Abstract
Maturation of Trypanosoma brucei mitochondrial mRNA involves massive posttranscriptional insertion and deletion of uridine residues. This RNA editing utilizes an enzymatic complex with seven major proteins, band I through band VII. We here use RNA interference (RNAi) to examine the band II and band V proteins. Band II is found essential for viability; it is needed to maintain the normal structure of the editing complex and to retain the band V ligase protein. Previously, band III was found essential for certain activities, including maintenance of the editing complex and retention of the band IV ligase protein. Thus, band II and band V form a protein pair with features analogous to the band III/band IV ligase pair. Since band V is specific for U insertion and since band IV is needed for U deletion, their parallel organization suggests that the editing complex has a pseudosymmetry. However, unlike the essential band IV ligase, RNAi to band V has only a morphological but no growth rate effect, suggesting that it is stimulatory but nonessential. Indeed, in vitro analysis of band V RNAi cell extract demonstrates that band IV can seal U insertion when band V is lacking. Thus, band IV ligase is the first activity of the basic editing complex shown able to serve in both forms of editing. Our studies also indicate that the U insertional portion may be less central in the editing complex than the corresponding U deletional portion.
Trypanosomes are anciently diverging parasitic protozoa that exhibit fascinating biological properties, most notably a remarkable processing of mitochondrial transcripts. This RNA editing involves insertion and deletion of uridine residues (U's) at specific sites (reviewed in references 1, 4, 14, and 38) and can be massive, generating over three-fourths of the codons in highly edited mRNAs. It is directed by separate short guide RNAs (gRNAs) that are complementary to edited sequence and thus mismatch the preedited mRNA at each site of editing (5). The first gRNA overlaps the 3′ end of the editing domain, so it hybridizes with the pre-mRNA to form an “anchor duplex,” with the abutting mismatch defining the first editing site. This (5′) single-strand-(3′) double-strand juncture directs endonucleolytic cleavage of the pre-mRNA (9, 10). Then at the 3′ end of the upstream fragment, U's are added or removed. Next the mRNA is rejoined by RNA ligase, and the anchor duplex “zips up” to the next mismatch, completing one editing cycle. As editing progresses upstream along the mRNA, it concomitantly corrects any errors that may have been introduced (8).
The comparable reactions of U insertion and U deletion could most simply be envisioned to use all common activities: the same endonuclease, a terminal-U-transferase (TUTase) that, in reverse, acts as a 3′-U-exonuclease (3′-U-exo), and the same ligase (15, 38). However, endonuclease activities for U insertion and U deletion have different responses to adenosine nucleotides (9) and to gRNA features (11, 20; unpublished data), the 3′-U-exo has none of the characteristics of a reverse TUTase (8, 32), and two editing RNA ligases have different abilities to ligate in U deletion (12, 18). These data suggested that all steps of U insertion and U deletion cycles may utilize distinct activities and that therefore the editing complex could have separate U deletional and U insertional halves (9-13).
A Trypanosoma brucei ∼20S complex contains the aforementioned RNA ligases (31, 32, 34). This complex remains together through chromatography on all examined resins (30, 32, 37), nondenaturing gel electrophoresis (32), and immunoprecipitation (28, 29), and it actively catalyzes U insertion and U deletion editing cycles (10, 12, 32). We find that all the enzymatic activities inferred in the editing cycles (gRNA-directed endonuclease, TUTase, 3′-U-exo, and RNA ligase) copurify with this complex (10, 32, 37). Our purified complex contains a simple pattern with seven major, approximately equimolar proteins and no detectable gRNA or mRNA (32, 37). These proteins are temporarily designated band I (largest) through band VII (smallest), with band IV and band V the RNA ligases (see reference 12 for introduction of a function-based nomenclature). Even though this purification method yields the most active preparations at supporting full-cycle U insertion and U deletion reactions yet reported (10, 11, 12), proteins in addition to these seven also appear important in RNA editing cycles (2, 3, 23, 40).
Other investigators have reported purified preparations containing 15 to 20 major proteins, plus gRNA and mRNA, that support several partial editing reactions and discernible full-cycle U deletion (23, 24, 28, 35). Only a few of the copurifying proteins appear to be contaminants (39), and at least six coimmunoprecipitate in a common complex (29); notably, they are our bands II through VII. These laboratories designate their proteins by molecular mass of the cytoplasmic precursors, which are variably larger than the active mitochondrial proteins (28).
Efforts to understand the individual proteins of the editing complex began with the ligases, identifiable by their autoadenylylation and deadenylylation (32, 34). Gene knockout showed that band IV (TbMP52) is essential for viability (18, 33, 36). Genetic and biochemical analyses demonstrated that this protein is needed to ligate in U deletion (12, 18) and has a structural role in the editing complex (18). Biochemical analysis showed that band V functions to ligate U insertion (12), but the literature has remained unclear on whether band IV could also ligate U insertion.
Other proteins of the editing complex have been examined by RNA interference (RNAi), inducibly expressing cognate double-strand RNA (dsRNA) to diminish the mRNA (26, 41). RNAi showed that band III is essential for viability, for cleavage at U deletion and U insertion sites, and for maintaining band IV in the editing complex (19). Another T. brucei protein shown essential by RNAi is a 108-kDa TUTase (2), although it may not be the TUTase that adds U residues in U insertion (3). RNAi has also been used to study band II and band V of the editing complex. While preparing a manuscript describing our results, Drozdz et al. (13) reported that RNAi to band II (TbMP81) appears to do the following: (i) slow the growth of procyclic trypanosomes by about half, (ii) not alter the sedimentation of the depleted editing complex, (iii) inhibit each assayed step of editing to various extents, most strongly the TUTase step, and (iv) diminish the amount of unadenylylated band V (TbMP48) (although this ligase result may be expected from a diminished TUTase product [13] that serves to deadenylylate band V ligase in vivo [12] and thus is not diagnostic for ligase abundance). Drozdz et al. (13) additionally report that RNAi to band V (TbMP48) has no growth effect on uncloned procyclic trypanosomes.
We here report our RNAi studies of band II and band V, with notably different results from those previously published (13). We start by cloning the gene for band II, demonstrating that this protein is an authentic member of the simple ligase-containing editing complex and determining its abundance. RNAi analyses then demonstrate that band II is absolutely essential for viability of procyclic trypanosomes and is critical for maintaining the normal ∼20S structure of the editing complex. Assessing total band V, independent of its adenylylation level, demonstrates that band II is needed to retain the band V ligase protein. We also show that band II is not needed to retain any of the other major proteins of the simple editing complex. This requirement is reminiscent of the structurally essential band III being needed to retain band IV ligase (19), suggesting a somewhat parallel organization within the editing complex for securing the ligases associated with U insertion and U deletion. Clonal cell lines showed that band V RNAi indeed does not affect the growth rate. However, we observe a morphological effect from band V depletion, suggesting a positive role of the U insertion-specific band V ligase. Analyses using a quantitative in vitro assay (12) demonstrate that, upon band V RNAi, the mRNA is sealed by band IV ligase in U insertion. This makes band IV the first activity found able to serve both forms of editing.
MATERIALS AND METHODS
Cloning the band II gene.
The band II gene was cloned much as for band IV (33) and band III (19), sequencing three tryptic peptides (Wistar protein facility, Philadelphia, Pa.) (Fig. 1A) of band II, which was gel isolated from the seven-major-protein RNA editing complex (32, 37). PCR of genomic (virtually intron-less) trypanosome DNA utilized degenerate primers (II-1a, 5′-GARGARGGNG ARGTNGTNGA RCC-3′; II-1b, 5′-GGYTCNACNA CYTCNCCYTC YTC-3′; II-2a, 5′-CAYGGNGARG ARYTNACNGA RGAYCA-3′; and II-2b, 5′-TGRTCYTCNG TNARYTCYTC NCCRTG-3′, marked in Fig. 1A by heavy underlines). The fragment from II-1b and II-2a, prepared with 2 mM MgCl2, was TA cloned into pCR2.1 (Invitrogen) to probe a T. brucei bloodstream form cDNA phage library (19). The longest clone was sequenced, and oligonucleotides to its upstream portion (5′-CGGGTGAGGC GTCTCATGAA CGATAC-3′; Fig. 1A) and to the miniexon (5′-CGCTATTATT AGAACAGTTTT CTGTACTATA TTG-3′) primed reverse transcriptase PCR (RT-PCR) of total cell RNA. Database searches for sequence used BLAST 2 (http://www.ncbi.nlm.nih.gov/BLAST) and GeneDB (http://www.genedb.org) with the low-complexity filter enabled.
