RNAi.
Animals arrested at various larval stages.
Protruding vulva coupled with sterility phenotype.
High incidence of male (Him) phenotype; males resulted from self-fertilization of hermaphrodites.
Hermaphrodites with normal morphology. WT, wild type.
Embryos were put on RNAi feeding plates for examination of postembryonic phenotypes.
NA, not applicable.
To investigate the postembryonic phenotype caused by scc-3 RNAi, we placed wild-type embryos on scc-3 RNAi feeding plates (see Materials and Methods) and observed larval lethality and Pvl-Ste phenotypes which are similar to the postembryonic phenotypes seen with evl-14 RNAi (Table 1). The Pvl-Ste worms observed following scc-3 RNAi had defects similar to those of the scc-3(ku263) animals.
Embryonic lethality can be the result of meiotic and/or mitotic defects. To address the nature of the embryonic lethality associated with scc-3 RNAi, we used differential interference contrast optics to examine scc-3 RNAi embryos and generated time-lapse images for further analyses. All (n > 30) of the embryos we examined displayed abnormal mitosis (Fig. 7 and data not shown). Unlike wild-type animals, RNAi-treated embryos often contained nuclei with abnormal shapes which were often fragmented in daughter cells after a division (Fig. 7H), suggesting abnormal chromosome segregation. In some cases, blastomeres in RNAi embryos divided along abnormal axes (Fig. 7E to H).
FIG. 7.
scc-3 RNAi embryos presented abnormal mitosis at the early stage. Time-lapse images of ABa cell division of a wild-type (WT) embryo (A to D) and an scc-3 RNAi embryo (E to H) are shown. All images are oriented with the anterior portion to the left and the dorsal portion to the top. In the wild type the ABa cell divided with a left-right orientation, and its daughter cell, ABal (D), had one perfectly round nucleus. The sibling of ABal, ABar, was out of the focal plane beneath ABal. In the RNAi-treated embryo, the ABa cell divided with abnormal orientation and morphology. Note that the plane of division was dorsoventral. (H) An abnormally shaped and fragmented nucleus apparently formed in its daughter cell (indicated by the white outline). Bar, 10 μm.
The embryonic lethality observed in RNAi-treated animals may have been due to defects in meiosis prior to embryogenesis. We therefore examined the oocytes from RNAi-treated animals that produced embryonic lethal F1 progeny. Of 55 oocytes from 10 scc-3 RNAi-treated worms, 2 displayed aneuploidy (seven DAPI-staining spots), indicating a low level of penetrance of meiotic defects. For evl-14 RNAi, 0 of 51 examined oocytes from 10 RNAi-treated worms displayed aneuploidy. However, evl-14 RNAi did cause a Him phenotype of low penetrance that was a result of nondisjunction occurred during meiosis. Therefore, in addition to mitotic defects, meiotic defects may have also contributed to the embryonic phenotypes observed in scc-3 and evl-14 RNAi-treated worms.
DISCUSSION
EVL-14/PDS-5 and SCC-3 function in both mitosis and meiosis.
Sister chromatid cohesion is essential for both mitosis and meiosis. Some proteins involved in cohesion function strictly in meiosis or in mitosis, while some others function in both. The meiotic germ line defects in evl-14 and scc-3 mutants indicated that both genes are required for meiosis. That evl-14 and scc-3 mutants had cell division defects in the vulva and somatic gonad and that scc-3 RNAi embryos displayed mitotic defects suggest that they are required for mitosis. evl-14 and scc-3 are the only C. elegans genes identified so far with significant similarities to the yeast pds5 and scc3, respectively.
SCC-3 is essential for sister chromatid cohesion.
In budding yeast, all cohesin subunits are essential for establishing and maintaining sister chromatid cohesion and they colocalize to chromatin in an interdependent manner. Our observations of scc-3 mutants showed that (i) normal SCs were undetectable and sister chromatids separated in pachytene nuclei, (ii) 24 completely separated chromosomes at diakinesis were present, and (iii) no localization of REC-8 to the chromosomes was detectable. These results strongly suggest that scc-3 mutant animals were severely compromised for sister chromatid cohesion. Unlike the results seen with scc-3 mutants, extensive splitting of sister chromatids in the pachytene zone was rare in REC-8 RNAi worms (34). One possible explanation for this is that REC-8 is the meiosis-specific cohesin subunit, so its mitotic counterparts may provide a partial function in early meiotic prophase. However, when double RNAi was applied to rec-8 and apc-11, complete detachment of sister chromatids occurred during metaphase I arrest (10), which indicated that loosely associated sister chromatids in REC-8-depleted worms can be disrupted by microtubule-related prometaphase force.