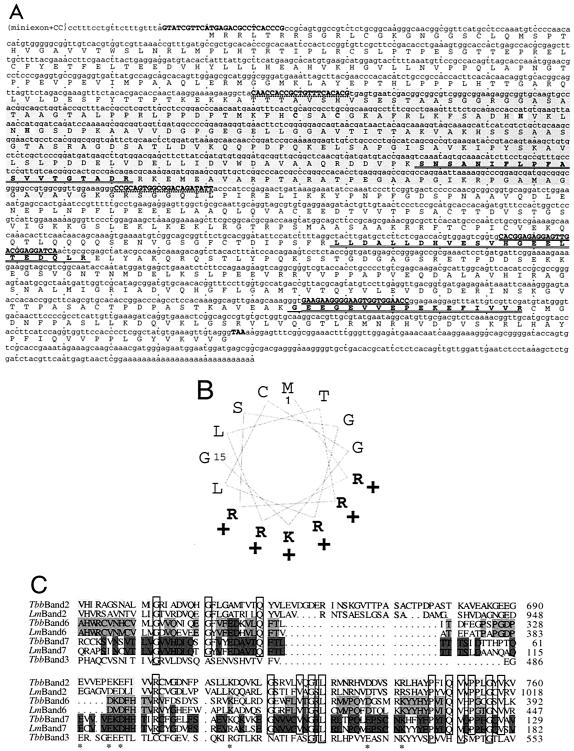
FIG. 1.
Band II sequence. (A) Upper line, the nucleotide sequence (the longest cDNA, plus in parentheses the RT-PCR-determined 5′ end). Oligonucleotide primers are indicated in boldface and uppercase (pairs have common underlining). The dsRNA region, start and stop codons, and zinc finger are indicated by highlighting. Lower line, the protein sequence. The three sequenced tryptic fragments are boldface amino acids; residues that confirmed the initial PCR fragment are singly underlined, and residues that additionally confirmed the cDNA are doubly underlined. (B) Helical wheel representation of the first 15 amino acids, indicating an amphipathic alpha helix. (C) C-terminal region of T. brucei band II (TbMP81), compared with the analogous region of T. brucei band VI (TbMP52), VII (TbMP18), and III (TbMP63) proteins and to L. major homologs by use of T-Coffee (27). Highlighting indicates cross-species conservation. Boxes and stars show identity and charge conservation, respectively between the different proteins. The L. major band III homolog is not included, as it lacks part of this C-terminal region.
RNAi cell lines.
RNAi plasmid pZJM:B2 is vector pZJM (41) carrying 712 bp of band II (highlighted in Fig. 1A), PCR amplified from genomic DNA using oligonucleotides 5′-ATTCTCGAGC AACCACCGCT GTTTCACACG-3′ and 5′-TCGAAGCTTA ATATCTGTCC GCCACTGCGG-3′ (indicated by a dotted underscore in Fig. 1A, plus indicated XhoI and HindIII sites for cloning). RNAi plasmid pZJM:B5 was prepared similarly, carrying 700 bp of band V, PCR amplified with 5′-AATTCTCGAG TACGTCACTT CCGGCGAACG-3′ and 5′-TCGAAGCTTC AGCGGGTAGT TGCCCATACC-3′. pZJM directs phleomycin resistance, ribosomal DNA integration, and double-strand expression of the insert from two tetracycline (Tet)-inducible T7 promoters. Twenty-five micrograms of NotI-linearized plasmid was electroporated into T. brucei 29.13 procyclic cells in Cunningham's media plus 15% fetal bovine serum (Gibco-BRL), 15 μg of G418 (Mediatech)/ml, and 50 μg of hygromycin (Sigma)/ml; after 24 h, 2.5 μg of phleomycin (Sigma)/ml was added (43, 44). After about a week, remaining cells were cloned by extreme dilution, distributing ∼10 live cells in 20 ml of conditioned Cunningham's media to 96 wells (M. Klingbeil, personal communication) (19). After ∼12 days at 27°C, growing wells were provided 1.8 ml of conditioned media and were subsequently cultured normally. Plasmid insertion was verified by PCR with the downstream primer from pZJM:B2 or pZJM:B5 and 5′-GGTACCCCGG ATATAGTTCC TCCTTTCAGC-3′ to the vector. For induction, cultures were split, were either provided 1 μg of Tet (Sigma)/ml or not, and were counted each day on a Reichert hemocytometer. They were diluted in Cunningham's media to remain <5 × 106 cells/ml, consistent with log-phase growth (diluting noninduced cultures ∼1:10 every 2 days). Every 2 days induced cultures were supplemented with 1 μg of Tet/ml. Untransfected 29.13 procyclic trypanosomes were also maintained in the log phase.
Northern blots.
For each lane, 2.5 × 107 cell equivalents of total RNA that was Trizol (Invitrogen) purified, following the manufacturer's directions, in 5 μl of diethyl pyrocarbonate-water) was heated at 65°C for 15 min, snap-chilled on ice, provided loading dye (10 μl of formamide, 3.5 μl of formaldehyde [37% {vol/vol}], 2 μl of morpholinepropanesulfonic acid 10× buffer [Quality Biological], and 1 μl of saturated bromophenol blue), and run on a formaldehyde-1% agarose gel at 5 V/cm. After soaking of the gel in 1 M ammonium acetate (J. T. Baker), the RNA was transferred onto a HyBond-XL nylon membrane (Amersham Pharmacia Biotech), washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), baked 1 h at 80°C, and probed in Rapid-hyb buffer (Amersham) for 5 h at 65°C by using 106 counts of pZJM insert per min per ml that was gel purified after HindIII-XhoI digestion and was labeled by random priming (Invitrogen) with [32P]dATP (ICN).
Extract preparation, sedimentation, adenylylation, and U insertion reactions.
Small-scale cell extracts were rapidly prepared as described elsewhere (19, 33), suspended at 109 cell equivalents/ml in MRB10 (25 mM Tris-HCl, pH 8.0, 10 mM Mg[OAc]2, 10 mM KCl, 1 mM EDTA, 0.5 mM dithiothreitol, and 5% glycerol), and standardized for protein concentration with the method described by Bradford. Large-scale mitochondrial extracts were prepared and resolved by centrifugation on 10 to 30% glycerol gradients (where sedimentation distance is approximately linear in S) by using thyroglobulin (19S) and catalase (11S) sedimentation markers as described earlier (18, 32, 37). Editing reactions were as described earlier, treating with phosphatase prior to loading the gel (33). Adenylylation assays (18) (3 μg of protein of small-scale extracts) always include an initial deadenylylation step, unless specifically indicated, without which those ligase molecules already adenylated in the extract (selectively band V [32]) are not detected (32).
Western blot analysis.