Our observations that scc-3 mutants had approximately four separated FISH signals at the pachytene stage and were devoid of normal SCs support the theory that sister chromatid cohesion is required for chromosome synapsis as well. This explains our observation of 24 DAPI-positive structures (homologs and sisters were all separated) in scc-3 mutant oocytes later at diakinesis.
The presence of extra chromosomal fragments in scc-3 mutant animals and scc-3 RNAi-treated animals, but not in scc-3 RNAi-treated spo-11 worms, suggested that SCC-3 is required for the repairing of DSBs. In REC-8-depleted worms, similar chromosomal fragments that originated from DSBs were found at diakinesis (34). However, only a small number of extra chromosome fragments were observed in mutant or RNAi-treated oocytes at diakinesis. We thus cannot exclude the possibility that the scc-3 gene also plays a role in generating DSBs, and such a role in the mutant or in the RNAi-treated animals was compromised but not completely eliminated. Under this scenario, complete elimination of scc-3 activity would have prevented the occurrence of DSBs and chromosomal fragmentation.
Our observations of the lack of normal SCs in scc-3(ku263) animals, of approximately four separated FISH signals at the pachytene stage of oogenesis, and of 24 (and sometimes more) DAPI-positive structures in scc-3 mutant oocytes argue that the product of this gene is essential for sister chromatid cohesion in C. elegans.
The present model of the cohesin complex suggests that it is a large proteinaceous ring which can hold two DNA molecules (sister chromatids) together (3, 16, 20). Most of the circumference of the ring consists of the long arms of the Smc1/Smc3 heterodimer held together at one end by a hinge (the Smc dimer interface) and at the other end (at the Smc “heads”) by Scc1 and Scc3. Recently it has been shown that each of the two halves of Scc1 (separated after cleavage by separase) binds to one of the Smc heads, suggesting that Scc1 stabilizes the closed ring configuration (16, 20). The mechanism by which this ring is assembled and maintained is not well understood. But the evidence presented here suggests that SCC-3 is important for establishing cohesion. Further analysis is needed to determine the role of Scc3 in this process.
EVL-14/PDS-5 is important for maintaining sister chromatid cohesion in late prophase.
Our observations of the meiotic region of the gonad in evl-14 mutants and in Pvl-Ste worms treated with evl-14 RNAi showed that (i) no obvious defects were present in pachytene nuclei, (ii) REC-8 localization to the chromosomes did not depend on PDS-5, (iii) more than six DAPI-positive structures were present in some diakinesis nuclei, and (iv) three or four FISH signals were detectable in all diakinesis nuclei. In addition, RNAi against the evl-14 gene caused a Him phenotype, suggesting nondisjunction of the sex chromosome. RNAi against three scc1 homologues in C. elegans, coh-1, coh-2, and rec-8, also produced a Him phenotype (34). A Him phenotype can result from defects in sister chromatid cohesion, which is consistent with the evl-14 mutant phenotypes. Therefore, it seems that evl-14 is not necessary for establishing sister chromatid cohesion but is important for the maintenance of cohesion in late prophase. The role for evl-14 in this process is more like that of an assistant which strengthens the sister chromatid cohesion. Although the defects observed in this study and the sequence similarity between EVL-14 and yeast Pds5 suggest strongly a role of evl-14 in sister chromatid cohesion, the phenotypes in the evl-14 mutants and evl-14 RNAi-treated animals are also consistent with a role for the protein in other cellular functions that affect genome stability or segregation fidelity.
Acknowledgments
We thank Matt Siebert for RT-PCR work on evl-14, J. Loidl for anti-REC-8 antiserum, Iva Greenwald for evl-14 alleles, Yuji Kohara for cDNA clones, Bill Wood, Jinwei Zhao, and Qian Hu for the use of equipment, and Yo Suzuki, Dan Starr, Lois Edgar, and Quinn Crawford for comments on the manuscript. Some of the strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources of the NIH.
This study was supported by grants from Shanghai Municipal Government (Division of Sci. and Tech), China's NNSF (Oversea Young Scholar Collaborative Research Award), and the NIH of the United States (RO1, GM37869). J.Y. was supported by an NIH predoctoral training grant. I.A. was a research associate and M.H. is an associate investigator of HHMI.
REFERENCES
- 1.Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberts, and J. Watson. 1994. Molecular biology of the cell, 3rd ed., p. 1017. Garland Publishing, Inc., New York, N.Y.