Polyclonal antibodies were generated in mice (as described in reference 19) or rabbits (Covance) to recombinant band II protein expressed with a C-terminal His6 tag with the pET29a vector (Novagen) in Escherichia coli BL21 cells and were purified as described earlier (16, 19). Antibodies to other editing proteins were similarly prepared (19; S. F. O'Hearn et al., unpublished data); antibody to Trypanosoma cruzi mitochondrial protein lipoamide dehydrogenase (LipDH) was a gift of R. L. Krauth-Siegel. Western blotting was basically conducted as described earlier (19), at 4°C, by using 3 μg of protein of small-scale extracts or similar ligase amounts of purified editing complex, sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis, membrane washing in TNT buffer (10 mM Tris, pH 8.0, 150 mM NaCl, and 0.05% Tween 20), and blocking with added ∼3% (wt/vol) bovine albumin fraction V (Sigma). The secondary antibody was alkaline phosphatase-conjugated goat anti-immunoglobulin G (Sigma, added 1:5,000), detected with 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium tablets (Sigma) or with horseradish peroxidase-conjugated goat anti-immunoglobulin G (Santa Cruz, added 1:5,000) and developed with the ECL-Plus chemiluminescent detection kit following the manufacturer's instructions (Amersham) (19).
Nomenclature for proteins of the T. brucei RNA editing complex.
The 48-kDa ligase has been called band IV (32), TbMP52 (28), p52 (25), REL1 (for RNA editing ligase [36]), and DREL (for deletion-requiring RNA editing ligase [12]). The 46-kDa ligase of this complex has been called band V (32), TbMP48 (28), p48 (25), REL2 (36), and IREL (for insertion-specific RNA editing ligase [12]). (Because the REL1 acronym is already used for a different well-known protein, its use appears contrary to recommendations for trypanosome protein nomenclature [6].) Other multiply designated proteins of the editing complex mentioned in this paper are band I (32) or TbMP99 (39), band II (32) or TbMP81 (29), band III (32) or TbMP63 (29), band VI (32) or TbMP42 (29), and band VII (32) or TbMP18 (29).
RESULTS
The band II gene and protein.
We cloned the gene for band II, the second largest of seven major proteins in the T. brucei RNA editing complex (32, 37), by using tryptic fragment sequence data (boldface peptides in Fig. 1A; see references 19 and 33). Since that gene was not in the public database, degenerate primers (heavily underlined oligonucleotides in Fig. 1A) were used for genomic PCR. The resulting fragment, verified by its encoding the underlined amino acids, was used to probe a cDNA library, and the identified clones encode all the sequenced tryptic fragments (56 additional amino acids, doubly underlined). RT-PCR showed that only two C residues separate the miniexon sequence, characteristic of all nucleus-encoded trypanosome mRNAs, from the longest cloned cDNA (top line, Fig. 1A). Therefore, the first AUG of the cloned cDNA initiates the coding region, a 2,286-nucleotide open reading frame for a 762-amino-acid protein (81 kDa). This same gene was also identified to encode one of 20 proteins (TbMP81) coisolated with editing activities (28, 29). The predicted protein contains a presumptive 5′ mitochondrial targeting sequence (Fig. 1B), a C2H2 zinc finger (reverse highlighted in Fig. 1A), and no other obvious motifs except a C-terminal region that is highly conserved with other editing proteins (Fig. 1C) (18, 19, 29).
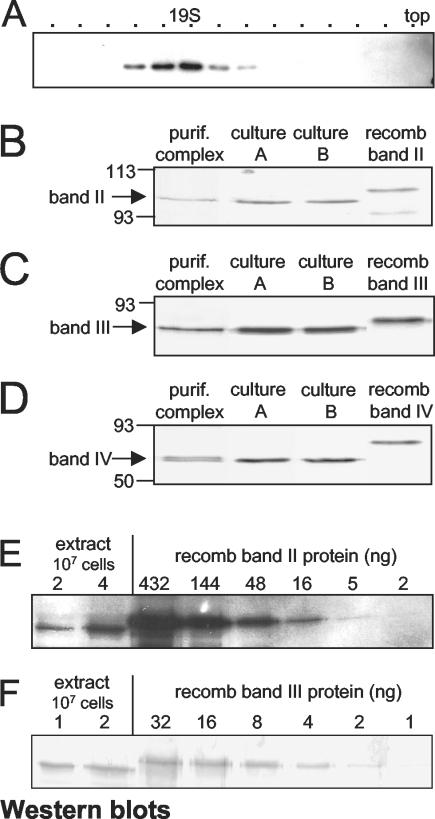
Polyclonal antibodies generated to recombinant band II protein expressed from the cloned cDNA recognize band II in crude trypanosome extract, the ∼20S complex, and the purified editing complex (Fig. 2A and B). Virtually all band II protein is present in the editing complex (Fig. 2A and data not shown), and accordingly, upon purification of the editing complex, band II becomes enriched to approximately the same extent as band III (Fig. 2C) and band IV (Fig. 2D), two other integral components of the simple editing complex (19, 29, 33). Comparing the band II (Fig. 2E) and band III (Fig. 2F) signals from extract with those from titrated recombinant protein indicates that both band II and band III constitute ∼1/1,000 of the extract protein or ∼1/10,000 of total trypanosome protein. This implies that there are several thousand editing complexes per trypanosome, a reasonable number given the large extent of editing. These data also verify (32) that band II is an inherent component of the editing complex and does not represent a transient, adventitious, or substoichiometric association.
FIG. 2.
Band II and other editing proteins. Western blotting of sodium dodecyl sulfate-polyacrylamide gel electrophoresis. (A) Fractions from control trypanosome mitochondrial extract, resolved by glycerol gradient centrifugation and detected using antibody to band II. (B to D) Antibody to band II (B), band III (C), or band IV (D) was used to detect editing complex purified to the seven major proteins (32) (lanes “purif. complex”), small-scale extracts of two clonal 29.13 cell lines (ligase amounts were similar to those loaded from purified complex) (lanes “culture A” and “culture B”), and the respective recombinant protein (lanes “recomb band”). The recombinant proteins are larger than the corresponding native ones by the His6 tag plus the mitochondrial targeting sequence. The antibodies do not appreciably detect either purified mitochondrial or recombinant versions of the other editing proteins. (E and F) Titrated amounts of small-scale trypanosome extract and recombinant band II (E) or band III (F) were detected using antibodies to the respective proteins. The recombinant protein was quantified as described by Bradford; ∼2/3 was full length. The trypanosome extract was 5 μg of protein/107 cells; trypanosomes contain ∼50 μg of protein/107 cells (17).
Band II protein is essential.
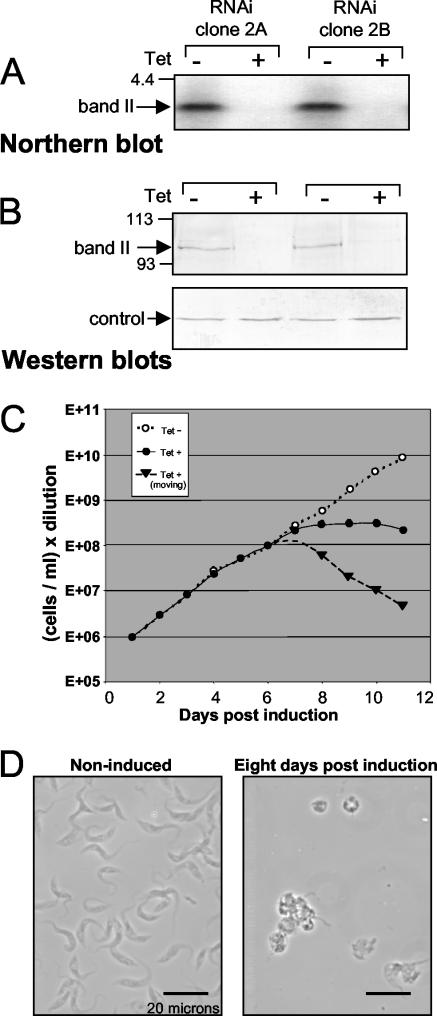
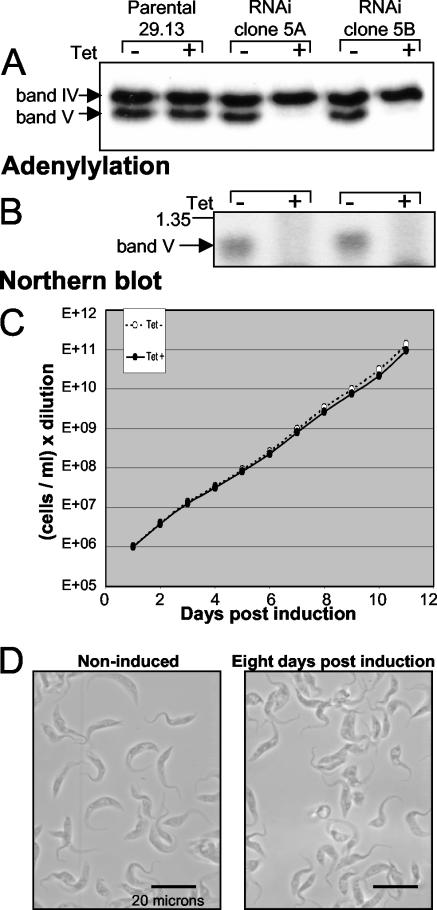
To examine the role of band II protein, a 0.7-kb segment of the band II coding region (highlighted in Fig. 1A) was cloned between opposing Tet-regulated T7 promoters (41) and transfected into procyclic 29.13 cells, which produce T7 RNA polymerase and Tet repressor (43, 44). From the transformed cell populations, clonal lines were established by limiting dilution, enabling study of the RNAi effect unencumbered by the occasional nonresponsive cell. When aliquots of representative lines are induced for band II RNAi expression by Tet administration, band II dsRNA accumulates, band II mRNA virtually disappears (Fig. 3A and data not shown), and band II protein becomes greatly depleted (Fig. 3B, upper panel). Constant abundance of LipDH, a mitochondrial protein not involved in RNA editing, suggests that the induced extracts remain otherwise intact (Fig. 3B, lower panel). One week after induction, cultures have ceased expanding (Fig. 3C). Microscopic examination reveals that the remaining cells have stopped their normally rapid undulations and round up (Fig. 3D), indicative of sick or dying trypanosomes. The fraction of normally elongated and moving cells decreases to ∼1% (Fig. 3C, dotted line), and trypan blue exclusion verifies that ∼99% of the remaining cells are dead. Thus, band II is essential for procyclic trypanosomes to grow and even to remain viable.
FIG. 3.
Band II RNAi. (A) Northern blot of band II-specific RNA from two clonal band II RNAi cell lines, noninduced or 24 h postinduction. (B) Western blot of band II-specific protein from the two clonal band II RNAi cell lines, noninduced or 5 days postinduction. (C) Growth curve of band II RNAi cells, noninduced (white dots) or induced, showing total cells (dark dots) and the cells that remain healthy-looking, i.e., elongated and undulating (triangles). (D) Typical view of band II RNAi cells, noninduced or 8 days postinduction.
When inducing heterogeneous cultures of initial transformants (41, 43, 44) rather than clonal lines, the dying band II RNAi cells are overtaken, sometimes quite rapidly, by minor cell types that retain band II mRNA (O'Hearn, unpublished observation). The induced culture can even appear to continue growing at a reduced rate, making band II erroneously appear stimulatory but not essential. We have additionally noted that, without intentional induction, band II RNAi cells can exhibit cell death, evidently due to variable low expression of the dsRNA (O'Hearn, unpublished). Both these observations emphasize the trypanosome's critical need for band II protein.
Band II serves to retain band V protein.
Because band III of the editing complex is needed to retain band IV ligase (19), we examined whether band II might have an analogous role. Clonal band II RNAi cell lines were split, induced or not, and prior to the onset of growth inhibition, extracts were prepared from the paired cultures. Band II silencing does diminish a ligase, but it is band V, not band IV (Fig. 4A, left panel). Note that all our normal adenylylation assays begin with a deadenylylation step, which is critical to score actual ligase abundance (18, 19, 32, 33). In contrast, assays performed without this step show variable labeling of band V, sometimes virtually none (e.g., compare right panels of Fig. 4A and B), because band V is preferentially adenylylated already in cell extract preparations (12, 32). Constant abundance of the LipDH control mitochondrial protein (Fig. 4C) verifies that our extracts and reactions are comparable. Thus, band II RNAi dramatically diminishes active band V.
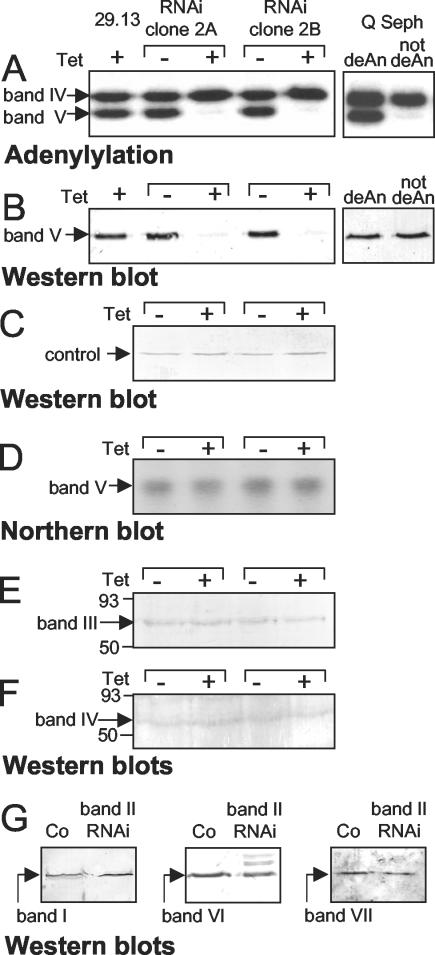
FIG. 4.
Effect of band II RNAi on the ligases and other editing proteins. Cultures of two clonal band II RNA cell lines and 29.13 control cells were induced or noninduced, and on day 5 small-scale extracts were prepared in parallel. (A) They were assayed by adenylylation (following deadenylylation, to detect all ligases). The right two lanes of panels A and B both show a partly purified trypanosome extract fraction (after Q-Sepharose) (32), assayed without or with deadenylylation (deAn), either by adenylylation (A) or Western blotting for band V (B). In this preparation, band V is very highly preadenylylated and thus is virtually not detected unless first deadenylylated. (B and C) Western blotting for band V protein and LipDH control protein, respectively. (D) Northern blotting for band V sequences. (E and F) Western blotting for band III and band IV proteins, respectively. (G) Similar extracts from separate clones of band II RNA cells and control (29.13) cells, enriched by velocity centrifugation, were assayed by Western blotting for band I, VI, and VII proteins.
Adenylylation analyses cannot address whether band II RNAi causes band V protein to be physically lost or to remain present but lose adenylylation catalytic activity. To distinguish, we performed Western analysis with antibody specific for band V protein. Cells induced for band II RNAi indeed lack the vast majority of band V protein that is present in control cells (Fig. 4B, left panel, and C). Thus, band V protein is physically lost upon band II RNAi. To determine whether this occurs at the protein or RNA level, Northern analysis confirmed control abundance of band V mRNA (Fig. 4D). Indeed, the scenario that band II dsRNA fortuitously destroys band V mRNA had seemed unlikely, since the two genes do not have appreciable sequence similarity. We conclude that band II RNAi causes loss of band V protein at the protein level. This implies that band II protein is needed to retain the band V protein in the editing complex and that free band V protein is short-lived, analogous to band IV requiring band III protein (19, 33).
Other major proteins of the editing complex.
Since band V protein is lost when band II protein is depleted, we examined whether other major proteins of the editing complex are also affected. Western analyses on cultures induced or not for band II RNAi show that band I, III, IV, VI, and VII proteins remain present (Fig. 4E through G). Thus, none of the other major protein of the basic editing complex except band II and band V are lost upon band II RNAi.