- 2.Albertson, D. G., A. M. Rose, and A. M. Villeneuve. 1997. Chromosome organization, mitosis, and meiosis, p. 47-78. In D. L. Riddle, T. Blumenthal, B. J. Meyer, and J. R. Priess (ed.), C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 3.Anderson, D. E., A. Losada, H. P. Erickson, and T. Hirano. 2002. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 156:419-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoshechkin, I., and M. Han. 2002. The C. elegans evl-20 gene is a homolog of the small GTPase ARL2 and regulates cytoskeleton dynamics during cytokinesis and morphogenesis. Dev. Cell 2:579-591. [DOI] [PubMed] [Google Scholar]
- 5.Boer, B. G. W., S. Sookhareea, P. Dufourcq, and M. Labouesse. 1998. A tissue-specific knock-out strategy reveals that lin-26 is required for the formation of the somatic gonad epithelium in Caenorhabditis elegans. Development 125:3213-3224. [DOI] [PubMed] [Google Scholar]
- 6.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buonomo, S. B., R. K. Clyne, J. Fuchs, J. Loidl, F. Uhlmann, and K. Nasmyth. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103:387-398. [DOI] [PubMed] [Google Scholar]
- 8.Carramolino, L., B. C. Lee, A. Zaballos, A. Peled, I. Barthelemy, Y. Shav-Tal, I. Prieto, P. Carmi, Y. Gothelf, G. Gonzalez de Buitrago, M. Aracil, G. Marquez, J. L. Barbero, and D. Zipori. 1997. SA-1, a nuclear protein encoded by one member of a novel gene family: molecular cloning and detection in hemopoietic organs. Gene 195:151-159. [DOI] [PubMed] [Google Scholar]
- 9.Ciosk, R., M. Shirayama, A. Shevchenko, T. Tanaka, A. Toth, and K. Nasmyth. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5:243-254. [DOI] [PubMed] [Google Scholar]
- 10.Davis, E. S., L. Wille, B. A. Chestnut, P. L. Sadler, D. C. Shakes, and A. Golden. 2002. Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I. Genetics 160:805-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dernburg, A. F., K. McDonald, G. Moulder, R. Barstead, M. Dresser, and A. M. Villeneuve. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94:387-398. [DOI] [PubMed] [Google Scholar]
- 12.Dernburg, A. F., and J. W. Sedat. 1998. Mapping three-dimensional chromosome architecture in situ. Methods Cell. Biol. 53:187-233. [DOI] [PubMed] [Google Scholar]
- 13.DeVeaux, L. C., and G. R. Smith. 1994. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes Dev. 8:203-210. [DOI] [PubMed] [Google Scholar]
- 14.Fay, D. S., and M. Han. 2000. Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development. Development 127:4049-4060. [DOI] [PubMed] [Google Scholar]
- 15.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 16.Gruber, S., C. H. Haering, and K. Nasmyth. 2003. Chromosomal cohesin forms a ring. Cell 112:765-777. [DOI] [PubMed] [Google Scholar]
- 17.Gu, T., S. Orita, and M. Han. 1998. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell. Biol. 18:4556-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gumienny, T. L., E. Lambie, E. Hartwieg, R. H. Horvitz, and M. O. Hengartner. 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126:1011-1022. [DOI] [PubMed] [Google Scholar]
- 20.Haering, C. H., J. Lowe, A. Hochwagen, and K. Nasmyth. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9:773-788. [DOI] [PubMed] [Google Scholar]
- 21.Hanna-Rose, W., and M. Han. 1999. COG-2, a Sox domain protein necessary for establishing a functional vulval-uterine connection in Caenorhabditis elegans. Development 126:169-179. [DOI] [PubMed] [Google Scholar]
- 22.Hartman, T., K. Stead, D. Koshland, and V. Guacci. 2000. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 151:613-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvitz, H. R., and P. W. Sternberg. 1991. Multiple intercellular signalling systems control the development of the Caenorhabditis elegans vulva. Nature 351:535-541. [DOI] [PubMed] [Google Scholar]
- 24.Iwasaki, K., J. McCarter, R. Francis, and T. Schedl. 1996. emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J. Cell Biol. 134:699-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein, F., P. Mahr, M. Galova, S. B. Buonomo, C. Michaelis, K. Nairz, and K. Nasmyth. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98:91-103. [DOI] [PubMed] [Google Scholar]