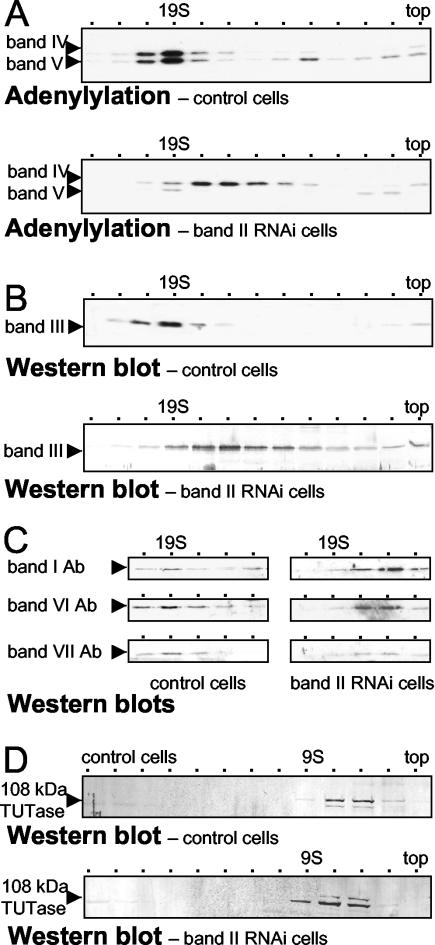
Because Fig. 4 scored whole extract, it could not reveal if band II RNAi caused release of additional proteins from the band II-depleted editing complex, so long as they remained stable in the mitochondria. To examine this, complexes from both control and band II RNAi extract were separated from potentially freed proteins by velocity centrifugation and various proteins were analyzed by adenylylation and Western blotting. As shown previously (8, 19), editing complex from mitochondrial extract that we prepare from control trypanosomes by the standard protocol (9, 31) forms a single ∼20S peak (Fig. 5A, upper panel, and C, left panel); it sediments very slightly faster than a ∼19S thyroglobulin marker (see also references 18, 19, 30, 32, and 34). This ∼20S peak is seen when scoring band IV and band V (19, 32) (Fig. 5A, upper panel), band II (Fig. 2A), band III (19) (Fig. 5B, upper panel), and bands I, VI, and VII (Fig. 5C, left), supporting the idea that these proteins are all in a common complex (32). In contrast, the depleted complex from band II RNAi cells sediments strikingly more slowly and more broadly (Fig. 5A to C). That same sedimentation profile is seen for band IV (Fig. 5A, lower panel), band III (Fig. 5B, lower panel), and bands I, VI and VII (Fig. 5C, right panel), i.e., for all the remaining members of the original seven major editing proteins. Their common sedimentation profile indicates that those five remaining proteins still form a mutual complex upon band II RNAi, without the contribution of bands II and V. Its ∼15S sedimentation is determined by using the parallel protein markers and more precisely by using an internal 20S standard. Taking advantage of RNAi being <100% efficient, the small remaining amount of intact editing complex provides an ∼20S sedimentation marker in the same gradient as the depleted complex. This intact complex is seen as the residual band V along with a correspondingly small amount of band IV (Fig. 5A, lower panel) and the residual band II (data not shown). We conclude that, when band II is diminished and band V becomes comparably depleted, the five other major proteins of the basic editing complex remain associated. Furthermore, and contrary to Drozdz et al. (13), this depleted complex sediments significantly more slowly than does intact editing complex.
FIG. 5.
Sedimentation analysis of editing components from band II RNA cells. In parallel, mitochondrial extracts were prepared from a clonal band II RNA cell line and 29.13 control cells at 5 days postinduction and were resolved by glycerol gradient centrifugation. (A) Adenylylation assays of fractions 4 to 16 (top) from the control cells (upper) and band II RNAi cells (lower), following deadenylylation. ∼19S peaks in fraction 7. (B) Fractions 4 to 16 (top) from the control cells (upper) and band II RNAi cells (lower) were detected by using antibody to band III protein. (C) Fractions 7 to 10 from the control cells (left) and band II RNAi cells (right), including the ∼20S and ∼15S regions, were analyzed by using antibodies (Ab) to each of the other major proteins of the basic editing complex. (D) Fractions 4 to 16 (top) from the control cells (above) and band II RNAi cells (below) were detected by using antibody to the 108-kDa TUTase. Because this TUTase near the top fraction (last fraction collected) had different volumes in these two gradient, it appears in slightly shifted fractions.
In contrast to the associated proteins of the basic editing complex, a 108-kDa TUTase (2) sediments distinctly, at ≤9S (Fig. 5D, upper panel) (3). Its sedimentation is not affected by band II RNAi (Fig. 5D, lower panel), providing additional evidence that this TUTase is not in the basic editing complex.
RNAi to band V allows growth but affects morphology.
Since band II RNAi causes loss of band V ligase (Fig. 4), which acts to seal mRNA specifically in U insertion (12), might band V be essential and its retention the primary role of band II? To assess this ligase, a 700-bp fragment from the band V gene, which was identified earlier (25, 33, 36), was inserted into the Tet-regulated RNAi vector (41) and clonal lines were again isolated from transfected cells. Upon induction, band V protein was lost (Fig. 6A), as efficiently as with band II RNAi (Fig. 4A and data not shown) but now due to loss of band V mRNA (Fig. 6B). However, the clonal band V RNAi lines continued to grow at the control rate and remained depleted of band V mRNA and protein even after long-term culture (Fig. 6C and data not shown). Thus, procyclic trypanosomes are viable with ≥90% reduced levels of band V protein. Nonetheless, failure to inhibit growth is a negative result and therefore cannot establish whether band V is truly dispensable for editing or was intially present in substantial excess.
FIG. 6.
Band V RNAi. (A) Adenylylation assays, following deadenylylation, of small-scale extract from 29.13 and clonal band V RNAi cell lines, noninduced or 5 days postinduction. (B) Northern blot of band V-specific RNA from the RNAi cell lines, noninduced or 24 h postinduction. A 1.35-kb marker is indicated. (C) Growth curve of band V RNAi cells, noninduced (white dots) or induced (dark dots). (D) Typical view of band V RNAi cells, noninduced or 8 days postinduction.
Notably, microscopic examination of induced band V RNAi cells indicates that they are not entirely healthy. While control cells are elongated and undulate rapidly, induced band V RNAi cultures generally contain cells that are abnormally short, wide, or round and are relatively immobile (Fig. 6D). The prevalence of these abnormal morphologies suggests that band V ligase serves a positive role in procyclic trypanosomes.
The ability of trypanosomes to grow with greatly depleted band V has enticing implications for editing. Studies to date have shown that band IV seals U deletion (12, 18) while band V seals U insertion but not U deletion (12), and no protein of the simple seven-major-protein complex has been proven dispensable or able to catalyze both kinds of editing. Thus, if band V is not necessary to ligate U insertion and if band IV can ligate both forms of editing, both would be quite novel. To address this, we assayed extracts of band V RNAi cells by using an in vitro editing system that has been extensively characterized for ligase utilization (12). Specifically, by incremental activation of first band V and then band IV from control cell mitochondrial extract of purified complex, the amount of ligated U insertion product was found to increase in proportion with band V activation and to become maximal without band IV activation (12). Thus, extracts of induced band V RNAi cells, which have ≤10% of the normal amounts of band V (Fig. 6A and B), should seal ≤10% of the normal amount of U insertion by using their band V. Any additional U insertion product should therefore be sealed by a different ligase. Multiple studies find that band IV and band V are the only RNA ligases in T. brucei mitochondria (24, 32, 33, 36). Thus, if these band II RNAi extracts ligate more than 10% of the U insertion product of control extract, that extra should have been sealed by band IV.
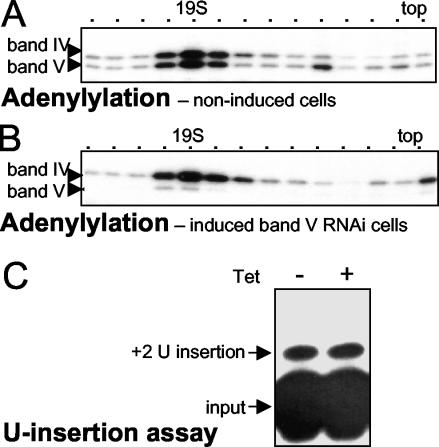
To meaningfully conduct this experiment, nonspecific inhibitors of in vitro U insertion known to be present in crude extract were partly removed by velocity centrifugation (10, 21). This was performed in parallel with mitochondrial extract from induced and noninduced band V RNAi clonal cell lines. Similarly sedimenting fractions had similar amounts of band IV ligase but, of course, very different amounts of band V ligase (Fig. 7A and B). Western analysis confirmed that the ∼20S peaks from control and band V RNAi extracts had similar amounts of all six nontargeted major proteins of the editing complex: bands I, II, III, IV, VI, and VII (data not shown). Furthermore, the residual intact ∼20S complex of the band V RNAi extract, represented by the small remaining amount of band V, cosedimented with the depleted complex, represented by the bulk of band IV and the other editing proteins (Fig. 7B and data not shown). Thus, in contrast to band II protein (Fig. 5), band V protein had minimal effect on sedimentation of the depleted editing complex.
FIG. 7.
Effect of band V RNAi on sedimentation and U insertion. In parallel, mitochondrial extracts were prepared from a clonal band V RNA cell line, noninduced or 5 days postinduction, and were resolved by glycerol gradient centrifugation. (A and B) Adenylylation assays, following deadenylylation, of fractions from the noninduced (A) and induced (B) band V RNAi cells. (C) The peak ∼20S fractions, which had similar amounts of band IV ligase, were analyzed for full-round U insertion.
Comparable ∼20S fractions were then used for in vitro U insertion reactions (Fig. 7C). Notably, as much U insertion product was generated from band V RNAi preparations as from their parallel control preparations (Fig. 7C). Importantly, these reactions were confirmed to be in the approximately linear range for extract amount (data not shown), indicating that the signal intensities are meaningful. Since ≤10% of the U insertion signal from the band V RNAi preparation should have been sealed by the remaining band V ligase (12; see above), the other ≥90% of its U insertion product will have been sealed by band IV, the ligase remaining in the depleted editing complex. This makes band V the first protein of the simple editing complex demonstrated to be dispensable for the editing cycles and band IV the first protein demonstrated to be able to catalyze both forms of editing. Nonetheless, the propensity of induced band V RNAi cells for altered morphology indicates that they are not totally normal, suggesting that band IV may be less effective at ligating U insertion than is band V.
DISCUSSION
RNAi to band II and band V.
We report analysis of band II and band V, two of the seven major proteins of the basic T. brucei RNA editing complex. In addition to the major message discussed in separate sections below, our data yield several other noteworthy conclusions and inferences. First, antibody prepared to recombinant band II protein confirms band II as a bona fide constituent of the ∼20S editing complex, both in crude extract and when purified to the seven major protein profile (Fig. 2). No substantial amount of band II protein was found apart from the editing complex (Fig. 2A), as also shown for bands III (19), IV, and V (30-33). Consistent with their being in a common complex, all these examined proteins show similar enrichment in the purified editing complex (Fig. 2B through D), ∼200-fold relative to extract or ∼2,000-fold relative to the trypanosome (Fig. 2E and F), supportive of previous inferences from activity assays (32, 37). This abundance corresponds to several thousand editing complexes per cell, which is in line with other RNA-processing activities.
Second, band II begins with a putative mitochondrial targeting sequence (Fig. 1B) (29) that appears cleaved upon mitochondrial import (Fig. 2B and E), as also occurs with bands III, IV, V, and VII (19, 28, 29, 32). This suggests that these editing proteins are independently imported into the mitochondrion and only subsequently assembled into the editing complex.
Third, RNAi analysis showed that band II is absolutely essential for viability of procyclic trypanosomes, since multiple independent clonal lines of band II RNAi transformants stop growing, round up, and die following induction, leaving ≤1% viable-looking cells (Fig. 3C and D and data not shown). When uncloned band II RNAi transformants are induced, aberrant cell types that retain band II mRNA overtake the cultures and can camouflage the lethality of the authentic band II RNAi (data not shown). Use of such uncloned RNAi transformants likely explains why a previous study (13) showed band II (TbMP81) to be only ∼2-fold stimulatory for growth of procyclic trypanosomes rather than inferring that a protein is essential if its depletion increases cell doubling time (13); we, however, reserve that conclusion for demonstration of actual lethality. Indeed, if band II was stimulatory (13), while band III (19) and band IV (18, 33, 36) are essential, band II would seem less important for editing. By showing ∼99% cell death upon band II RNAi, we submit that these proteins are all critical for procyclic growth and maintenance of viability (Fig. 3).
Fourth in contrast to the requirement for bands II, III, and IV of the editing complex, cells at least as depleted for band V ligase by its RNAi grow at a normal rate (Fig. 6C) (13). We submit that this lack of growth effect is considerably more meaningful when observed using clonal band V RNAi cell lines (Fig. 6C), where RNAi of an essential gene is lethal (Fig. 3) than when using uncloned transformants (13) where an essential gene can appear only ∼2-fold stimulatory (see above). Nonetheless, it remains negative data. While it may be tempting to draw conclusions (13) from such negative results, these data cannot prove that band V is dispensable, rather than the residual band V being sufficient. In fact, in vivo data (Fig. 6) (13, 18, 33, 36) do not even demonstrate if essential editing requires less band V than band IV, because band IV's role in RNA repair (18) could define its critical abundance. Nonetheless, the indication that cells might accomplish editing without band V (13) (Fig. 6C) has intriguing implications for ligase utilization. V—to seal U insertion. Previous biochemical and genetic studies had shown band V ligating only U insertion, not U deletion, and band IV ligating U deletion (12, 18); an additional role for band IV in ligating U insertion has been suggested (12) but not rigorously addressed. To that end, we here make use of an in vitro system where, without band IV activation, maximal levels of U insertion are ligated by band V and proportionately lower levels are ligated when band V is only partially activated (12). Thus, the residual band V in band V RNAi cell extract (≤10%; Fig. 6A and 7B) could ligate only 10% of the amount of U insertion as in parallel control extract (12). Obtaining equal amounts of U insertion with the two extracts demonstrates that most U insertion from band V RNAi cells is sealed by their remaining ligase, band IV. Band V is therefore the only component of the simple editing complex shown to be nonessential in catalyzing editing cycles, since band IV seals U insertion when band V is depleted. Nonetheless, it remains to be determined whether band IV also seals U insertion in wild-type editing complex or whether band IV serves this function only when band V is lacking from the editing complex.
Sixth, our data indicate that band IV may not fully replace band V in vivo. Microscopic examination reveals that band V RNAi cells appear prone to altered morphology (Fig. 6D), suggesting that band V augments the vitality of procyclic trypanosomes in some manner. It may be that band IV is not a totally effective substitute for band V ligase in sealing U insertion in vivo, possibly due to speed and/or accuracy issues that would not be apparent in the in vitro system.
Complexes lacking band II also lack band V.