- 26.Krawchuk, M. D., L. C. DeVeaux, and W. P. Wahls. 1999. Meiotic chromosome dynamics dependent upon the rec8+, rec10+ and rec11+ genes of the fission yeast Schizosaccharomyces pombe. Genetics 153:57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, J. Y., and T. L. Orr-Weaver. 2001. The molecular basis of sister-chromatid cohesion. Annu. Rev. Cell Dev. Biol. 17:753-777. [DOI] [PubMed] [Google Scholar]
- 28.Losada, A., T. Yokochi, R. Kobayashi, and T. Hirano. 2000. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 150:405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarter, J., B. Bartlett, T. Dang, and T. Schedl. 1997. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev. Biol. 181:121-143. [DOI] [PubMed] [Google Scholar]
- 30.Mello, C. C., J. M. Kramer, D. Stinchcomb, and V. Ambros. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 32.Newman, A. P., J. G. White, and P. W. Sternberg. 1995. The Caenorhabditis elegans lin-12 gene mediates induction of ventral uterine specialization by the anchor cell. Development 121:263-271. [DOI] [PubMed]
- 33.Panizza, S., T. Tanaka, A. Hochwagen, F. Eisenhaber, and K. Nasmyth. 2000. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 10:1557-1564. [DOI] [PubMed] [Google Scholar]
- 34.Pasierbek, P., M. Jantsch, M. Melcher, A. Schleiffer, D. Schweizer, and J. Loidl. 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15:1349-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petronczki, M., M. F. Siomos, and K. Nasmyth. 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112:423-440. [DOI] [PubMed] [Google Scholar]
- 36.Pezzi, N., I. Prieto, L. Kremer, L. A. Perez Jurado, C. Valero, J. Del Mazo, A. C. Martinez, and J. L. Barbero. 2000. STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking the Williams-Beuren syndrome deletion. FASEB J. 14:581-592. [DOI] [PubMed] [Google Scholar]
- 37.Ross, K. E., and O. Cohen-Fix. 2002. Separase: a conserved protease separating more than just sisters. Trends Cell Biol. 12:1-3. [DOI] [PubMed] [Google Scholar]
- 38.Schedl, T. 1997. Developmental genetics of the germ line, p. 244. In D. L. Riddle, T. Blumenthal, B. J. Meyer, and J. R. Press (ed.), C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 39.Seydoux, G., C. Savage, and I. Greenwald. 1993. Isolation and characterization of mutations causing abnormal eversion of the vulva in Caenorhabditis elegans. Dev. Biol. 157:423-436. [DOI] [PubMed] [Google Scholar]
- 40.Siomos, M. F., A. Badrinath, P. Pasierbek, D. Livingstone, J. White, M. Glotzer, and K. Nasmyth. 2001. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 11:1825-1835. [DOI] [PubMed] [Google Scholar]
- 41.Sulston, J., and J. Hodgkin. 1988. Methods, p. 599. In W. B. Wood (ed.), The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Sumara, I., E. Vorlaufer, C. Gieffers, B. H. Peters, and J. M. Peters. 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 151:749-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka, K., Z. Hao, M. Kai, and H. Okayama. 2001. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 20:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas, C., P. DeVries, J. Hardin, and J. White. 1996. Four-dimensional imaging: computer visualization of 3D movements in living specimens. Science 273:603-606. [DOI] [PubMed] [Google Scholar]
- 45.Tomonaga, T., K. Nagao, Y. Kawasaki, K. Furuya, A. Murakami, J. Morishita, T. Yuasa, T. Sutani, S. E. Kearsey, F. Uhlmann, K. Nasmyth, and M. Yanagida. 2000. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14:2757-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uhlmann, F. 2003. Chromosome cohesion and separation: from men and molecules. Curr. Biol. 13:R104-R114. [DOI] [PubMed] [Google Scholar]
- 48.Uhlmann, F., F. Lottspeich, and K. Nasmyth. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37-42. [DOI] [PubMed] [Google Scholar]
- 49.Uhlmann, F., D. Wernic, M. A. Poupart, E. V. Koonin, and K. Nasmyth. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103:375-386. [DOI] [PubMed] [Google Scholar]
- 50.Wang, S. W., R. L. Read, and C. J. Norbury. 2002. Fission yeast Pds5 is required for accurate chromosome segregation and for survival after DNA damage or metaphase arrest. J. Cell Sci. 115:587-598. [DOI] [PubMed] [Google Scholar]
- 51.Yoder, J. H., and M. Han. 2001. Cytoplasmic dynein light intermediate chain is required for discrete aspects of mitosis in Caenorhabditis elegans. Mol. Biol. Cell 12:2921-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]