Quantitative adenylylation analysis (Fig. 4A and 5A) and Western blotting (Fig. 4B) demonstrate another important message of our study: that without band II, band V ligase protein is lost. Thus, band II is needed to retain band V ligase, and this occurs on the protein level (Fig. 4D), analogous to the need for band III retain band IV ligase (19). To evaluate this contribution, it is important to appreciate that two kinds of adenylylation assays are used in the trypanosome editing literature. Both provide [α-32P]ATP for the ligase's initial charging reaction (E + ATP ⇋ AMP + PPi), but assays that begin with a deadenylylation step score actual ligase abundance (12, 18, 19, 32, 33) (Fig. 4 to 7), while assays without this decharging step (e.g., reference 13) detect only those ligase molecules that were not already adenylylated in the cell extract (32). This abbreviated assay therefore cannot be used to reliably estimate ligase abundance, especially band V, which has a very high affinity for ATP (12) and can become almost completely adenylylated upon manipulation of the cell extract (32). This preadenylylation causes band V to erroneously appear depleted or entirely lacking in the abbreviated assay (Fig. 4A and B, right panel) (32). Indeed, Drozdz et al. (13) detect only ∼1/5 of band V (TbMP48) in control extract (where the two ligases are approximately equimolar [32, 34, 37]), presumably because ∼4/5 was adenylylated and therefore not scored. Further complicating the abbreviated assay, the extent of band V preadenylylation can vary dramatically and sometimes unexpectedly (32; data not shown). Notably, in vivo band V becomes deadenylylated by activating its substrate, the TUTase product in U insertion (12), but since Drozdz et al. show that this TUTase activity is reduced by ∼90% upon band II (TbMP81) RNAi (13), there may be insufficient TUTase to appreciably deadenylylate band V (TbMP48) in vivo. Then band V could well be highly preadenylylated in band II RNAi cells, and one would expect the abbreviated assay to detect a minimal amount of this ligase in extract of band II RNAi cells. Drozdz et al. reporting this (13) could be a direct consequence of the reduced TUTase activity. Conclusion of decreased band V protein abundance upon band II RNAi (13) instead requires adenylylation assays with the de-adenylylation step (Fig. 4A) and/or Western blotting with a band V-specific antibody (Fig. 4B).
Note that, upon band II RNAi, band V becomes lost from the whole trypanosome (Fig. 4A and B), not just from the editing complex (Fig. 5A, lower panel). This implies that, when band II is not present to retain band V in the editing complex, the free band V ligase is unstable. That also is analogous to band IV ligase, which is unstable when separate from the editing complex (18, 33). In fact, trypanosomes could maintain approximately equimolar stoichiometry of the editing proteins, with virtually all in the editing complex, through such degradation of unbound protein. This would be akin to how yeast ensures that excess ribosomal proteins do not accumulate (42).
Role of band II.
It is also significant that band V protein is depleted as extensively in band V RNAi cells (Fig. 6A), which are viable (Fig. 6C), as in band II RNAi cells (Fig. 4A and B), which are not viable (Fig. 3C and D). Thus, band II must serve a separate essential role. We show that this role is not to maintain the other major proteins of the basic editing complex, since, upon band II RNAi, band I, III, IV, VI, and VII proteins remain present (Fig. 4A, E, and F) and associated (Fig. 5C and D). These findings add considerably to the data of Drozdz et al. (13), who show that, upon band II (TbMP81) RNAi, two of their ∼20 identified (28) major proteins were retained in the depleted complex (TbMP63/band III and TbMP42/band VI) but band IV (TbMP52) surprisingly appeared to sediment separately, and the other major proteins were not assessed.
Our sedimentation analyses indicate that band II serves a structural role in the editing complex. We use a mitochondrial extract protocol previously shown to yield an ∼20S complex of editing activities (8, 32) and here confirmed by using sedimentation markers; additionally the RNAi-depleted complex is sized relative to an internal ∼20S standard, the small amount of wild-type complex detected by the remaining band V and band II in that same gradient (Fig. 5A, lower panel). Complexes lacking both band II and band V have markedly reduced sedimentation, ∼15S (Fig. 5A through C). However, complexes lacking only band V show no appreciable change in sedimentation relative to the internal control of the residual complete ∼20S complex, since the bulk of band IV and the other remaining proteins cosediment with the small residual amount of band V (Fig. 7B and data not shown). The sedimentation decrease that occurs upon loss of band II is more than expected by mass reduction alone, implying that, without band II, the protein association is less compact. Band II thus appears to help organize the editing complex.
Since the editing complex binds the multiple activities for the U insertion and U deletion pathways, which are most likely arranged to channel substrate between sequentially used active centers, its function may well depend on a rigid organization. Therefore, a less compact structure of the depleted complex could contribute to the lethality upon band II RNAi. In contrast to our data, Drozdz et al. (13) report unaltered sedimentation of editing proteins upon band II (TbMP81) RNAi. An unchanged S value would appear to imply a more compact structure of the band II-depleted complex than of the wild-type editing complex, opposite of what our data indicate. In fact, Drozdz et al. do not report the actual S value of their control editing complexes (13), but they appeared to have much broader sedimentation profiles than the usual ∼20S complex (e.g., references 29, 30, 31, and 32), more like the ∼30-50S association of editing proteins obtained from that laboratory's earlier preparation of extract (7). If their present preparation also generates the ∼30-50S association, a component decreasing from ∼20S to ∼15S may be barely detectable. Furthermore, data indicating that band IV (TbMP52) sediments apart from the other editing proteins in their control extract (13) would suggest a complex quite unlike any previously reported (18, 19, 24, 29, 32-34).
When we first examined our band II sequence against the public database, it showed striking similarity to only three other listed proteins, all from T. brucei, all components of the seven-major-protein editing complex (band II, band III, band VI, and band VII) and all containing this similarity in the C-terminal ∼15 kDa (Fig. 1C; see also references 19 and 29). The improbability of this occurring by chance alone implies that this “editing complex sequence” confers an advantageous function. Subsequent inclusion of sequences from the related trypanosomatid Leishmania major revealed a band II equivalent (expectation [E] value of 10−58) with homology throughout but with the greatest identity in this C-terminal region (Fig. 1C). L. major also has equivalents of bands III, VI, and VII. (This band VI equivalent is presently identified in the database as cytochrome c1.) Therefore, these editing proteins, especially their C-terminal domains, have been maintained since at least the divergence of Trypanosoma and Leishmania, ∼108 years ago (22, 33). Among the T. brucei C-terminal sequences, band II is similar to band VI over nearly twice the length seen for the other protein pairs, while band III is most similar to band VII, possibly suggesting their respective derivations. However, band VI and band VII have the greatest identity, with an E value of 10−18, suggesting the strongest selective pressure for this element in these two proteins. If this sequence serves to associate and/or maintain these four editing complex proteins if the two ligases bind to them, it could explain the association of all but one of the major proteins of the basic editing complex.
Symmetry in the editing complex.
Many aspects of the basic editing complex appear symmetric. There are two RNA ligases, band IV and band V, the former needed for U deletion and the latter specific for U insertion (12, 18). Each ligase is tethered to the editing complex through a larger zinc finger-containing protein, band III (19) and band II (Fig. 4 and 5), respectively, and both of these serve additional essential roles (Fig. 3) (19). As noted above, band II may show an evolutionary relationship with band VI and band III with band VII. The band III/band IV/(band VII) and band II/band V/(band VI) associations may thus constitute two pseudosymmetric portions of the basic editing complex, consistent with our earlier suggestion that the editing complex could contain largely distinct portions for U deletion and U insertion (9). However, the new findings that loss of either band II or band V has much less effect on the sedimentation coefficient of the remaining ligase, and therefore on the structure of the remaining protein association, than does loss of either band III or band IV (Fig. 5B and 7B) (18, 19) add insight that the latter protein pair appears more critical in the overall organization of the editing complex. Furthermore, the demonstration that band IV ligase can replace band V ligase to seal U insertion, at least when band V is absent from the editing complex, suggests that U insertion and U deletion might not be catalyzed in rigidly segregated domains of the editing complex. The trypanosome editing field has certainly progressed rapidly from 2 years ago, when the importance of just one major protein of the T. brucei basic RNA editing complex was known (12, 18, 33, 36) and when there was insufficient information to raise meaningful speculations about the general requirement and organization of the other proteins.
ADDENDUM IN PROOF
Two recent publications should be noted in conjunction with this study. Gao and Simpson (J. Biol. Chem. 278:27570-27574, 2003) also provide evidence that band IV can seal U insertion. And Schnaufer et al. (Mol. Cell 12:307-319, 2003) provide evidence that the basic editing complex contains subcomplexes for U insertion and U deletion.
Acknowledgments
We thank Laura Rusché for preparing the purified editing complex and arranging for its tryptic analysis. We also thank Zefeng Wang, Jim Morris, Mark Drew, and Paul Englund for the pZJM vector and discussions on its use, Elisabetta Ullu for suggestions about RNAi, Michelle Klingbeil for information on cell cloning, Rahul Bakshi for the Northern blot protocol, R. Luise Krauth-Siegel for the lipoamide dehydrogenase antibody, and members of our laboratory for helpful discussions.
This research was funded by the NIH (GM34231).
REFERENCES
- 1.Alfonzo, J., O. Thiemann, and L. Simpson. 1997. The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res. 25:3751-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aphasizhev, R., S. Sbicego, M. Peris, S. Jang, L. Simpson, et al. 2002. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell 108:637-648. (Erratum, 110:133.) [DOI] [PubMed]
- 3.Aphasizhev, R., I. Aphasizheva, R. Nelson, G. Gao, L. Simpson, et al. 2003. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 22:913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benne, R., and D. Speijer. 1998. RNA editing types and characteristics, p. 551-554. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 5.Blum, B., N. Bakalara, and L. Simpson. 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60:189-198. [DOI] [PubMed] [Google Scholar]
- 6.Clayton, C., G. Cross, N. El-Sayed, K. Gull, K. Stuart, et al. 1998. Genetic nomenclature for Trypanosoma and Leishmania. Mol. Biochem. Parasitol. 97:221-224. [DOI] [PubMed] [Google Scholar]
- 7.Corell, R. A., L. K. Read, G. R. Riley, J. K. Nellissery, T. E. Allen, M. L. Kable, M. D. Wachal, S. D. Seiwert, P. J. Myler, and K. D. Stuart. 1996. Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol. Cell. Biol. 16:1410-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz-Reyes, J., and B. Sollner-Webb. 1996. Trypanosome U-deletional RNA editing involves gRNA-directed endonuclease cleavage, terminal U-exonuclease and RNA ligase activities. Proc. Natl. Acad. Sci. USA 93:8901-8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz-Reyes, J., L. N. Rusché, K. J. Piller, and B. Sollner-Webb. 1998. T. brucei RNA editing: adenosine nucleotides inversely affect U deletion and U insertion reactions at mRNA cleavage. Mol. Cell 1:401-409. [DOI] [PubMed] [Google Scholar]
- 10.Cruz-Reyes, J., L. Rusché, and B. Sollner-Webb. 1998. Trypanosoma brucei U insertion and U deletion activities co-purify with an enzymatic editing complex but are differentially optimized. Nucleic Acids Res. 26:3634-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz-Reyes, J., A. Zhelonkina, L. Rusché, and B. Sollner-Webb. 2001. Trypanosome RNA editing: simple guide RNA features enhance U deletion 100-fold. Mol. Cell. Biol. 21:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz-Reyes, J., A. Zhelonkina, C. Huang, and B. Sollner-Webb. 2002. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol. Cell. Biol. 22:4652-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drozdz, M., S. Palazzo, R. Salavati, J. O'Rear, C. Clayton, and K. Stuart. 2002. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 21:1791-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gott, J. M., and R. B. Emeson. 2000. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34:499-531. [DOI] [PubMed] [Google Scholar]
- 15.Hajduk, S. 1997. Defining the editing “reaction”. Trends Microbiol. 5:1-2. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Hereld, D., J. Krakow, J. Bangs, G. Hart, and P. Englund. 1986. A phospholipase C from Trypanosoma brucei which selectively cleaves the glycolipid on the variant surface glycoprotein. J. Biol. Chem. 261:13813-13819. [PubMed] [Google Scholar]
- 18.Huang, C. E., J. Cruz-Reyes, A. G. Zhelonkina, S. O'Hearn, E. Wirtz, and B. Sollner-Webb. 2001. Roles for ligases in the RNA editing complex of Trypanosoma brucei: band IV is needed for U-deletion and RNA repair. EMBO J. 20:4694-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, C., S. O'Hearn, and B. Sollner-Webb. 2002. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 22:3194-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igo, R. P., Jr., S. D. Lawson, and K. Stuart. 2002. RNA sequence and base pairing effects on insertion editing in Trypanosoma brucei. Mol. Cell. Biol. 22:1567-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kable, M. L., S. D. Seiwert, S. Heidmann, and K. Stuart. 1996. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science 273:1189-1195. [DOI] [PubMed] [Google Scholar]
- 22.Lake, J. A., V. F. de la Cruz, P. C. Ferreira, C. Morel, and L. Simpson. 1988. Evolution of parasitism: kinetoplastid protozoan history reconstructed from mitochondrial rRNA gene sequences. Proc. Natl. Acad. Sci. USA 85:4779-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madison-Antenucci, S., R. Sabatini, V. Pollard, and S. Hajduk. 1998. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J. 17:6368-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McManus, M., B. Adler, V. Pollard, and S. Hajduk. 2000. Trypanosoma brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Mol. Cell. Biol. 20:883-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McManus, M. T., M. Shimamura, J. Grams, and S. L. Hajduk. 2001. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA 7:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ngo, H., C. Tschudi, K. Gull, and E. Ullu. 1998. Double-stranded RNA introduces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:14687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 28.Panigrahi, A. K., S. P. Gygi, N. L. Ernst, R. P. Igo, Jr., S. S. Palazzo, A. Schnaufer, D. S. Weston, N. Carmean, R. Salvati, R. Aebersold, and K. D. Stuart. 2001. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:380-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panigrahi, A. K., A. Schnaufer, N. Carmean, R. P. Igo, Jr., K. D. Stuart, et al. 2001. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:6833-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piller, K., L. Rusché, J. Cruz-Reyes, and B. Sollner-Webb. 1997. Resolution of the RNA editing gRNA-directed endonuclease from two other endonucleases of Trypanosoma brucei mitochondria. RNA 3:279-290. [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard, V., M. Harris, and S. Hajduk. 1992. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 11:4429-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rusché, L., J. Cruz-Reyes, K. Piller, and B. Sollner-Webb. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16:4069-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rusché, L., C. Huang, K. Piller, M. Hemann, E. Wirtz, and B. Sollner-Webb. 2001. The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol. 21:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabatini, R., and S. Hajduk. 1995. RNA ligase and its involvement in guide RNA/mRNA chimera formation. J. Biol. Chem. 270:7233-7240. [DOI] [PubMed] [Google Scholar]
- 35.Salavati, R., A. Panigrahi, B. Morach, S. Palazzo, R. Igo, and K. Stuart. 2002. Endoribonuclease activities of Trypanosoma brucei mitochondria. Mol. Biochem. Parasitol. 120:23-31. [DOI] [PubMed] [Google Scholar]
- 36.Schnaufer, A., A. K. Panigrahi, B. Panicucci, R. P. Igo, Jr., E. Wirtz, R. Salavati, and K. Stuart. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291:2159-2162. (Erratum, 293:5537.) [DOI] [PubMed]
- 37.Sollner-Webb, B., L. Rusché, and J. Cruz-Reyes. 2001. Generally useful novel nuclease activities of the trypanosome RNA editing complex. Methods Enzymol. 341:154-174. [DOI] [PubMed] [Google Scholar]
- 38.Stuart, K., T. Allen, S. Heidmann, and S. Seiwert. 1997. RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev. 61:105-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuart, K., A. Panigrahi, A. Schnaufer, M. Drozdz, C. Clayton, and R. Salavati. 2002. Composition of the editing complex of Trypanosoma brucei. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357:71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, B., N. L. Ernst, S. S. Palazzo, A. K. Panigrahi, R. Salavati, and K. Stuart. 2003. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei. Eukaryot. Cell 2:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 42.Warner, J. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 43.Wirtz, E., M. Hoek, and G. Cross. 1998. Regulated processive transcription of chromatin by T7 RNA polymerase in Trypanosoma brucei. Nucleic Acids Res. 26:4626-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wirtz, E., S. Leal, C. Ochatt, and G. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]